Abstract
Glycoproteins are well represented among biomarkers for inflammatory and cancer diseases. Secreted and membrane-associated glycoproteins make excellent targets for noninvasive detection. In this review, we discuss clinically applicable markers of cancer diseases and methods for their analysis. High throughput discovery continues to supply marker candidates with unusual glycan structures, altered glycoprotein abundance, or distribution of site-specific glycoforms. Improved analytical methods are needed to unlock the potential of these discoveries in validated clinical assays. A new generation of targeted quantitative assays is expected to advance the use of glycoproteins in early detection of diseases, molecular disease classification, and monitoring of therapeutic interventions.
With the expansion of targeted therapeutics and individualized treatment regimens, markers for classification of diseases and treatment response are becoming increasingly important (1). Biomarkers represent surrogate measurements of correlated, perhaps difficult-to-measure, biological processes. Clinical biomarkers ideally represent one-to-one relationships with high probabilities of capturing true associations of a marker with a specific biological process. Such relationships are, however, rare in biology, which is one of the reasons for the lengthy standardized testing process required by regulatory agencies for their approval (2). Glycoproteins have high potential to serve as markers for a variety of biological processes; they represent a rich source of information because their nontemplate-based synthesis by complex enzymatic pathways reflects expression of protein-coding genes, the availability of activated monosaccharide substrates for synthesis of glycans, the activity of the enzymatic glycosylation machinery, and multiple downstream effectors (3).
In this review, we will primarily discuss O-GalNAc and N-glycoproteins as markers of cancer and associated infectious diseases. This reflects their prominence among established biomarkers associated with tissue- and disease-specific functions and the relative ease of detecting secreted and cell surface proteins (4–6). However, there are additional glycoconjugates whose assessment becomes increasingly feasible with newly developed analytical approaches (7, 8). Here, we summarize the current clinical use and future opportunities in the use of glycoproteins as disease markers. We use these examples to introduce the concept of “single protein-omics,” which we define as comprehensive characterization of all modified forms of a protein. We argue that detailed characterization of disease-specific modifications of a single protein may be a more effective biomarker discovery strategy than proteome-wide studies, which cannot capture the richness and diversity of individual protein forms.
Pathophysiology of Glycoproteins
Glycoprotein Biosynthesis and Structural Diversity of Glycans
Mucin-type O-glycosylation and N-glycosylation occur during protein passage through the endoplasmic reticulum and Golgi compartments (9, 10). Glycoproteins are secreted or localized to outer cell membranes and the inner membranes of cell compartments, including the endoplasmic reticulum, Golgi, and lysosomes. N-Linked glycosylation is initiated by oligosaccharyltransferase, which catalyzes the transfer of a dolichol-linked glycan precursor to protein asparagine residues typically within NX(S/T) (X ≠ P) sequons (11). Nascent N-linked glycans undergo trimming by glycosidases and extension of a common core (Manα1–6(Manα1–3)Manβ1–4GlcNAcβ1–4GlcNAcβ1-Asn) by glycosyltransferases (9). In a distinct pathway, O-GalNAc (mucin-type) protein glycosylation is initiated by a group of polypeptide N-acetylgalactosaminyltransferases that catalyze the linkage of GalNAc residues to serine or threonine residues, but no specific O-GalNAc glycosylation motif has been described (10, 12). Estimates suggest that over 50% of all proteins are glycosylated, but experimental evidence for the extent of protein glycosylation is incomplete (13). O-GalNAc glycoproteins are estimated to comprise 10% of proteins in the human proteome and 50% among those going through the secretory pathway (14). Two recent papers find, in various human tissues, ∼2,500 N-glycosylated proteins each, but the sets have only partial overlap (11, 15). It is likely that additional glycosylated proteins will be identified as detection methods improve.
Fucose, galactose, glucose, mannose, GlcNAc, GalNAc, and N-acetylneuraminic acid are the primary building blocks of human N- and O-GalNAc glycans. Over 200 glycosyltransferases and many additional regulatory proteins expressed in tissue-specific patterns and levels participate in glycan synthesis, using the seven monosaccharides to build a diverse set of glycan structures (16). The variety of glycan structures on human proteins is not entirely known, although some estimates of the numbers exist (17). Each glycosylation site is typically occupied by a few major and additional minor glycan forms (18, 19). In addition, the occupancy of each site, i.e. the percentage of a glycosylation site occupied by glycans, varies and can affect the quantity of glycoforms observed in a particular context. This micro-heterogeneity creates a large variety of site-specific glycoforms of potential biological interest. The search space for glycoprotein biomarkers is therefore quite large, and elucidation of the associations between site-specific glycoform variants and disease processes is one of the driving forces of current biomarker research.
Glycoproteins in Disease Context
Glycosylation is organism-, tissue-, and cell type-specific (20). This is closely tied to the varied functions that diverse protein glycoforms carry out in complex organisms (16). The activities of glycosyltransferases depend on factors, including their quantity, localization, and substrate availability (14, 16). These factors are modulated with respect to changes in the cellular environment; disturbance of this equilibrium in disease often leads to altered glycoprotein distribution. Major changes in glycoprotein distribution are associated with congenital disorders of glycosylation, and the resultant aberrantly glycosylated proteins serve as classic examples of glycomic and glycoproteomic markers of disease (21). More subtle changes in glycosylation accompany development of multifactorial diseases like inflammation and cancer (22, 23). Because tissue- and disease-specific modifications of outer membrane and secreted proteins are common, many N- and mucin-type O-glycoforms are good marker candidates and also serve as targets for imaging agents and therapeutics (24, 25). For example, glycosylation of immunoglobulin (26) and prostate-specific antigen (27), among many other proteins, represents well documented examples of disease-related alterations in site-specific protein glycoforms. Additionally, secreted glycoproteins are accessible in serum making them excellent candidates for the development of noninvasive serological assays (28).
Diseases can cause (or result from) the following: loss/gain of glycosylation sites leading to a change in their number per protein; altered glycosylation site occupancy; the type of glycans present and their distribution on particular sites; or the quantity of specific glycoproteins. Such changes can also impact downstream processes, e.g. binding of interacting proteins, which may be used to measure glycoprotein changes indirectly (29). Mutations of the NX(S/T) sequon occur frequently in disease context (30). Nonsynonymous genomic variants create or abolish NX(S/T) sites on several thousand human proteins and can be associated with cancer risk (31). Relocation of GalNAc transferases to the ER in cancer cells is associated with increased multiplicity of O-glycosylation (14), and Liu et al. (32) reported changes in site occupancy at several N-glycosylation sites in patients with ovarian cancer. We are not aware of established biomarker assays that measure changes in site multiplicity and occupancy outside of genotyping studies of cancer risk (33).
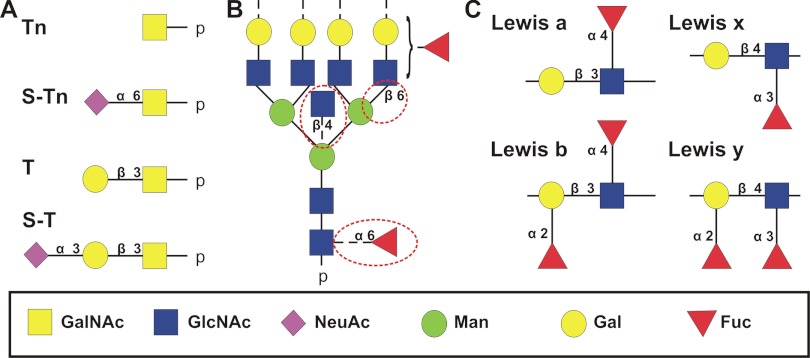
Glycoproteins derived from tumor cells and tissues carry, depending on disease context, truncated O-glycans, glycans with altered fucosylation, including Lewis antigens, N-glycans with bisecting GlcNAc, or increased β1–6GlcNAc branching (Fig. 1). Mucin-type glycoproteins are often incompletely glycosylated in cancer cells due to altered expression of T-synthase or Cosmc chaperone that stabilizes T-synthase (14, 34). The GalNAcα1-O-Ser/Thr and Galβ1–3-GalNAcα-O-Ser/Thr antigens and their sialylated counterparts (Fig. 1A) are detected on cell surface glycoproteins in breast and colon cancers (12, 35). The Lewis blood group epitopes, containing Fuc residues α1–3 or α1–4 linked to GlcNAc and/or Gal (Fig. 1C), and their sialylated forms, are found on membrane glycoproteins in a number of cancer cells (36). Increased N-acetylglucosaminyltransferase III, responsible for addition of a bisecting GlcNAc residue to N-glycans, results in decreased lung colonization by tumor cells but increases metastasis in the spleen (37). Addition of β1–6GlcNAc branches by N-acetylglucosaminyltransferase V increases in cancers of the breast (38), ovary (39), and liver (40) and is associated with enhanced tumor progression and metastasis (41). These changes are shown in Fig. 1B.
