Abstract
Lectin–glycan interactions have critical functions in multiple normal and pathological processes, but the binding partners and functions for many glycans and lectins are not known. An important step in better understanding glycan–lectin biology is enabling systematic quantification and analysis of the interactions. Glycan arrays can provide the experimental information for such analyses, and the thousands of glycan array datasets available through the Consortium for Functional Glycomics provide the opportunity to extend the analyses to a broad scale. We developed software, based on our previously described Motif Segregation algorithm, for the automated analysis of glycan array data, and we analyzed the entire storehouse of 2883 datasets from the Consortium for Functional Glycomics. We mined the resulting database to make comparisons of specificities across multiple lectins and comparisons between glycans in their lectin receptors. Of the lectins in the database, viral lectins were the most different from other organism types, with specificities nearly always restricted to sialic acids, and mammalian lectins had the most diverse range of specificities. Certain mammalian lectins were unique in their specificities for sulfated glycans. Simple modifications to a lactosamine core structure radically altered the types of lectins that were highly specific for the glycan. Unmodified lactosamine was specifically recognized by plant, fungal, viral, and mammalian lectins; sialylation shifted the binding mainly to viral lectins; and sulfation resulted in mainly mammalian lectins with the highest specificities. We anticipate that this analysis program and database will be valuable in fundamental glycobiology studies, detailed analyses of lectin specificities, and practical applications in translational research.
Glycans exert their functions and influence through specific interactions with glycan-binding proteins. These interactions are involved in the immune recognition of pathogens (1–3), pathogen infection (4–6), immune cell migration (7, 8), protein processing (9), regulation of cell-surface receptors (10, 11), sperm–egg binding (12), and other areas of biology. The abnormal production of certain glycans or glycan-binding proteins can have damaging consequences. The contribution of atypical glycan–lectin interactions to disease pathology has been established for several diseases but is unknown for others that display abnormal glycans or glycan-binding proteins. A difficulty in making that link often is a lack of information about the proteins that bind certain glycans, and about the glycans that are recognized by certain glycan-binding proteins. Therefore, an important goal in glycobiology research is to obtain more complete knowledge about the interactions between glycans and glycan-binding proteins.
A breakthrough technology for probing lectin–glycan interactions was the glycan microarray (13–19). Relative to previous technologies, glycan arrays significantly reduced the time and reagent costs of experiments and increased the number of different lectin–glycan interactions that could be practically probed. Glycan arrays are composed of diverse oligosaccharides, either produced synthetically or purified from natural sources, immobilized on a solid support. Researchers typically use the arrays to measure the binding of a glycan-binding protein, such as a lectin or glycan-binding antibody, to each glycan on the array. From such data, the researcher can learn much about the specificity of the glycan-binding protein.
Most laboratories would be incapable of constructing glycan arrays, mainly because of the difficulty of synthesizing or obtaining pure oligosaccharides. To provide increased access to this technology, the Consortium for Functional Glycomics (CFG)1 has since 2004 offered to run glycan microarray experiments using samples provided by individual investigators. Participating investigators have submitted a multitude of glycan-binding reagents to be analyzed by the CFG array, and the resulting data are freely available. These data have been extremely valuable to investigators studying the binding properties of particular lectins.
The huge repository of glycan array data presents another opportunity beyond the primary purpose of analyzing individual lectins. Broader studies of the compiled information from all the lectins might be possible—for example, to examine relationships between lectins or to identify lectins that recognize certain types of glycans. Such analyses previously were not possible because the glycan array data were provided in an uninterpreted form. To enable comparisons between glycans and lectins, each dataset needs to be interpreted to give a systematized representation of the specificity of the lectin. The data can be manually interpreted, but that approach would be impractical for so many datasets. Furthermore, manual interpretation is imprecise, particularly for proteins that bind multiple structures or have varying affinity depending on the presentation or overall context of a particular structure. For many proteins, the primary glycan-binding specificity is known, but details about the fine specificity, such as preferred presentations of binding determinants or potentially blocking side chains, are not clear. Objective and automated analyses are required.
Previously we introduced the Motif Segregation algorithm as an approach for systematizing and automating the analysis of glycan array data (20) and further developed the method using Outlier Motif analysis (21). We demonstrated the accuracy of Motif Segregation for extracting the primary binding specificities of a wide variety of glycan-binding proteins and the use of Outlier Motif analysis for a more detailed definition of binding specificity. More recently, we developed software that expands upon these algorithms. The new software enables the automated processing of glycan array datasets, which now makes it practical to interpret all of the datasets in the CFG repository and assemble the information into a database of lectin binding specificities. Such a database gives one the ability not only to extract the specificities of individual lectins, but also to perform global analyses and comparisons of lectin–glycan interactions.
Here we present the use of the software to develop a database of analyzed glycan array data and an initial exploration of questions that can now be investigated using the database. We began with broad comparisons of specificities across multiple lectins and comparisons between glycans in their lectin receptors. Detailed specificities are known for certain lectins, but it is less well known how representative those specificities are of a particular class of lectins. Likewise, alterations to glycan structures are known to shift lectin recognition, but experimental data providing details of those shifts are generally not available. The analyses of these problems provide insights into glycan–lectin biology and demonstrate the utility of this new tool.
EXPERIMENTAL PROCEDURES
Software Development and Data Source
The GlycoSearch software for analyzing glycan array data is written in Java and runs on various computer platforms, including Windows, Macintosh, and Linux platforms. GlycoSearch can directly input and process glycan array data in the CFG Microsoft Excel format, as well as other formats. GlycoSearch was initially developed to run one glycan array dataset at a time. We recently extended the program to process the entire volume of publicly available CFG glycan array data and store the results in an SQL database, thereby making it readily available for instant query. A custom script was used to download and process each glycan array spreadsheet from the CFG website. The entirety of the data, including the spreadsheet content, glycan measurement data, program results, and any additional metadata extracted from the CFG website, were all incorporated into and stored in an SQL database.
The CFG data are provided as Microsoft Excel files. Every recognizable spreadsheet was added to the database. The criteria for a “recognizable spreadsheet” were as follows: (i) the Excel file was readable; (ii) the spreadsheet contained glycan array measurement data in the specified CFG format; and (iii) the arrangement, number, and structure of glycans on the array corresponded to a known CFG glycan array version. A custom program was used to search and identify the data location within the content of each spreadsheet, instead of relying on a fixed data pattern. Datasets were excluded that contained (i) partial glycan measurement data (listing only a subset of the glycans on the array), (ii) data from early array version 1.0 (only data from version 2.0 or higher were used), or (iii) any Excel file that reported data integrity errors (such as a record data overrun).
Once the processing was complete (which required several days using a modern, 12-core, high-end server), both the measurement data and the analysis results were placed in a database for instant query without having to reprocess the data. The addition of new data as they become available is straightforward. A generalized query format was developed to support complex queries into the database using precise definitions of glycan attributes. GlycoSearch supports global and constrained queries and gives researchers the ability to specify which carbon linkage positions to consider, whether taken or free, and/or to specify the number of specific monosaccharide units such as fucose or sulfate residues. GlycoSearch further supports complex queries comprising any series of logical AND/OR/NOT operations on glycan query expressions. This flexible query format allows researchers to effectively search the database for potential lectins and/or proteins with high affinity to the specific glycan query pattern of interest.
Data Presentation
The extracted database queries were further analyzed and processed using Microsoft Excel 2010, MultiExperiment Viewer, and GraphPad Prism 6. The figures were prepared using Canvas XII and Canvas XIV from ACD Systems.
RESULTS
Constructing a Database of Lectin Specificities from Glycan Array Data
Our new software program, called GlycoSearch, uses the Motif Segregation (20) and Outlier Motif (21) algorithms, along with additional developments, to analyze glycan array data generated on any glycan array platform. In a typical glycan array experiment, a purified glycan-binding protein (such as a lectin or glycan-binding antibody, collectively referred to as lectins hereinafter) is incubated on a microarray of diverse oligosaccharides. The binding of the lectin to each glycan on the array is detected and quantified. A biotinylated lectin is often used, which allows for detection using dye-conjugated streptavidin followed by scanning for fluorescence. The output is a list of the glycans on the array and the numerical quantification of the fluorescence at each glycan (shown graphically in Fig. 1A). Such output files can be used directly by GlycoSearch.
Fig. 1.
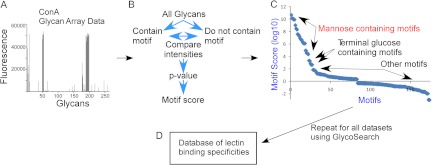
The development of the database of lectin-motif interactions. Quantified glycan array data (A) are processed using the GlycoSearch software and the motif segregation algorithm to produce motif scores for each experiment (B). The analysis of a glycan array experiment of the lectin concanavalin A (ConA) is given as an example, in which the top-scoring motifs contain mannose, followed by motifs with terminal glucose (C). All glycan array datasets were downloaded from the CFG website, processed, and assembled into a database (D). A variety of query types can be applied to the database.
The Motif Segregation algorithm was described in detail elsewhere (20) and is briefly reviewed here. Motifs, or component substructures of oligosaccharides, are predefined by the software. Examples include the blood group A antigen, terminal β-linked galactose, and internal lactosamine. The current version of GlycoSearch starts with over 200 such predefined motifs. For every glycan on an array, the software determines whether each motif is present or absent, and a matrix of the glycans and motifs (in the rows and columns of the matrix, respectively) is populated with 1s and 0s indicating the presence or absence, respectively, of each motif in each glycan. Glycan array data are provided to the program indicating the signal intensity at each glycan (corresponding to the amount of binding of a lectin). For each motif, the program statistically compares the signals of the glycans that contain the motif to those that do not contain the motif (Fig. 1B). Using the Mann–Whitney test, a p value is generated for each motif indicating the likelihood that the observed pattern of signals could be generated by chance. For ease of comparison between motifs, the software takes the logarithm (base 10) of the p value and adds a plus or minus sign, with plus indicating that motif-containing glycans have a higher average intensity, and minus indicating the opposite. This signed, logged p value is referred to as the motif score.
The motif score is a measure of the accuracy with which a motif describes the observed binding of a lectin. If a lectin always shows higher binding to glycans that contain the motif than to those that do not contain the motif, the score is high, but if exceptions occur (either binding is low when the motif is present or binding is high with the motif is missing), the score is lower. For example, when using glycan array data from the lectin concanavalin A, motifs containing mannose have high motif scores, and motifs with terminal glucose (a secondary specificity of concanavalin A) have slightly lower scores (Fig. 1C). The motif score also reflects statistical significance along with accuracy; that is, a motif that appears in only 10 of 500 glycans will have a lower motif score than one that appears in 20 of 500 glycans, even with perfect accuracy (all glycans with the motif have higher binding than all glycans without the motif). As such, the motif score does not provide a direct quantitative measurement of the affinity of the interaction; only the statistical significance with which a particular motif is strongly associated with the observed pattern of measurement data can be obtained.
The automated processing of glycan array datasets enabled the development of a database of lectins and their associated motif scores (Fig. 1D). We retrieved all glycan array datasets available as of August 2012, amounting to 2883 independent experiments, along with the metadata for each experiment (the type of lectin, the researcher submitting the sample, etc.). Each dataset was analyzed to evaluate 220 pre-defined motifs. (The complete list of motifs is in supplemental Table S2.) The motifs covered variations on the main features observed in N-linked and O-linked glycans and in glycolipids, which are the glycan types most heavily represented on the glycan arrays. The database of assembled motif scores makes possible many types of analyses and queries, such as comparing motif specificities between lectins and identifying lectins that bind specific motifs. Here we present initial, global analyses of this new information.