Fig. 1.
Glycan structures associated with cancer diseases. A, four truncated O-glycan structures; B, representative N-glycan linkages (circled); and C, four outer arm fucosylated structures that are commonly found in cancer diseases together with their sialylated forms. Tn, GalNAcα1-O-Ser/Thr; S-Tn, Neu5Acα2–6GalNAcα1-O-Ser/Thr; T, Galβ1–3-GalNAcα-O-Ser/Thr; S-T, Neu5Acα2–3-Galβ1–3-GalNAcα-O-Ser/Thr.
Quantitative differences in glycosylation are typically analyzed with lectins or carbohydrate-recognizing antibodies. Lectins bind specific carbohydrate epitopes; for example, Phaseolus vulgaris erythroagglutinin recognizes bisected glycans, P. vulgaris leukoagglutinin recognizes complex branched glycans, and Aleuria aurantia lectin recognizes core-fucosylated glycans (3). Recent studies using lectin arrays and multiaffinity lectin chromatography coupled to liquid chromatography-tandem mass spectrometry (LC-MS/MS) advance the use of lectins in biomarker discovery (42, 43). However, lectins typically have low affinity to single carbohydrate epitopes and display varying degrees of cross-reactivity. This complicates development of standardized lectin-based assays for use in biological samples that may contain carbohydrate epitopes differing in quantity by orders of magnitude. We are not aware of clinical assays using direct lectin recognition of carbohydrates. Nonetheless, lectin-based ELISAs have been developed, and engineered lectins with increased specificity and binding affinity should enhance biomarker discovery and reach clinical utility (44, 45).
Decades of research have led to the generation of many carbohydrate-recognizing antibodies, including several in current clinical use. It is interesting to note that many “tumor-specific” epitopes are carbohydrates. A recent study showed that 41% of antibodies to a cancer cell recognized carbohydrate epitopes (46). Such studies continue to identify new glycoconjugate-recognizing cancer-specific biomarker candidates and generate valuable detection reagents (47). Detailed characterization of the targets and validation of reagents for detection of specific carbohydrate structures are essential. A study of common antibodies to cancer antigen 19-9 (CA19-9)1, used in serological screening of pancreatic cancer, showed that some recognize the expected sialyl-Lewis A antigen but others cross-react with sialyl-Lewis C and N-glycolylneuraminic acid-containing epitopes (48). Table I summarizes antibodies to the important group of Lewis-type carbohydrates tested on the Consortium for Functional Glycomics arrays. The results confirm that some commonly used antibodies to the Lewis-type antigens cross-react with other structures in this assay (Table I). This is an important consideration for biomarker discovery and even more for the performance of clinical assays where the need for validation of the reagents cannot be overemphasized (34). Detailed mass spectrometric characterization of glycoprotein epitopes, development of engineered lectins, or optimization of carbohydrate-targeting immunoassays is expected to improve the quality of glycoprotein quantification (44, 49). Alternative quantitative methods complementary to immunoassays will further facilitate use of glycoconjugates in disease monitoring (50–52). In the next section we present examples of clinically validated glycan- and glycoprotein-based clinical assays, and we discuss how recent optimization of glycoform-specific measurements of these markers attempts to improve their performance, in line with the single protein-omic concept.
Table I. Antibodies toward Lewis carbohydrate antigens.
Antibodies to Lewis epitopes with clone name, isotype, and reference regarding its original production (if known) are shown. Specificity and cross-reactivity of the antibodies were extracted from literature and CFG Glycan Array screening results. The Lewis epitopes are abbreviated Le, with the superscript indicating specific identity (A, B, X, and Y); an S preceding the epitope indicates sialylation, and Di indicates a di-Lewis structure. BG-H1 and BG-H2 indicate blood group H1 and H2 glycan antigens. NA, not available.
| Epitope | Clone | Isotype | Ref. | Specificity and cross-reactivity |
|---|---|---|---|---|
| Lea | T174 | IgG1 | Sakamoto et al., 1986 (106) | Lea = SLea |
| 7LE | IgG1 | Daher et al., 1987 (107) | Lea, Lex | |
| Leb | T218 | IgM | Sakamoto et al., 1986 (106) | Leb > Lea > BG-H1 |
| 2–25LE | IgG1 | Bara et al., 1986 (108) | Leb ≫ Lea | |
| Lex | P12 | IgM | Richman, 1987 (109) | Lex = DiLex |
| 28 | IgM | Hogg et al., 1984 (110) | DiLex > Lex | |
| SH1 | IgG3 | Singhal et al., 1990 (111) | DiLex > Lex | |
| Ley | AH-6 | IgM | Abe et al., 1983 (112) | Ley |
| F3 | IgM | Lloyd et al., 1983 (113) | Ley > BG-H2 > Lex | |
| BR55 | IgG2a | NA | Ley ≫ DiLex > Lex |
Glycoproteins as Disease Markers
Glycoproteins are heavily represented among serological markers of cancer diseases approved by regulatory agencies (4–6). Most approved “glyco” markers use ELISA type assays that recognize a carbohydrate moiety, a combined glycan-protein epitope, or an (not always defined) amino acid sequence on the protein backbone. Full characterization of disease-related glycoconjugates remains challenging despite substantial methodological advances (6, 53–55). Regardless of the target or extent of characterization, these assays show clinical utility despite the limitations discussed below.
Carbohydrate Antigen 19-9 (CA19-9)
In 1979, Koprowski et al. (56) developed antibodies that recognized CA19-9, an antigen found primarily in serum from patients with colorectal and other gastrointestinal malignancies, including pancreatic cancer. CA19-9 was subsequently identified as a sialyl-Lewis A structure (57); later, mucins were identified as the most frequent carriers of the antigen (58). The CA19-9 ELISA used in clinical setting detects pancreatic cancer with an approximate sensitivity and specificity of 80 and 85%, respectively (59). CA19-9 also increases in bile duct obstruction, pancreatitis, and other benign conditions; the utility of the assay for pancreatic cancer screening is limited by its less than 1% positive predictive value (59). Recent studies confirm that mucins are major carriers of the sialyl-Lewis A antigen in serum of pancreatic cancer patients (60). In the same study, Haab and co-workers (60) found that MUC16 was elevated in 65% of pancreatic cancer patients, although MUC1 and MUC5A were elevated in ∼35% of cancer patients. Of the two, MUC5A displayed more frequent glycan alterations. A follow-up study that detected CA19-9 antigen specifically on MUC5A and MUC16 showed improved sensitivity of detection in patients without elevation in total CA19-9 (60). As clinical and biological knowledge of CA19-9 has progressed from its initial discovery in colorectal carcinoma cells, to development of CA19-9 immunoassays for detection of pancreatic cancer, to elucidation of the carbohydrate epitope, and finally to the determination of the major CA19-9 carrier proteins, the performance of detection has gradually improved. This suggests that targeted quantification of specific glycoproteins, or even targeting of site-specific glycoprotein modifications, could improve performance of the existing carbohydrate-based detection assays.