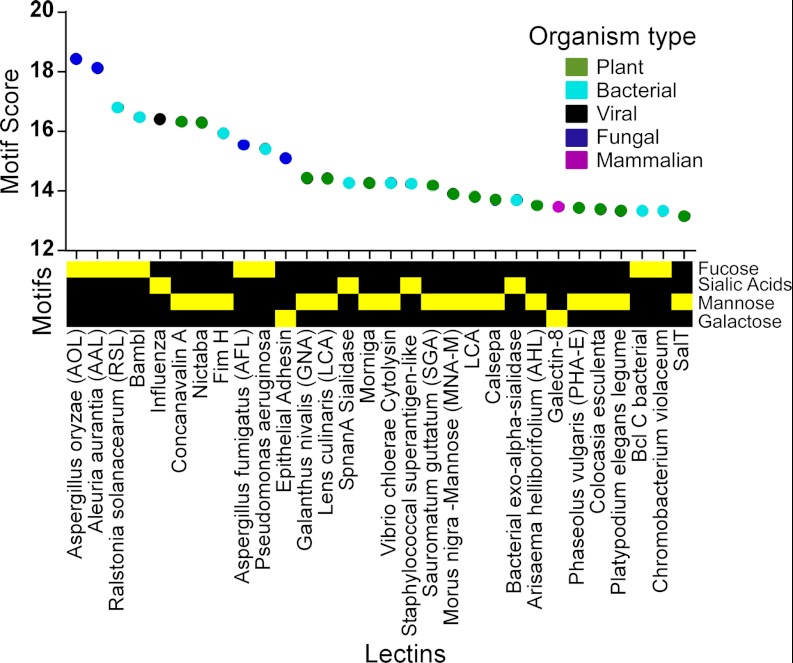
As the motif score reflects the accuracy with which the lectin binding is represented, we wondered what were the most accurately described interactions in the database and, more specifically, whether particular motifs were strongly represented among the highest scoring interactions. An extraction of the top-scoring interactions in the database showed an enrichment of simple monosaccharide motifs (Fig. 2). The top four motif scores, from 18.4 down to 16.5, were from the lectins AOL, AAL, RSL, and BambL, all known fucose binders, interacting with a simple fucose motif. Only three other motifs, sialic acid, mannose, and galactose, all of which are simple and common monosaccharides, were represented in the next 26 top scores (the complete information is provided in supplemental Table S1).
Fig. 2.
The top 30 motif scores. The motif score is indicated along the y-axis, the labels on the x-axis give the lectin name, and the color gives the organism category of each lectin. The motif type is indicated by the yellow squares below the lectin names. Only four motif types, all simple monosaccharides, are represented in the top 30.
This result suggests that a small set of motifs accounts for the best described interactions in the glycan array data and that those motifs tend to be simple monosaccharides. Lectins with complex binding preferences can have fine specificities that make an accurate definition of the motif difficult (21), so this result concurs with the concept that simple specificities are easier to accurately describe. Furthermore, we previously showed that the definition of higher complexity motifs to account for outliers in the glycan array data can result in higher motif scores (21), so this result also might indicate that the pre-defined motifs do not always accurately describe the lectin specificities. However, in the current analyses, precise comparisons of scores between motifs are not possible because of limitations of the Mann–Whitney test (see Discussion section).
We asked whether particular organism types tend to have the highest scoring interactions. No clear order was observed in the highest scoring lectins (Fig. 2), although plant, bacterial, and fungal lectins had most of the top scores. Only one mammalian and one viral lectin were among the top 30, suggesting that simple, well-described interactions are possible in mammal and viral lectins but are less common than among other organism types. This analysis is limited by the datasets available and the glycans on the array, and therefore it is not an unbiased survey of lectins in biology. A more systematic study will be required in order to address these observations.
Consistent Differences Exist between Types of Organisms in Lectin Specificity
We next asked whether consistent differences exist between the organism types in the motifs that they bind—for example, whether bacterial lectins in general bind different glycans than mammalian lectins. Such an analysis could provide insights into the nature of interactions between and within organisms. For this initial analysis, we relied on the category designations input by the researchers who submitted the lectins to the CFG. The designations were provided for 1191 of the 2883 (57%) datasets, comprising 477 plant, 55 bacterial, 344 human, 6 pig, 4 chicken, 163 mouse, 91 viral, 35 fungal, 1 antibody, and 2 toxin lectins. Future versions of the database will contain more complete information obtained through manual research and annotation.
The scores for each motif were averaged over all the lectins within each category type (fungi, plants, bacteria, viruses, and mammals) and clustered to allow visual comparison (Fig. 3A). Viral lectins had the narrowest specificities, confined primarily to sialylated motifs. Fungal and bacterial lectins showed high overall similarity, sharing high average scores for fucose motifs (as in Fig. 2) and for mannose-containing motifs. Mammalian lectins had the lower average scores for all motifs except sulfated glycans.
Fig. 3.
Differences in lectin specificities between organism categories. A, for each motif, the motif scores were averaged across all lectins within each category. Motifs with an average score below 0.88 in all organism categories were removed for clarity, and the remaining data were clustered. The categories are indicated by the column labels, and the motifs by the row labels. B, the average motif scores within each category were normalized to a maximum value of 10 (the average motif scores within a category all were divided by the maximum average in that category and multiplied by 10) to show the relative motif ranks within each category. C, for each motif, the maximum individual motif score was extracted from each category. For example, for the motif “Terminal Galβ1,4-” the program found the best score among all fungal lectins, then among all viral lectins, etc. Motifs with no scores above 7.4 in any category were removed for clarity.
To obtain a view of the relative preferences within each category, we normalized the motif scores within each category to a common maximum value (Fig. 3B). This view again shows the narrow specificity of viral lectins and the similarities among fungal, bacterial, and plant lectins. The mammalian lectins share with those categories a relative preference for mannose-containing motifs and a higher relative preference for galactose and lactosamine motifs. Mammalian lectins share with viral lectins some relative preference for sialylated motifs, but they are unlike all the other categories in their strong relative preference for sulfated motifs.
Individual lectins might have specificities that are different from the averages within each category. To examine this possibility, we extracted the best motif score from each category for each motif (Fig. 3C). As observed above (Fig. 2), individual plant, fungal, and bacterial lectins had the highest scores for fucose motifs. The top individual mammalian lectins had scores similar to those of the other categories for most motifs, in contrast to the lower scores observed using averages over all mammalian lectins (Fig. 3A), suggesting that simple and well-described specificities are possible but not common in mammalian lectins. The top sialic-acid-binding lectins are fungal and viral, and certain mammalian lectins bind almost as well to sialic acid. Several differences between the categories are apparent that could give insights into the nature of interactions between the organisms.
Consistent differences between the categories also could be identified through statistical comparison of the lectin motif scores of different categories. For each motif, the motif scores from all lectins in a category were compared (via t test) to the motif scores from all lectins in another category (supplemental Fig. S1). This analysis confirmed the significance of the differences observed above. Virus lectins were the most different from the other categories, always showing higher scores for sialic acids and lower scores for the other motifs. Bacterial, plant, and fungal were the most similar to one another, and mammalian lectins had significant differences with each group.
Terminal Modifications to a Core Glycan Shift the Type of Lectin Recognition
Another type of analysis begins with the glycans rather than the lectins. N-linked and O-linked glycans typically undergo alterations as cells respond to stimuli or change differentiation states. Common modifications include fucosylation, sialylation, and sulfation of outer-arm lactosamine. In some cases the modifications create high-affinity ligands for cell-surface receptors, and in other cases the modifications block interactions with receptors. We hypothesized that changes in terminal modifications to lactosamine would significantly alter the types of lectins that recognized the structure.
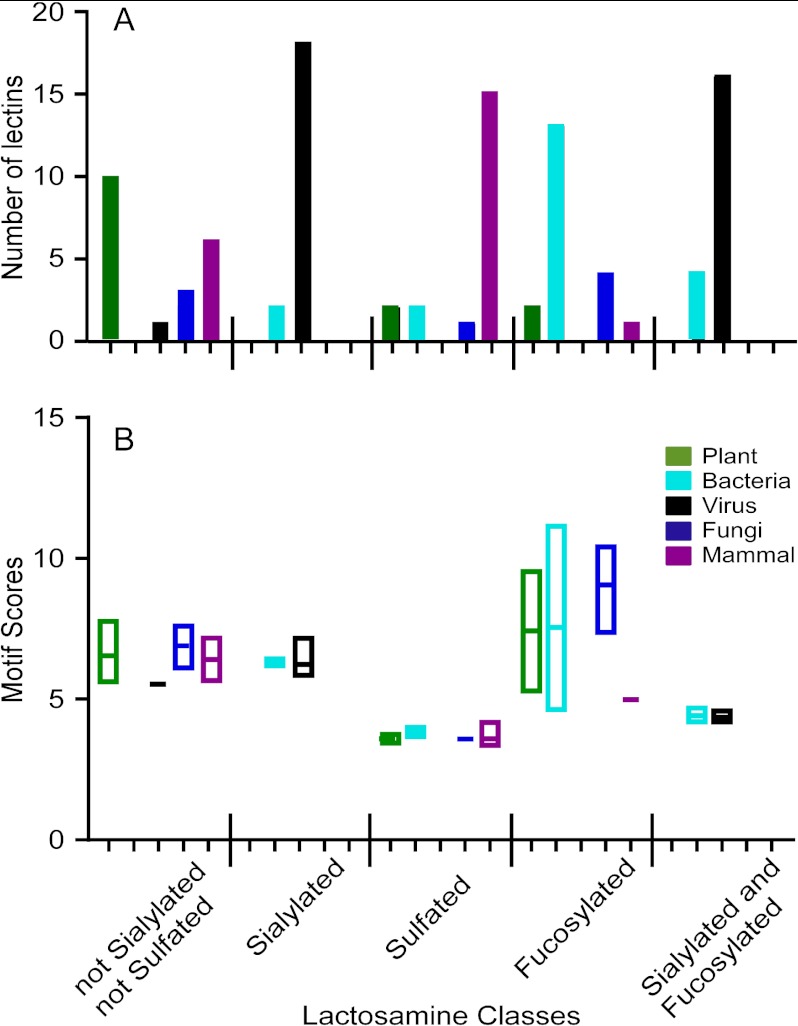
To test that hypothesis, we defined a series of related structures beginning with lactosamine and differing by the addition of one or two monosaccharides. These structures were lactosamine, fucosylated lactosamine, sialylated lactosamine, sulfated lactosamine, and combinations of these modifications (for example, fucosylated and sialylated). The database was queried to return the lectins with the highest scores for each of these motifs. This query selected from all lectins in the database, not just those with annotation indicating the organism type, and the resulting selections were manually annotated to specify the organism type.
Among the top 20 lectins for each motif, we tallied the number of lectins represented from each type of organism. This analysis revealed a major shift in the types of lectins binding the lactosamine variants (Fig. 4A). The unmodified lactosamine was best recognized by plant lectins, followed by representatives from the other types, excluding bacteria. The addition of sialic acid shifted the type almost exclusively to viral lectins, along with a small representation of bacterial lectins, whereas the addition of fucose shifted the dominant type to bacterial lectins and excluded viral lectins. (The manual annotation of the search results identified many viral lectins that previously were unspecified, in contrast to the analysis of Fig. 3, which relied on the available annotation.) Using lactosamine that was sialylated and fucosylated, only viral and bacterial lectins were represented in the top 20. A completely different result was obtained when the lactosamine was sulfated. Sulfation resulted in primarily mammalian lectins achieving the top scores, with some representation from the other types, except viruses. This result corresponds with the above analysis of lectin preferences within each category (Fig. 3).