Cancer Antigen 125 (CA125)
CA125, used to screen for ovarian cancer, was discovered in 1981 when Bast et al. (61) developed a monoclonal antibody to the CA125 epitope, which was detected in multiple ovarian carcinoma cell lines. It is expressed in the epithelial tissue of healthy ovarian, breast, colon, gallbladder, kidney, lung, pancreas, prostate, and stomach; it is elevated in a number of diseases, in particular ovarian carcinoma (62). CA125 has 50% sensitivity for early stage disease, compared with 90% for late stage ovarian cancer, at ∼95% specificity. However, Skates (63) has shown improved detection of ovarian cancer by serial assessment of CA125 incorporated in a risk of ovarian cancer algorithm together with additional demographic and clinical criteria. This strategy seems to be particularly effective in combination with follow-up imaging. The individual base-line level of CA125 justifies the serial serological testing and, in combination with the risk of ovarian cancer algorithm risk stratification, represents an important concept for improved biomarker screening.
CA125 is a heavily glycosylated version of MUC16, but the exact epitope was not identified when the protein was initially characterized (64). A detailed study of CA125 expressed in the OVCAR-3 cell line showed that this complex glycoprotein contains O-glycans with branched type I and type II cores, some of which are decorated with the Lewis X antigen (65). The antigen also contains high mannose and complex bisecting N-glycans in agreement with studies that observed increased P. vulgaris erythroagglutinin lectin binding to serum proteins of ovarian cancer patients (66). Treatment of the CA125 antigen with peptide:N-glycosidase F (which selectively removes N-glycans) reduced antibody binding but did not conclusively define the CA125 epitope. However, the distribution of N-glycans in CA125 is similar to that of gp120 derived from lymphoblastoid cells infected with HIV-1. Nagata et al. (66) suggest a functional impact of these CA125 glycoforms on immune response, and potential therapeutic use of monoclonal antibodies to CA125 is currently being explored (67).
Carcinoma Antigen 15-3 (CA15-3) and Cancer Antigen 27-29 (CA27-29)
CA15-3 and CA27-29 are MUC1 antigens elevated in breast cancer. MUC1 is a transmembrane protein overexpressed in many cancers; the protein contains a heavily O-glycosylated variable number of tandem repeat regions (68). Post-translational modifications of the MUC1 cytoplasmic tail modulate oncogenic regulatory pathways in response to signals from the tumor microenvironment (69). The CA15-3 epitope of MUC1 is recognized by antibodies DF3 and 115D8 via a sandwich ELISA (70, 71). The DF3 antibody recognizes an 8-amino acid epitope (Asp-Thr-Arg-Pro-Ala-Pro-Gly-Ser) that may be exposed as a result of altered glycosylation in breast carcinoma, whereas the 115D8 antibody is thought to recognize a glycosylated form of the same 8-amino acid repeat (71). In cancer, shorter O-glycans often dominate, which is thought to expose regions of the peptide backbone otherwise shielded by the glycans (72).
The CA27-29 epitope recognized by the B27.29 antibody is an 8-amino acid sequence that partially overlaps with the epitope on MUC1 recognized by CA15-3 (73). Initial studies on CA27-29 indicated that it was found at lower levels in healthy patients and higher levels in patients with breast cancer, compared with CA15-3 (74). A follow-up study concluded that the CA27-29 assay does not have high enough sensitivity and specificity to detect stage I and II breast cancer, but it can be used to monitor patients treated for breast cancer (75).
CA15-3 is elevated in 65–90% of patients with stage IV breast cancer and a smaller fraction of patients with stages I-III disease; it is also elevated in some benign breast tumors, liver disease, pulmonary malignancies, gastrointestinal/colonic malignancies, and ovarian cancer (76). Like CA27-29, CA15-3 is used to monitor patients undergoing treatment for breast cancer, but it is not a viable marker for early detection of breast cancer. To improve the diagnostic potential of MUC1 glycoforms in breast cancer patients, Blixt et al. (77) synthesized an array of specific MUC1 glycopeptides to screen autoantibodies in the serum of a breast cancer patient (n = 395) and benign disease controls (n = 108). Autoantibodies to the glycoforms GlcNAcβ1–3GalNAc-MUC1 and NeuAcα2,6GalNAc-MUC1 were significantly associated with reduced incidence and increased time to metastasis. Subsequent studies showed that autoantibody-mediated cell toxicity may be responsible for the observed increase in survival (78). The ability to identify improved biomarker targets and synthesize specific glycopeptide candidates to aid in detection opens new research avenues that could eventually establish reliable clinical tests that measure autoantibodies to cancer-specific glycoepitopes or the epitopes themselves.
α-Fetoprotein and AFP-L3
α-Fetoprotein (AFP) is a glycoprotein produced by fetal liver tissue and by a variety of tumors, including hepatocellular carcinoma (HCC), hepatoblastoma, and nonseminomatous germ cell tumors of the ovary and testis (79). AFP is also elevated during pregnancy, viral hepatitis, and cirrhosis of the liver. It is used clinically to monitor patients receiving treatment for HCC and as a marker for the detection of HCC in the high risk groups of cirrhosis or chronic hepatitis patients with approximate sensitivity of 50% and specificity of 85% (80). AFP is N-glycosylated at a single site (Asn-251), and core fucosylation was observed to increase in HCC. Fucosyltransferase 8 (FUT8), which catalyzes core fucosylation of N-glycans, was reported to increase in plasma of patients with HCC (81), but additional factors, including GDP-fucose substrate availability and loss of liver cell polarity, contribute to the increase in AFP-L3 (82). AFP-L3, an assay measuring AFP with core-fucosylated N-glycans, improves specificity for the detection of HCC in all stages of HCC (83); however the sensitivity of detection decreases. An optimized high sensitivity AFP-L3 assay was used for detection of HCC in a population of cirrhotic patients with AFP <20 ng/ml (84). The assay improved sensitivity of detection of HCC from 7% for the conventional AFP-L3 assay to 42% at a specificity of 85%. The survival rate of the patients with high AFP-L3 prior to treatment was significantly lower (p < 0.001) compared with the low AFP-L3 patients. The results demonstrate how optimized assays targeting specific protein glycoforms can achieve improved clinical applicability.
Chorionic Gonadotropin
Human chorionic gonadotropin (hCG) is a hydrophilic, heterodimeric protein composed of a 92-amino acid α subunit (hCGα) and a 145-amino acid β subunit (hCGβ). In healthy pregnant women, hCG is expressed in placental tissue, and hCG is measured in commercial pregnancy tests via sandwich ELISA. The β subunit of human chorionic gonadotropin (hCGβ) is elevated in a number of cancers, including germ line cancers such as choriocarcinoma and testicular cancer (85). Elevated levels of hCGβ in combination with AFP and lactate dehydrogenase are used to diagnose and stage testicular cancer, with serum levels of hCGβ <1000 ng/ml indicating a good prognosis in testicular cancer patients (stage I) and higher levels found in stage II (1000 to 10,000 ng/ml) and stage III (>10,000 ng/ml) (86). Most hCG and hCGβ immunoassays in clinical use are sandwich ELISA-styles (87).
hCGs contain 28–42% carbohydrate by mass with two N-glycosylation sites on the α subunit and four O-glycosylation sites and two N-glycosylation sites on the β subunit (85). The α subunit of hCG is encoded by one gene, although eight copies of the hCGβ gene exist. The amino acid sequences of hCGβ subunits present in healthy individuals are identical; other hCGβ forms are rarely expressed. Of the five described hCG forms, two act as endocrine hormones targeting the luteinizing hormone/choriogonadotropin receptor, and the remaining three are TGFβ receptor antagonists involved in autocrine signaling (85).