Fig. 4.
Differences between lactosamine classes in lectin recognition. For each motif defining a modification of lactosamine, the lectins with the top 20 motif scores for that motif were extracted from the database. Replicate experiments of the same lectin were removed. Within each list of 20 lectins, the number of representatives from each organism type was tallied, as indicated in panel A. The range of motif scores is indicated for each group of lectins in panel B.
Within each lactosamine class, considerable overlap in the range of scores was observed between the organism types (Fig. 4B), indicating that no single organism type completely dominates binding to a class of structures. The motif score ranges were very different between the lactosamine classes, with fucosylated lactosamine giving the highest scores (similar to above) and sulfated giving the lowest. This change is likely due to a change in motif representation on the arrays (see “Discussion”) and a change in selectivities for the defined motifs. For example, lectins binding sulfated motifs likely are not purely selective for sulfated glycans, whereas fucose-binding lectins tend to be more selective.
To examine whether the lectins returned from the searches of the motifs defining each lactosamine class were indeed specific for those motifs, we examined the individual lectins with the top scores for each class (Table I; complete information is presented in supplementary Table S2). In each case, the top-scoring lectins from each lactosamine class had specificities corresponding to their lactosamine class. For example, the lectins that were extracted from the search using sialylated lactosamine had highest scores for the motif “Neu5Ac anywhere,” and lectins pulled out from the sulfated lactosamine search had highest scores for the “Sulfated glycans” motif. These motifs correspond to the previously determined specificities of the lectins, if available. This analysis confirms the accuracy of the searches and furthermore demonstrates the use of the tool for identifying lectins with defined specificities.
Table I. The top three lectins for each lactosamine class.
| Class | Score of searched motifa | Primscreen IDb | Lectin | Organism type | Top motifs | Motif score |
|---|---|---|---|---|---|---|
| Not sialylated or sulfated | 7.76 | 4661 | Ricinus communis agglutinin (RCA I-10) | Plant | Terminal Galb1-4 | 9.59 |
| N-Glycan Complex | 9.34 | |||||
| Type 2 Chain (Galb1-4GlcNAcb) | 8.85 | |||||
| 7.53 | 3775 | Macrolepiota procera lectin (MPL-2) | Fungal | Terminal Galb | 10.57 | |
| Terminal Galactose AnyLinkage | 9.98 | |||||
| Poly LacNAc OR Neo Poly LacNAc Terminal | 8.98 | |||||
| 7.22 | 4714 | Solanum tuberosum lectin (STL-10) | Plant | Type 2 Chain (Galb1-4GlcNAcb) | 13.00 | |
| Poly LacNAc (Galb1-4GlcNAcb1–3Galb … ) | 8.48 | |||||
| Internal Poly LacNAc | 7.71 | |||||
| Sialylated | 7.15 | 5211 | A/OK/5386/2010 H3N2 | Viral | Neu5Aca Anywhere | 15.73 |
| Neu5Ac Anywhere | 15.35 | |||||
| Sialic Acids AnyLinkage | 14.98 | |||||
| 6.60 | 5235 | A/OK/5342/2010 H3N2 | Viral | Neu5Aca Anywhere | 14.37 | |
| Neu5Ac Anywhere | 13.81 | |||||
| Sialic Acids AnyLinkage | 13.29 | |||||
| 6.54 | 5218 | A/Shorebird/DE/300/2009 | Viral | Neu5Aca Anywhere | 16.42 | |
| Neu5Ac Anywhere | 15.86 | |||||
| Sialic Acids AnyLinkage | 15.65 | |||||
| Sulfated | 4.16 | 1704 | Surfactant protein A | Mammalian | Sulfated Glycans | 10.09 |
| 6′ Sulfo | 8.43 | |||||
| 3′ Sulfo | 6.84 | |||||
| 3.99 | 1640 | Chemotaxis inhibitory protein of Staphylococcus aureus | Bacterial | Sulfated Glycans | 8.72 | |
| 6′ Sulfo | 7.39 | |||||
| 3′ Sulfo | 6.14 | |||||
| 3.85 | 1667 | Mouse E selectin | Mammalian | Sulfated Glycans | 9.40 | |
| 6′ Sulfo | 7.90 | |||||
| Neu5Aca Anywhere | 6.44 | |||||
| Fucosylated | 11.68 | 3363 | Ralstonia solanacearum lectin (RSL) | Bacterial | Fucose Anywhere | 16.80 |
| Fuca Anywhere | 16.67 | |||||
| Fuca1-2 | 11.94 | |||||
| 11.14 | 3340 | BambL lectin | Bacterial | Fucose Anywhere | 16.48 | |
| Fuca Anywhere | 16.34 | |||||
| Fuca1-2 | 11.58 | |||||
| 10.40 | 4217 | Aspergillus oryzae lectin (AOL) | Fungal | Fucose Anywhere | 18.33 | |
| Fuca Anywhere | 18.24 | |||||
| Fuca1-2 | 10.16 | |||||
| Sialylated and fucosylated | 4.68 | 5109 | Staphylococcal superantigen-like protein (SSL0) | Bacterial | Sialic Acids AnyLinkage | 12.57 |
| Neu5Aca Anywhere | 12.08 | |||||
| Neu5Ac Anywhere | 11.74 | |||||
| 4.58 | 1773 | PR8 E158K: mutation in the HA gene | Viral | Neu5Aca Anywhere | 10.83 | |
| Neu5Ac Anywhere | 10.70 | |||||
| Sialic Acids AnyLinkage | 10.25 | |||||
| 4.56 | 2491 | HN protein from paramyxovirus | Viral | Sialic Acids AnyLinkage | 5.58 | |
| Terminal Neu5Aca2-3 | 5.39 | |||||
| Neu5Aca2-3 Anywhere | 5.39 |
a Score of the motif used in the search, corresponding to the indicated lactosamine class.
b The unique identifier of the dataset in the CFG website, where more information about the sample and the experiment can be found.