Hyperglycosylated hCG is detected in testicular and ovarian cancers originating from germ cells and contains larger N- and O-linked mucin-type glycans with increased branching compared with forms found in healthy individuals (88). Site-specific differences in glycosylation have been studied in urine from patients with choriocarcinoma and testicular cancer and in pregnant women. Increased triantennary N-linked oligosaccharides at Asn-30 and fucosylated glycans at Asn-13 of the β subunit were detected in patients with testicular cancer and choriocarcinoma. In testicular cancer patients, O-glycans at Ser-138 displayed higher levels of core-2 structures compared with pregnant women (88). The site-specific glycoforms described above represent promising targets for detection of testicular and other germ line cancers.
Single Protein-omics and Targeted Quantification of Site-specific Glycoforms
Historically, many tumor-specific antibodies have been shown to bind carbohydrate epitopes (89, 90). The examples of immunoassays discussed in the preceding section demonstrate that glycoprotein analytes are of substantial clinical utility for disease monitoring, but their sensitivity and specificity for the detection of early stage cancers is often lower than desired (59, 61). Improved characterization of the targets, understanding of mechanisms of action, development of improved reagents, optimization of analytical assays, consideration of appropriate clinical variables, repeated testing addressing the variable individual base line of markers, or efficient marker combinations will improve diagnostic performance.
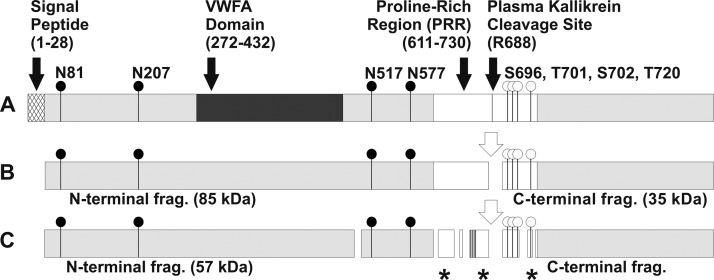
Proteins and their co- and post-translational modifications are rich sources of information regarding the cellular and extracellular environment. Glycoproteins are complex biopolymers with multiple forms expected to capture information relevant to specific disease conditions. Fig. 2A presents serum glycoprotein with UniProtKB/SwissProt entry Q14624 as an example of a potential biomarker candidate. This protein contains 22% carbohydrate by weight with four N-glycosylation sites and several potential O-glycosylation sites (Fig. 2B) (91). It is cleaved by plasma kallikrein into two fragments, which undergo additional processing into multiple peptides quantifiable in serum by LC-MS-MRM methods (92). The potential glycoforms and related proteolytic products are shown in Fig. 2C. The complexity of potential analytes that arise from this single protein is immediately evident from this figure. Examination of all its forms is a complex task that rivals the complexity of classic proteomic experiments covering entire proteomes. A 35-kDa fragment of the protein enriched by champedak galactose-binding lectin is enriched in sera of patients with different carcinomas (93), suggesting that characterization of its glycoform may indeed be relevant to examine in the disease context.
Fig. 2.
Schematic of glycoforms and proteolytic fragments of UniProtKB/SwissProt entry Q14624. A, full-length protein with signal peptide, domains (labeled), N-glycosylation sites (black circles), and proposed O-glycosylation sites (white circles); B, cleavage of mature protein by plasma kallikrein; C, additional internal cleavage of protein fragments. Asterisks indicate proteolytic fragments (frag.) detected in serum.
It can be argued that such single protein-omics, covering all forms of a protein, have a better chance to reveal biomarkers of disease than an examination of complete proteomes, which cannot cover the richness of individual protein forms. It is inviting to speculate that incorporation of detailed structure-function understanding into the biomarker discovery process will allow selection of relevant and quantifiable glycopeptide targets. The single protein-omics approach is reminiscent of classic experiments in which proteins were isolated from biological materials for their full characterization. This approach will have limitations as any given peptide, especially in a modified form, may not be analyzable. However, the growing arsenal of innovative workflows and analytical options opens many opportunities for single protein-omic analyses.
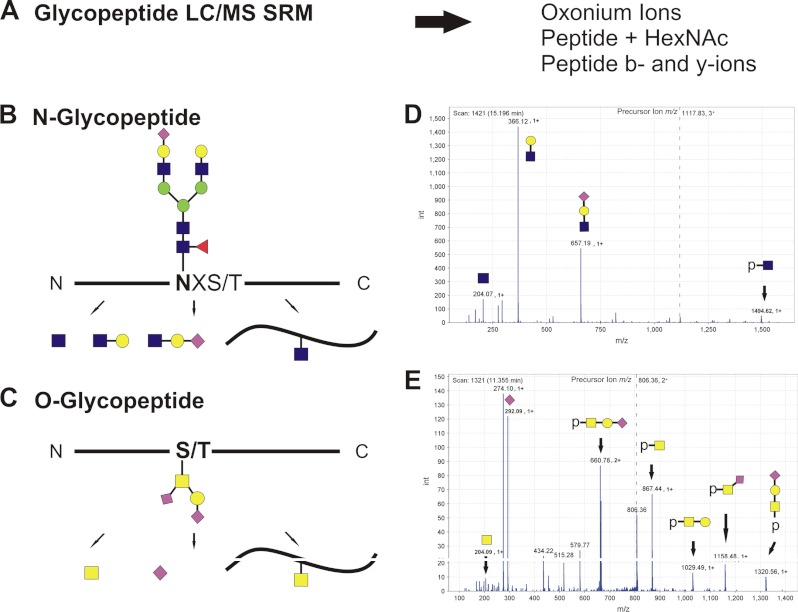
There are multiple examples of site-specific glycoproteomic discovery workflows that identify biomarker candidates (19, 25, 39, 94). The biomarker discovery phase is typically followed by development of non-MS-based screening methods, but it is important to note that the site-specific glycoforms can be quantified by optimized LC-MS-MRM assays (Fig. 3A). These methods target glycopeptide-specific ions, including oxonium ions, peptide b- and y-ions, and intact peptide ions (Fig. 3, B–D). Optimization of mass spectrometric MRM assays for specific classes of glycopeptides will be needed, and clinical applications of such assays may require synthesis of isotopically labeled standards. Recent studies suggest that such methods are, in general, feasible; analysis of single proteins (e.g. immunoglobulins) by LC-MS-MRM methods was reported, and additional examples are likely to follow (51, 95–97). Quantifications of some of the glycoforms may be limited to the analysis of isolated proteins, and not all glycoforms will be within methodological limits of detection, but successful applications can be expected. Site-specific quantification of relevant glycoforms will complement clinical immunoassays, as immunoassays are not readily available for such targets. Verification of site-specific glycopeptide candidates by LC-MS-MRM may be a productive approach to select candidates for targeted development of immunoaffinity reagents.
Fig. 3.
Toward quantitative measurement of site-specific glycoforms of proteins. A, one strategy for quantitative measurement of glycopeptides is LC-MS-MRM. Optimized quantification of appropriate oxonium ions, peptide-glycan fragments, or peptide b and y ions is expected to facilitate detection of specific types of glycopeptides. B, schematic of N-glycopeptide fragmentation. C, schematic of O-glycopeptide fragmentation. D, spectrum of N-glycopeptide LPTQNITFQTE-HexNAc4Hex5NeuAc1Fuc1 fragmented via CID. E, spectrum of O-glycopeptide ASFSPR-HexNAcHexNeuAc2 fragmented using CID.
In conclusion, novel discovery strategies, including analysis of autoantibodies (77, 98), arrays (42, 99), glycopeptide enrichment coupled with mass spectrometric identification (39, 43, 100), innovations in cellular glycobiology (101, 102), and chemoenzymatic methods (103–105) continue to expand the pipeline of glycoproteomic biomarker candidates. Detailed characterization of the relevant site-specific glycoforms and targeted quantification of protein glycoforms by innovative immunoassays and alternative methods, including mass spectrometric quantification, are expected to enhance current clinical diagnostic options.
Acknowledgments
We acknowledge the Consortium for Functional Glycomics for providing public access to glycan array results for the antibodies discussed in this paper.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grants U01 CA168926 and RO1 CA135069 (to R.G.).