DISCUSSION
Systematic comparisons of specificity between multiple lectins or comparisons between glycans and their lectin partners previously were not possible because of the lack of the necessary experimental analyses. The producers and users of glycan array data have long recognized the latent potential of the data for such bioinformatics studies but did not have the data in an interpretable and readily usable format. A new software program for the automated analysis of glycan array data was used to process the entire repository of CFG glycan array data, resulting in a database of lectin-motif interactions. The database enabled several types of analyses that provided provocative insights into glycan–lectin biology. For example, we observed that sulfate recognition is primarily found in mammalian lectins and that modifications to a core glycan result in shifts in the type of organism recognizing the glycan. These observations provide routes for further investigation, and they demonstrate the utility of the database and analysis program. We anticipate that these resources will be highly valuable to the glycobiology research community.
Because the metadata were available for many of the lectins, we could examine questions relating to differences and similarities between organism types in terms of their lectin specificities. Viral lectins had the least diversity in specificity, showing nearly exclusive preference for sialic acids. Lectins from the other categories also can have high specificity for sialic acids (Fig. 3), but they have more diversity in specificities. The competition for sialic acid motifs among lectins of various organism types might play a major role in microorganism–host biology. Both viral and bacterial pathogens can use sialic acids for attachment to host epithelial surfaces (22), and the binding to sialic acids might be in competition with host sialic-acid-binding lectins such as the siglecs (1). The interplay among host recognition of self-associated molecular patterns, sialic acids on invaders, and potential mimics of sialic acid on host antigens (23) might work to determine the outcome of microbial colonization (24).
Mammalian lectins were different from the other lectins in a few respects. Although the average motif scores were lowest for most motifs (Fig. 3A), individual mammalian lectins had scores comparable to those of other organism types for most motifs (Fig. 3C). This result might indicate a greater diversity of specificities among mammalian lectins. For example, because most viral lectins have good specificity for sialic acids, the average score for the sialic acid motif is high among viral lectins, whereas because mammalian lectins bind many different motifs, the average is not high for any motif. The normalized averages, reflecting relative scores within each category, also showed a broader diversity of specificities for mammalian lectins than for any other category (Fig. 3B). This greater diversity might reflect the greater general complexity of the mammalian species and the need for a greater number of functions for the lectins. Each function of the mammalian lectins carries unique requirements in terms of glycan specificity. Mammalian lectins involved in innate immune recognition, immune regulation, protein quality control and clearance, lymphocyte homing, receptor regulation, and others have specificities covering most of the major motif types. In contrast, the more limited functions of the lectins of the other organism types might require fewer specificities.
The motif for which mammalian lectins had the highest average and individual scores was sulfated glycans. Although individual examples of plant and bacterial lectins appear to bind sulfated glycans (Fig. 4), the requirements of sulfation for binding are not as well characterized as for certain mammalian lectins such as L-selectin (25) and siglec-8 (26). The sulfation of glycans could restrict binding uniquely to lectins of the host, thereby restricting the glycan–lectin signaling to internal routes. Given that some pathogens display sialylated glycans that mimic those of the host (23), sulfation might prevent the development of autoimmunity based on the cross-over of immunity from foreign antigens to self-antigens.
Plant, bacterial, and fungal lectins showed many similarities but also had distinctions. They shared, on average, preferences for motifs based on fucose, mannose, and lactosamine (Figs. 3A and 3B). These similarities in specificities might reflect general, shared functional features among these species. Plants had a greater number of lectins with high selectivity for lactosamine (Fig. 4), perhaps owing to a great need for broad immune protection against foreign invaders (27).
The evaluation of a series of lactosamine modifications showed a significant change in the type of lectin that binds the structures. Unmodified lactosamine had the broadest parity of lectin binding among the organism types (Figs. 4A and 3B), suggesting that lactosamine interactions are broadly used in biology. Multiple beneficial plant and bacterial species use such interactions in the human gut, and some plants use lactosamine interactions in immune defense (27). As noted above, sulfation shifted binding primarily to mammalian lectins, and fucosylation shifted to mainly bacterial lectins, as the most specific binders. The results are restricted by the lectins that are present in the database, so a more comprehensive and unbiased look at lectin binding would be required in order to pursue these findings. However, the results show the significant effects on lectin binding of glycan modifications and demonstrate a useful approach to this type of question.
These observations raise the possibility of characterizing whether certain lectin–glycan interactions are endogenous, occurring primarily within the organism, or exogenous, occurring between organisms. In order to thoroughly investigate that topic, we would need detailed information on the glycomes of each organism; this is available for certain tissue types but is still is being generated for most. The question of interactions between bacteria and their hosts is particularly interesting, because they live in either a symbiotic or an antagonistic relationship with each other. Having detailed information on both the lectin specificities and the glycomes of the various bacteria and hosts could lead to insights into those relationships.
The present study has some limitations in the comparisons between motif scores. A relatively small set of monosaccharides accounted for the top scores (Fig. 2), probably because simple specificities are easier to accurately describe and therefore result in higher motif scores. Lectins with more complex specificities would be harder to describe and thus would not be accurately described by the pre-defined motifs. In addition, complex lectins might more frequently require other factors for optimal binding, such as certain multivalent presentations of the glycans (28, 29), particular peptide backbones, or other protein cofactors. Complex glycan motifs recognized by certain lectins might not be present on the arrays, as the arrays contain only a fraction of the glycome (30). These limitations could be addressed in future developments, such as new glycan array data with expanded glycan repertoires or varying densities (25, 29) and the continued definition of motifs that account for complex specificities (21). In addition, we will continue to develop the analysis algorithms, for example, by building in additional statistical analyses or other, recently described approaches for analyzing glycan array data (31, 32). The incorporation of new motifs, enhanced analysis methods, and new data from more complex glycan arrays should allow more accurate information to be obtained from the searches.