1 The abbreviations used are:
- CA19-9
- cancer antigen 19-9
- sialyl-Lewis A
- Neu5Acα2–3Galβ1–3(Fucα1–4)GlcNAc
- CA125
- cancer antigen 125
- CA15-3
- carcinoma antigen 15–3
- CA27-29
- cancer antigen 27–29
- AFP
- α-fetoprotein
- HCC
- hepatocellular carcinoma
- FUT8
- fucosyltransferase 8
- hCG
- human chorionic gonadotropin
- MRM
- multiple reaction monitoring.
REFERENCES
- 1. Taube S. E., Clark G. M., Dancey J. E., McShane L. M., Sigman C. C., Gutman S. I. (2009) A perspective on challenges and issues in biomarker development and drug and biomarker codevelopment. J. Natl. Cancer Inst. 101, 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gutman S., Kessler L. G. (2006) The US Food and Drug Administration perspective on cancer biomarker development. Nat. Rev. Cancer 6, 565–571 [DOI] [PubMed] [Google Scholar]
- 3. Varki A., Cummings R., Esko J., Freeze H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. (2009) Essentials of Glycobiology, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 16–17 & 20–21 [PubMed] [Google Scholar]
- 4. Ludwig J. A., Weinstein J. N. (2005) Biomarkers in cancer staging, prognosis, and treatment selection. Nat. Rev. Cancer 5, 845–856 [DOI] [PubMed] [Google Scholar]
- 5. Angata T., Fujinawa R., Kurimoto A., Nakajima K., Kato M., Takamatsu S., Korekane H., Gao C. X., Ohtsubo K., Kitazume S., Taniguchi N. (2012) Integrated approach toward the discovery of glyco-biomarkers of inflammation-related diseases. Ann. N.Y. Acad. Sci. 1253, 159–169 [DOI] [PubMed] [Google Scholar]
- 6. Pan S., Chen R., Aebersold R., Brentnall T. A. (2011) Mass spectrometry-based glycoproteomics–from a proteomics perspective. Mol. Cell. Proteomics 10, R110.003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zaia J. (2009) On-line separations combined with MS for analysis of glycosaminoglycans. Mass Spectrom. Rev. 28, 254–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li L., Zhang F., Zaia J., Linhardt R. J. (2012) Top-down approach for the direct characterization of low molecular weight heparins using LC-FT-MS. Anal. Chem. 84, 8822–8829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Helenius A., Aebi M. (2001) Intracellular functions of N-linked glycans. Science 291, 2364–2369 [DOI] [PubMed] [Google Scholar]
- 10. Bennett E. P., Mandel U., Clausen H., Gerken T. A., Fritz T. A., Tabak L. A. (2012) Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zielinska D. F., Gnad F., Wiśniewski J. R., Mann M. (2010) Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 [DOI] [PubMed] [Google Scholar]
- 12. Brockhausen I. (2006) Mucin-type O-glycans in human colon and breast cancer: glycodynamics and functions. EMBO Rep. 7, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Apweiler R., Hermjakob H., Sharon N. (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 [DOI] [PubMed] [Google Scholar]
- 14. Gill D. J., Clausen H., Bard F. (2011) Location, location, location: new insights into O-GalNAc protein glycosylation. Trends Cell Biol. 21, 149–158 [DOI] [PubMed] [Google Scholar]
- 15. Kaji H., Shikanai T., Sasaki-Sawa A., Wen H., Fujita M., Suzuki Y., Sugahara D., Sawaki H., Yamauchi Y., Shinkawa T., Taoka M., Takahashi N., Isobe T., Narimatsu H. (2012) Large scale identification of N-glycosylated proteins of mouse tissues and construction of a glycoprotein database, GlycoProtDB. J. Proteome Res. 11, 4553–4566 [DOI] [PubMed] [Google Scholar]
- 16. Dennis J. W., Nabi I. R., Demetriou M. (2009) Metabolism, cell surface organization, and disease. Cell 139, 1229–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krambeck F. J., Bennun S. V., Narang S., Choi S., Yarema K. J., Betenbaugh M. J. (2009) A mathematical model to derive N-glycan structures and cellular enzyme activities from mass spectrometric data. Glycobiology 19, 1163–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang D., Hincapie M., Rejtar T., Karger B. L. (2011) Ultrasensitive characterization of site-specific glycosylation of affinity-purified haptoglobin from lung cancer patient plasma using 10 μm i.d. porous layer open tubular liquid chromatography-linear ion trap collision-induced dissociation/electron transfer dissociation mass spectrometry. Anal. Chem. 83, 2029–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mann B. F., Goetz J. A., House M. G., Schmidt C. M., Novotny M. V. (2012) Glycomic and proteomic profiling of pancreatic cyst fluids identifies hyperfucosylated lactosamines on the N-linked glycans of overexpressed glycoproteins. Mol. Cell. Proteomics 11, M111.015792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brooks S. A., Carter T. M., Royle L., Harvey D. J., Fry S. A., Kinch C., Dwek R. A., Rudd P. M. (2008) Altered glycosylation of proteins in cancer: what is the potential for new anti-tumour strategies. Anticancer Agents Med. Chem. 8, 2–21 [DOI] [PubMed] [Google Scholar]
- 21. Wada Y. (2006) Mass spectrometry for congenital disorders of glycosylation, CDG. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 838, 3–8 [DOI] [PubMed] [Google Scholar]
- 22. Alavi A., Axford J. S. (2008) Sweet and sour: the impact of sugars on disease. Rheumatology 47, 760–770 [DOI] [PubMed] [Google Scholar]
- 23. Lau K. S., Dennis J. W. (2008) N-Glycans in cancer progression. Glycobiology 18, 750–760 [DOI] [PubMed] [Google Scholar]
- 24. Mechref Y., Hu Y., Garcia A., Hussein A. (2012) Identifying cancer biomarkers by mass spectrometry-based glycomics. Electrophoresis 33, 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wandall H. H., Blixt O., Tarp M. A., Pedersen J. W., Bennett E. P., Mandel U., Ragupathi G., Livingston P. O., Hollingsworth M. A., Taylor-Papadimitriou J., Burchell J., Clausen H. (2010) Cancer biomarkers defined by autoantibody signatures to aberrant O-glycopeptide epitopes. Cancer Res. 70, 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zauner G., Selman M. H., Bondt A., Rombouts Y., Blank D., Deelder A. M., Wuhrer M. (2013) Glycoproteomic analysis of antibodies. Mol. Cell. Proteomics, e-pub January 16, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vermassen T., Speeckaert M. M., Lumen N., Rottey S., Delanghe J. R. (2012) Glycosylation of prostate-specific antigen and its potential diagnostic applications. Clin. Chim. Acta 413, 1500–1505 [DOI] [PubMed] [Google Scholar]
- 28. Arnold J. N., Saldova R., Hamid U. M., Rudd P. M. (2008) Evaluation of the serum N-linked glycome for the diagnosis of cancer and chronic inflammation. Proteomics 8, 3284–3293 [DOI] [PubMed] [Google Scholar]
- 29. Carlsson M. C., Cederfur C., Schaar V., Balog C. I., Lepur A., Touret F., Salomonsson E., Deelder A. M., Fernö M., Olsson H., Wuhrer M., Leffler H. (2011) Galectin-1-binding glycoforms of haptoglobin with altered intracellular trafficking, and increase in metastatic breast cancer patients. PLoS One 6, e26560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vogt G., Chapgier A., Yang K., Chuzhanova N., Feinberg J., Fieschi C., Boisson-Dupuis S., Alcais A., Filipe-Santos O., Bustamante J., de Beaucoudrey L., Al-Mohsen I., Al-Hajjar S., Al-Ghonaium A., Adimi P., Mirsaeidi M., Khalilzadeh S., Rosenzweig S., de la Calle Martin O., Bauer T. R., Puck J. M., Ochs H. D., Furthner D., Engelhorn C., Belohradsky B., Mansouri D., Holland S. M., Schreiber R. D., Abel L., Cooper D. N., Soudais C., Casanova J. L. (2005) Gains of glycosylation comprise an unexpectedly large group of pathogenic mutations. Nat. Genet. 37, 692–700 [DOI] [PubMed] [Google Scholar]
- 31. Pompach P., Chandler K. B., Lan R., Edwards N., Goldman R. (2012) Semi-automated identification of N-glycopeptides by hydrophilic interaction chromatography, nano-reverse-phase LC-MS/MS, and glycan database search. J. Proteome Res. 11, 1728–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu Z., Cao J., He Y., Qiao L., Xu C., Lu H., Yang P. (2010) Tandem 18O stable isotope labeling for quantification of N-glycoproteome. J. Proteome Res. 9, 227–236 [DOI] [PubMed] [Google Scholar]
- 33. Cui Y., Shu X. O., Cai Q., Jin F., Cheng J. R., Cai H., Gao Y. T., Zheng W. (2005) Association of breast cancer risk with a common functional polymorphism (Asp327Asn) in the sex hormone-binding globulin gene. Cancer Epidemiol. Biomarkers Prev. 14, 1096–1101 [DOI] [PubMed] [Google Scholar]
- 34. Ju T., Otto V. I., Cummings R. D. (2011) The Tn antigen-structural simplicity and biological complexity. Angew. Chem. Int. Ed. Engl. 50, 1770–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Itzkowitz S. H., Yuan M., Montgomery C. K., Kjeldsen T., Takahashi H. K., Bigbee W. L., Kim Y. S. (1989) Expression of Tn, sialosyl-Tn, and T antigens in human colon cancer. Cancer Res. 49, 197–204 [PubMed] [Google Scholar]
- 36. Kim Y. S., Yuan M., Itzkowitz S. H., Sun Q. B., Kaizu T., Palekar A., Trump B. F., Hakomori S. (1986) Expression of LeY and extended LeY blood group-related antigens in human malignant, premalignant, and nonmalignant colonic tissues. Cancer Res. 46, 5985–5992 [PubMed] [Google Scholar]
- 37. Miwa H. E., Song Y., Alvarez R., Cummings R. D., Stanley P. (2012) The bisecting GlcNAc in cell growth control and tumor progression. Glycoconj. J. 29, 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo H. B., Johnson H., Randolph M., Nagy T., Blalock R., Pierce M. (2010) Specific post-translational modification regulates early events in mammary carcinoma formation. Proc. Natl. Acad. Sci. U.S.A. 107, 21116–21121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abbott K. L., Lim J. M., Wells L., Benigno B. B., McDonald J. F., Pierce M. (2010) Identification of candidate biomarkers with cancer-specific glycosylation in the tissue and serum of endometrioid ovarian cancer patients by glycoproteomic analysis. Proteomics 10, 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mehta A., Norton P., Liang H., Comunale M. A., Wang M., Rodemich-Betesh L., Koszycki A., Noda K., Miyoshi E., Block T. (2012) Increased levels of tetra-antennary N-linked glycan but not core fucosylation are associated with hepatocellular carcinoma tissue. Cancer Epidemiol. Biomarkers Prev. 21, 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Taniguchi N., Miyoshi E., Ko J. H., Ikeda Y., Ihara Y. (1999) Implication of N-acetylglucosaminyltransferases III and V in cancer: gene regulation and signaling mechanism. Biochim. Biophys. Acta 1455, 287–300 [DOI] [PubMed] [Google Scholar]
- 42. Koshi Y., Nakata E., Yamane H., Hamachi I. (2006) A fluorescent lectin array using supramolecular hydrogel for simple detection and pattern profiling for various glycoconjugates. J. Am. Chem. Soc. 128, 10413–10422 [DOI] [PubMed] [Google Scholar]
- 43. Fanayan S., Hincapie M., Hancock W. S. (2012) Using lectins to harvest the plasma/serum glycoproteome. Electrophoresis 33, 1746–1754 [DOI] [PubMed] [Google Scholar]
- 44. Romano P. R., Mackay A., Vong M., DeSa J., Lamontagne A., Comunale M. A., Hafner J., Block T., Lec R., Mehta A. (2011) Development of recombinant Aleuria aurantia lectins with altered binding specificities to fucosylated glycans. Biochem. Biophys. Res. Commun. 414, 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Powlesland A. S., Quintero-Martinez A., Lim P. G., Pipirou Z., Taylor M. E., Drickamer K. (2010) Engineered carbohydrate-recognition domains for glycoproteomic analysis of cell surface glycosylation and ligands for glycan-binding receptors. Methods Enzymol. 480, 165–179 [DOI] [PubMed] [Google Scholar]
- 46. Gildersleeve J. C., Wang B., Achilefu S., Tu Z., Xu M. (2012) Glycan array analysis of the antigen repertoire targeted by tumor-binding antibodies. Bioorg. Med. Chem. Lett. 22, 6839–6843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Palma A. S., Liu Y., Childs R. A., Herbert C., Wang D., Chai W., Feizi T. (2011) The human epithelial carcinoma antigen recognized by monoclonal antibody AE3 is expressed on a sulfoglycolipid in addition to neoplastic mucins. Biochem. Biophys. Res. Commun. 408, 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Partyka K., Maupin K. A., Brand R. E., Haab B. B. (2012) Diverse monoclonal antibodies against the CA19-9 antigen show variation in binding specificity with consequences for clinical interpretation. Proteomics 12, 2212–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yue T., Haab B. B. (2009) Microarrays in glycoproteomics research. Clin. Lab. Med. 29, 15–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Doherty M., McManus C. A., Duke R., Rudd P. M. (2012) High-throughput quantitative N-glycan analysis of glycoproteins. Methods Mol. Biol. 899, 293–313 [DOI] [PubMed] [Google Scholar]
- 51. Toyama A., Nakagawa H., Matsuda K., Sato T. A., Nakamura Y., Ueda K. (2012) Quantitative structural characterization of local N-glycan microheterogeneity in therapeutic antibodies by energy-resolved oxonium ion monitoring. Anal. Chem. 84, 9655–9662 [DOI] [PubMed] [Google Scholar]
- 52. Vanderschaeghe D., Laroy W., Sablon E., Halfon P., Van Hecke A., Delanghe J., Callewaert N. (2009) GlycoFibroTest is a highly performant liver fibrosis biomarker derived from DNA sequencer-based serum protein glycomics. Mol. Cell. Proteomics 8, 986–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ruhaak L. R., Zauner G., Huhn C., Bruggink C., Deelder A. M., Wuhrer M. (2010) Glycan labeling strategies and their use in identification and quantification. Anal. Bioanal. Chem. 397, 3457–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kolarich D., Jensen P. H., Altmann F., Packer N. H. (2012) Determination of site-specific glycan heterogeneity on glycoproteins. Nat. Protoc. 7, 1285–1298 [DOI] [PubMed] [Google Scholar]
- 55. North S. J., Hitchen P. G., Haslam S. M., Dell A. (2009) Mass spectrometry in the analysis of N-linked and O-linked glycans. Curr. Opin. Struct. Biol. 19, 498–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Koprowski H., Herlyn M., Steplewski Z., Sears H. F. (1981) Specific antigen in serum of patients with colon carcinoma. Science 212, 53–55 [DOI] [PubMed] [Google Scholar]