We foresee this program and database being useful not just for global searches and analyses, but also for deep analyses of individual datasets. GlycoSearch reports all the significant motifs and motif combinations, including weaker secondary motifs and binding inhibitors, from among the pre-defined set of 220 motifs. The program also provides a list of outlier glycans—those with signals that do not fit the predicted binding of the lectin (21). This initial analysis might provide directions for further investigations of the data allowing us to better understand the lectin specificity. GlycoSearch enables such investigations by supporting the addition of new, user-defined motifs of any complexity. GlycoSearch can parse and interpret any additional user-defined motifs, automatically determine which glycans on the array have the motifs, and accurately compute the resulting motif scores for the newly defined (not previously seen) user motifs. Completely flexible motif definitions and search parameters will allow the pursuit of very specific questions relating to complex, fine specificities of lectins. This capability should be increasingly useful as glycan arrays become available with more and more glycans, and as lectins are analyzed with specificities not corresponding to the pre-defined motifs. An intriguing option is to define motifs based on structural characteristics, rather than simple nomenclature. Such motifs could reveal structural features that influence the binding of certain lectins and that are not identifiable from the simple nomenclature.
Another topic that can be investigated using the flexible analysis of GlycoSearch is the independence of motifs. A lectin potentially could bind two distinct motifs, as in wheat-germ agglutinin binding of motif GlcNAc and sialic acid. In order to discern whether each motif independently contributes to lectin binding, special analyses are required. For example, we could constrain a comparison to only glycans that do not contain the first motif and either do or do not contain the second motif, thereby removing the variable of the first motif. The flexibility of the motif definitions and comparisons in GlycoSearch enable this type of analysis. Furthermore, the current version of GlycoSearch includes a new development in which pairwise combinations of motifs and exclusions of motifs are each tested for potential improvements to the motif score. If the motif score is improved using such combinations, the two motifs are inferred to be independent contributors to the lectin binding. The results from these analyses have been incorporated into the database.
This database and analysis program promises to be a useful resource for the glycobiology community. The database enables analyses and comparisons of glycan–lectin interactions across unrelated experiments, and the GlycoSearch program enables detailed studies of individual datasets. Data from other glycan array platforms could be incorporated into the database, such as arrays composed of natural glycans (33) or designed to probe virus specificities (34–36), as the motif score output of GlycoSearch is independent of the platform. With further refinement of the annotation in the database, development of the analysis algorithms and motif definitions, and incorporation of new data, we expect increasing usefulness in glycobiology studies. Future studies in systems biology also could benefit from broader, accessible information on protein–glycan interactions. Furthermore, we foresee practical uses of these tools, such as identifying lectins with defined specificities that could be used as analytical reagents, or modeling the effects of drugs targeting particular glycan–lectin interactions (37).
Supplementary Material
Acknowledgments
This work used publicly available data provided by the Consortium for Functional Glycomics.
Footnotes
* We gratefully acknowledge support of this work by a Bridging Grant from the Consortium for Functional Glycomics, the Alliance of Glycobiologists for Detection of Cancer (1U01CA168896), and the Van Andel Research Institute.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CFG
- Consortium for Functional Glycomics.
REFERENCES
- 1. Crocker P. R., Paulson J. C., Varki A. (2007) Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7, 255–266 [DOI] [PubMed] [Google Scholar]
- 2. Rabinovich G. A., van Kooyk Y., Cobb B. A. (2012) Glycobiology of immune responses. Ann. N. Y. Acad. Sci. 1253, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Kooyk Y., Rabinovich G. A. (2008) Protein-glycan interactions in the control of innate and adaptive immune responses. Nat. Immunol. 9, 593–601 [DOI] [PubMed] [Google Scholar]
- 4. Marth J. D., Grewal P. K. (2008) Mammalian glycosylation in immunity. Nat. Rev. Immunol. 8, 874–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rudd P. M., Elliott T., Cresswell P., Wilson I. A., Dwek R. A. (2001) Glycosylation and the immune system. Science 291, 2370–2376 [DOI] [PubMed] [Google Scholar]
- 6. Stevens J., Blixt O., Paulson J. C., Wilson I. A. (2006) Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat. Rev. Microbiol. 4, 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rosen S. D. (2004) Ligands for l-selectin: homing, inflammation, and beyond. Annu. Rev. Immunol. 22, 129–156 [DOI] [PubMed] [Google Scholar]
- 8. Zarbock A., Ley K., McEver R. P., Hidalgo A. (2011) Leukocyte ligands for endothelial selectins: specialized glycoconjugates that mediate rolling and signaling under flow. Blood 118, 6743–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aebi M., Bernasconi R., Clerc S., Molinari M. (2010) N-glycan structures: recognition and processing in the ER. Trends Biochem. Sci. 35, 74–82 [DOI] [PubMed] [Google Scholar]
- 10. Partridge E. A., Le Roy C., Di Guglielmo G. M., Pawling J., Cheung P., Granovsky M., Nabi I. R., Wrana J. L., Dennis J. W. (2004) Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science 306, 120–124 [DOI] [PubMed] [Google Scholar]