- 57. Magnani J. L., Nilsson B., Brockhaus M., Zopf D., Steplewski Z., Koprowski H., Ginsburg V. (1982) A monoclonal antibody-defined antigen associated with gastrointestinal cancer is a ganglioside containing sialylated lacto-N-fucopentaose II. J. Biol. Chem. 257, 14365–14369 [PubMed] [Google Scholar]
- 58. Magnani J. L., Steplewski Z., Koprowski H., Ginsburg V. (1983) Identification of the gastrointestinal and pancreatic cancer-associated antigen detected by monoclonal antibody 19-9 in the sera of patients as a mucin. Cancer Res. 43, 5489–5492 [PubMed] [Google Scholar]
- 59. Ballehaninna U. K., Chamberlain R. S. (2012) The clinical utility of serum CA19-9 in the diagnosis, prognosis, and management of pancreatic adenocarcinoma: An evidence-based appraisal. J. Gastrointest. Oncol. 3, 105–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yue T., Partyka K., Maupin K. A., Hurley M., Andrews P., Kaul K., Moser A. J., Zeh H., Brand R. E., Haab B. B. (2011) Identification of blood-protein carriers of the CA19-9 antigen and characterization of prevalence in pancreatic diseases. Proteomics 11, 3665–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bast R. C., Jr., Feeney M., Lazarus H., Nadler L. M., Colvin R. B., Knapp R. C. (1981) Reactivity of a monoclonal antibody with human ovarian carcinoma. J. Clin. Invest. 68, 1331–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gentry-Maharaj A., Menon U. (2012) Screening for ovarian cancer in the general population. Best Pract. Res. Clin. Obstet. Gynaecol. 26, 243–256 [DOI] [PubMed] [Google Scholar]
- 63. Skates S. J. (2012) Ovarian cancer screening: development of the risk of ovarian cancer algorithm (ROCA) and ROCA screening trials. Int. J. Gynecol. Cancer 22, S24–S26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yin B. W., Lloyd K. O. (2001) Molecular cloning of the CA125 ovarian cancer antigen: identification as a new mucin, MUC16. J. Biol. Chem. 276, 27371–27375 [DOI] [PubMed] [Google Scholar]
- 65. Kui Wong N., Easton R. L., Panico M., Sutton-Smith M., Morrison J. C., Lattanzio F. A., Morris H. R., Clark G. F., Dell A., Patankar M. S. (2003) Characterization of the oligosaccharides associated with the human ovarian tumor marker CA125. J. Biol. Chem. 278, 28619–28634 [DOI] [PubMed] [Google Scholar]
- 66. Nagata A., Hirota N., Sakai T., Fujimoto M., Komoda T. (1991) Molecular nature and possible presence of a membranous glycan-phosphatidylinositol anchor of CA125 antigen. Tumour Biol. 12, 279–286 [DOI] [PubMed] [Google Scholar]
- 67. Ehlen T. G., Hoskins P. J., Miller D., Whiteside T. L., Nicodemus C. F., Schultes B. C., Swenerton K. D. (2005) A pilot phase 2 study of oregovomab murine monoclonal antibody to CA125 as an immunotherapeutic agent for recurrent ovarian cancer. Int. J. Gynecol. Cancer 15, 1023–1034 [DOI] [PubMed] [Google Scholar]
- 68. Singh R., Bandyopadhyay D. (2007) MUC1: a target molecule for cancer therapy. Cancer Biol. Ther. 6, 481–486 [DOI] [PubMed] [Google Scholar]
- 69. Behrens M. E., Grandgenett P. M., Bailey J. M., Singh P. K., Yi C. H., Yu F., Hollingsworth M. A. (2010) The reactive tumor microenvironment: MUC1 signaling directly reprograms transcription of CTGF. Oncogene 29, 5667–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gang Y., Adachi I., Ohkura H., Yamamoto H., Mizuguchi Y., Abe K. (1985) CA15-3 is present as a novel tumor marker in the sera of patients with breast cancer and other malignancies. Gan To Kagaku Ryoho 12, 2379–2386 [PubMed] [Google Scholar]
- 71. Price M. R., Rye P. D., Petrakou E., Murray A., Brady K., Imai S., Haga S., Kiyozuka Y., Schol D., Meulenbroek M. F., Snijdewint F. G., von Mensdorff-Pouilly S., Verstraeten R. A., Kenemans P., Blockzjil A., Nilsson K., Nilsson O., Reddish M., Suresh M. R., Koganty R. R., Fortier S., Baronic L., Berg A., Longenecker M. B., Hilgers J. (1998) Summary report on the ISOBM TD-4 Workshop: analysis of 56 monoclonal antibodies against the MUC1 mucin. San Diego, Calif., November 17–23, 1996. Tumour Biol. 19, 1–20 [DOI] [PubMed] [Google Scholar]
- 72. Burchell J. M., Mungul A., Taylor-Papadimitriou J. (2001) O-Linked glycosylation in the mammary gland: changes that occur during malignancy. J. Mammary Gland Biol. Neoplasia 6, 355–364 [DOI] [PubMed] [Google Scholar]
- 73. Klee G. G., Schreiber W. E. (2004) MUC1 gene-derived glycoprotein assays for monitoring breast cancer (CA15-3, CA 27.29, BR): are they measuring the same antigen? Arch. Pathol. Lab. Med. 128, 1131–1135 [DOI] [PubMed] [Google Scholar]
- 74. Gion M., Mione R., Leon A. E., Dittadi R. (1999) Comparison of the diagnostic accuracy of CA27.29 and CA15.3 in primary breast cancer. Clin. Chem. 45, 630–637 [PubMed] [Google Scholar]
- 75. Gion M., Mione R., Leon A. E., Lüftner D., Molina R., Possinger K., Robertson J. F. (2001) CA27.29: a valuable marker for breast cancer management. A confirmatory multicentric study on 603 cases. Eur. J. Cancer 37, 355–363 [DOI] [PubMed] [Google Scholar]
- 76. Brooks M. (2009) Breast cancer screening and biomarkers. Methods Mol. Biol. 472, 307–321 [DOI] [PubMed] [Google Scholar]
- 77. Blixt O., Bueti D., Burford B., Allen D., Julien S., Hollingsworth M., Gammerman A., Fentiman I., Taylor-Papadimitriou J., Burchell J. M. (2011) Autoantibodies to aberrantly glycosylated MUC1 in early stage breast cancer are associated with a better prognosis. Breast Cancer Res. 13, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lavrsen K., Madsen C. B., Rasch M. G., Woetmann A., Odum N., Mandel U., Clausen H., Pedersen A. E., Wandall H. H. (2012) Aberrantly glycosylated MUC1 is expressed on the surface of breast cancer cells and a target for antibody-dependent cell-mediated cytotoxicity. Glycoconj. J., e-Pub August 10, 2012 [DOI] [PubMed] [Google Scholar]
- 79. Lange P. H., Hakala T. R., Fraley E. E. (1975) Serum α-fetoprotein and β-human chorionic gonadotropin levels in patients with non-seminomatous germ cell testicular cancer. Minn. Med. 58, 813–815 [PubMed] [Google Scholar]
- 80. Lopez L. J., Marrero J. A. (2004) Hepatocellular carcinoma. Curr. Opin. Gastroenterol. 20, 248–253 [DOI] [PubMed] [Google Scholar]
- 81. Mita Y., Aoyagi Y., Suda T., Asakura H. (2000) Plasma fucosyltransferase activity in patients with hepatocellular carcinoma, with special reference to correlation with fucosylated species of α-fetoprotein. J. Hepatol. 32, 946–954 [DOI] [PubMed] [Google Scholar]
- 82. Miyoshi E., Moriwaki K., Nakagawa T. (2008) Biological function of fucosylation in cancer biology. J. Biochem. 143, 725–729 [DOI] [PubMed] [Google Scholar]
- 83. Marrero J. A., Feng Z., Wang Y., Nguyen M. H., Befeler A. S., Roberts L. R., Reddy K. R., Harnois D., Llovet J. M., Normolle D., Dalhgren J., Chia D., Lok A. S., Wagner P. D., Srivastava S., Schwartz M. (2009) α-fetoprotein, des-γ carboxyprothrombin, and lectin-bound α-fetoprotein in early hepatocellular carcinoma. Gastroenterology 137, 110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Toyoda H., Kumada T., Tada T., Kaneoka Y., Maeda A., Kanke F., Satomura S. (2011) Clinical utility of highly sensitive Lens culinaris agglutinin-reactive α-fetoprotein in hepatocellular carcinoma patients with α-fetoprotein <20 ng/ml. Cancer Sci. 102, 1025–1031 [DOI] [PubMed] [Google Scholar]