- 11. Rana N. A., Haltiwanger R. S. (2011) Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of notch receptors. Curr. Opin. Struct. Biol. 21, 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pang P. C., Chiu P. C., Lee C. L., Chang L. Y., Panico M., Morris H. R., Haslam S. M., Khoo K. H., Clark G. F., Yeung W. S., Dell A. (2011) Human sperm binding is mediated by the sialyl-LewisX oligosaccharide on the zona pellucida. Science 333, 1761–1764 [DOI] [PubMed] [Google Scholar]
- 13. Bathe O. F., Shaykhutdinov R., Kopciuk K., Weljie A. M., McKay A., Sutherland F. R., Dixon E., Dunse N., Sotiropoulos D., Vogel H. J. (2010) Feasibility of identifying pancreatic cancer based on serum metabolomics. Cancer Epidemiol. Biomarkers Prev. 20, 140–147 [DOI] [PubMed] [Google Scholar]
- 14. Lo J. F., Yu C. C., Chiou S. H., Huang C. Y., Jan C. I., Lin S. C., Liu C. J., Hu W. Y., Yu Y. H. (2010) The epithelial-mesenchymal transition mediator s100a4 maintains cancer initiating cells in head and neck cancers. Cancer Res. 71, 1912–1923 [DOI] [PubMed] [Google Scholar]
- 15. Drickamer K., Taylor M. E. (2002) Glycan arrays for functional glycomics. Genome Biol. 3, REVIEWS1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yue T., Haab B. B. (2009) Microarrays in glycoproteomics research. Clin. Lab. Med. 29, 15–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Manimala J. C., Roach T. A., Li Z., Gildersleeve J. C. (2006) High-throughput carbohydrate microarray analysis of 24 lectins. Angew Chem. Int. Ed. Engl. 45, 3607–3610 [DOI] [PubMed] [Google Scholar]
- 18. Blixt O., Head S., Mondala T., Scanlan C., Huflejt M. E., Alvarez R., Bryan M. C., Fazio F., Calarese D., Stevens J., Razi N., Stevens D. J., Skehel J. J., van Die I., Burton D. R., Wilson I. A., Cummings R., Bovin N., Wong C. H., Paulson J. C. (2004) Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee S. M., Chan R. W., Gardy J. L., Lo C. K., Sihoe A. D., Kang S. S., Cheung T. K., Guan Y. I., Chan M. C., Hancock R. E., Peiris M. J. (2010) Systems-level comparison of host responses induced by pandemic and seasonal influenza A H1N1 viruses in primary human type I-like alveolar epithelial cells in vitro. Respir. Res. 11, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Porter A., Yue T., Heeringa L., Day S., Suh E., Haab B. B. (2010) A motif-based analysis of glycan array data to determine the specificities of glycan-binding proteins. Glycobiology 20, 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maupin K. A., Liden D., Haab B. B. (2011) The fine specificity of mannose-binding and galactose-binding lectins revealed using outlier-motif analysis of glycan array data. Glycobiology 22, 160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pieters R. J. (2011) Carbohydrate mediated bacterial adhesion. Adv. Exp. Med. Biol. 715, 227–240 [DOI] [PubMed] [Google Scholar]
- 23. Carlin A. F., Uchiyama S., Chang Y. C., Lewis A. L., Nizet V., Varki A. (2009) Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil siglec-9 and dampen the innate immune response. Blood 113, 3333–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Varki A. (2011) Since there are pamps and damps, there must be samps? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology 21, 1121–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Uchimura K., Gauguet J. M., Singer M. S., Tsay D., Kannagi R., Muramatsu T., von Andrian U. H., Rosen S. D. (2005) A major class of L-selectin ligands is eliminated in mice deficient in two sulfotransferases expressed in high endothelial venules. Nat. Immunol. 6, 1105–1113 [DOI] [PubMed] [Google Scholar]
- 26. Bochner B. S., Alvarez R. A., Mehta P., Bovin N. V., Blixt O., White J. R., Schnaar R. L. (2005) Glycan array screening reveals a candidate ligand for siglec-8. J. Biol. Chem. 280, 4307–4312 [DOI] [PubMed] [Google Scholar]
- 27. Rudiger H., Gabius H. J. (2001) Plant lectins: occurrence, biochemistry, functions and applications. Glycoconj. J. 18, 589–613 [DOI] [PubMed] [Google Scholar]
- 28. Oyelaran O., Li Q., Farnsworth D. W., Gildersleeve J. C. (2009) Microarrays with varying carbohydrate density reveal distinct subpopulations of serum antibodies. J. Proteome Res. 8, 3529–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y., Li Q., Rodriguez L. G., Gildersleeve J. C. (2010) An array-based method to identify multivalent inhibitors. J. Am. Chem. Soc. 132, 9653–9662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cummings R. D. (2009) The repertoire of glycan determinants in the human glycome. Mol. Biosyst. 5, 1087–1104 [DOI] [PubMed] [Google Scholar]
- 31. Xuan P., Zhang Y., Tzeng T. R., Wan X. F., Luo F. (2011) A quantitative structure-activity relationship (QSAR) study on glycan array data to determine the specificities of glycan-binding proteins. Glycobiology 22, 552–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cholleti S. R., Agravat S., Morris T., Saltz J. H., Song X., Cummings R. D., Smith D. F. (2012) Automated motif discovery from glycan array data. OMICS 16, 497–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Song X., Lasanajak Y., Xia B., Heimburg-Molinaro J., Rhea J. M., Ju H., Zhao C., Molinaro R. J., Cummings R. D., Smith D. F. (2011) Shotgun glycomics: a microarray strategy for functional glycomics. Nat. Methods 8, 85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Song X., Yu H., Chen X., Lasanajak Y., Tappert M. M., Air G. M., Tiwari V. K., Cao H., Chokhawala H. A., Zheng H., Cummings R. D., Smith D. F. (2011) A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. J. Biol. Chem. 286, 31610–31622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nycholat C. M., McBride R., Ekiert D. C., Xu R., Rangarajan J., Peng W., Razi N., Gilbert M., Wakarchuk W., Wilson I. A., Paulson J. C. (2012) Recognition of sialylated poly-n-acetyllactosamine chains on N- and O-linked glycans by human and avian influenza A virus hemagglutinins. Angew Chem. Int. Ed. Engl. 51, 4860–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Childs R. A., Palma A. S., Wharton S., Matrosovich T., Liu Y., Chai W., Campanero-Rhodes M. A., Zhang Y., Eickmann M., Kiso M., Hay A., Matrosovich M., Feizi T. (2009) Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat. Biotechnol. 27, 797–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fuster M. M., Esko J. D. (2005) The sweet and sour of cancer: glycans as novel therapeutic targets. Nat. Rev. Cancer 5, 526–542 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.