- 85. Cole L. A. (2012) hCG, five independent molecules. Clin. Chim. Acta 413, 48–65 [DOI] [PubMed] [Google Scholar]
- 86. Salem M., Gilligan T. (2011) Serum tumor markers and their utilization in the management of germ-cell tumors in adult males. Expert Rev. Anticancer Ther. 11, 1–4 [DOI] [PubMed] [Google Scholar]
- 87. Sturgeon C. M., Berger P., Bidart J. M., Birken S., Burns C., Norman R. J., Stenman U. H. (2009) Differences in recognition of the 1st WHO international reference reagents for hCG-related isoforms by diagnostic immunoassays for human chorionic gonadotropin. Clin. Chem. 55, 1484–1491 [DOI] [PubMed] [Google Scholar]
- 88. Valmu L., Alfthan H., Hotakainen K., Birken S., Stenman U. H. (2006) Site-specific glycan analysis of human chorionic gonadotropin β-subunit from malignancies and pregnancy by liquid chromatography– electrospray mass spectrometry. Glycobiology 16, 1207–1218 [DOI] [PubMed] [Google Scholar]
- 89. Hakomori S. (1984) Tumor-associated carbohydrate antigens. Annu. Rev. Immunol. 2, 103–126 [DOI] [PubMed] [Google Scholar]
- 90. Feizi T. (1985) Carbohydrate antigens in human cancer. Cancer Surv. 4, 245–269 [PubMed] [Google Scholar]
- 91. Nishimura H., Kakizaki I., Muta T., Sasaki N., Pu P. X., Yamashita T., Nagasawa S. (1995) cDNA and deduced amino acid sequence of human PK-120, a plasma kallikrein-sensitive glycoprotein. FEBS Lett. 357, 207–211 [DOI] [PubMed] [Google Scholar]
- 92. van den Broek I., Sparidans R. W., Schellens J. H., Beijnen J. H. (2008) Liquid chromatography/tandem mass spectrometric method for the quantification of eight proteolytic fragments of ITIH4 with biomarker potential in human plasma and serum. Rapid Commun. Mass Spectrom. 22, 2915–2928 [DOI] [PubMed] [Google Scholar]
- 93. Mohamed E., Abdul-Rahman P. S., Doustjalali S. R., Chen Y., Lim B. K., Omar S. Z., Bustam A. Z., Singh V. A., Mohd-Taib N. A., Yip C. H., Hashim O. H. (2008) Lectin-based electrophoretic analysis of the expression of the 35-kDa inter-α-trypsin inhibitor heavy chain H4 fragment in sera of patients with five different malignancies. Electrophoresis 29, 2645–2650 [DOI] [PubMed] [Google Scholar]
- 94. Pompach P., Brnakova Z., Sanda M., Wu J., Edwards N., Goldman R. (2013) Site-specific glycoforms of haptoglobin in liver cirrhosis and hepatocellular carcinoma. Mol. Cell. Proteomics, e-Pub February 6, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Song E., Pyreddy S., Mechref Y. (2012) Quantification of glycopeptides by multiple reaction monitoring liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 26, 1941–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ahn Y. H., Lee J. Y., Lee J. Y., Kim Y. S., Ko J. H., Yoo J. S. (2009) Quantitative analysis of an aberrant glycoform of TIMP1 from colon cancer serum by L-PHA-enrichment and SISCAPA with MRM mass spectrometry. J. Proteome Res. 8, 4216–4224 [DOI] [PubMed] [Google Scholar]
- 97. Sanda M., Pompach P., Brnakova Z., Wu J., Makambi K., Goldman R. (2013) Quantitative LC-MS-MRM analysis of site-specific glycoforms of haptoglobin in liver disease. Mol. Cell Proteomics, e-Pub February 6, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Padler-Karavani V., Hurtado-Ziola N., Pu M., Yu H., Huang S., Muthana S., Chokhawala H. A., Cao H., Secrest P., Friedmann-Morvinski D., Singer O., Ghaderi D., Verma I. M., Liu Y. T., Messer K., Chen X., Varki A., Schwab R. (2011) Human xeno-autoantibodies against a non-human sialic acid serve as novel serum biomarkers and immunotherapeutics in cancer. Cancer Res. 71, 3352–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Park S., Gildersleeve J. C., Blixt O., Shin I. (2012) Carbohydrate microarrays. Chem. Soc. Rev., e-pub November 28, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ahn Y. H., Shin P. M., Oh N. R., Park G. W., Kim H., Yoo J. S. (2012) A lectin-coupled, targeted proteomic mass spectrometry (MRM MS) platform for identification of multiple liver cancer biomarkers in human plasma. J. Proteomics 75, 5507–5515 [DOI] [PubMed] [Google Scholar]
- 101. Pinto R., Carvalho A. S., Conze T., Magalhães A., Picco G., Burchell J. M., Taylor-Papadimitriou J., Reis C. A., Almeida R., Mandel U., Clausen H., Söderberg O., David L. (2012) Identification of new cancer biomarkers based on aberrant mucin glycoforms by in situ proximity ligation. J. Cell. Mol. Med. 16, 1474–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Steentoft C., Vakhrushev S. Y., Vester-Christensen M. B., Schjoldager K. T., Kong Y., Bennett E. P., Mandel U., Wandall H., Levery S. B., Clausen H. (2011) Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat. Methods 8, 977–982 [DOI] [PubMed] [Google Scholar]
- 103. Chaubard J. L., Krishnamurthy C., Yi W., Smith D. F., Hsieh-Wilson L. C. (2012) Chemoenzymatic probes for detecting and imaging fucose-α(1–2)-galactose glycan biomarkers. J. Am. Chem. Soc. 134, 4489–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Rabuka D., Rush J. S., deHart G. W., Wu P., Bertozzi C. R. (2012) Site-specific chemical protein conjugation using genetically encoded aldehyde tags. Nat. Protoc. 7, 1052–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Boltje T. J., Buskas T., Boons G. J. (2009) Opportunities and challenges in synthetic oligosaccharide and glycoconjugate research. Nat. Chem. 1, 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sakamoto J., Furukawa K., Cordon-Cardo C., Yin B. W., Rettig W. J., Oettgen H. F., Old L. J., Lloyd K. O. (1986) Expression of Lewisa, Lewisb, X, and Y blood group antigens in human colonic tumors and normal tissue and in human tumor-derived cell lines. Cancer Res. 46 (3), 1553–1561 [PubMed] [Google Scholar]
- 107. Daher N., Bara J., Moubarak M. D. (1987) Non A non B and Lewis related antigens in normal human prostates: an immunohistological study of 20 anti-glycoconjugate monoclonal antibodies. Rev. Fr. Transfus. Immunohematol. 30 (5), 681–684 [DOI] [PubMed] [Google Scholar]
- 108. Bara J., Gautier R., Daher N., Zaghouani H., Decaens C. (1986) Monoclonal antibodies against oncofetal mucin M1 antigens associated with precancerous colonic mucosae. Cancer Res. 46 (8), 3983–3989 [PubMed] [Google Scholar]
- 109. Richman P. I., Bodmer W. F. (1987) Monoclonal antibodies to human colorectal epithelium: markers for differentiation and tumour characterization. Int. J. Cancer 39 (3), 317–328 [DOI] [PubMed] [Google Scholar]
- 110. Hogg N., MacDonald S., Slusarenko M., Beverley P. C. (1984) Monoclonal antibodies specific for human monocytes, granulocytes and endothelium. Immunology 53 (4), 753–767 [PMC free article] [PubMed] [Google Scholar]
- 111. Singhal A. K., Orntoft T. F., Nudelman E., Nance S., Schibig L., Stroud M. R., Clausen H., Hakomori S. (1990) Profiles of Lewisx-containing glycoproteins and glycolipids in sera of patients with adenocarcinoma. Cancer Res. 50 (5), 1375–1380 [PubMed] [Google Scholar]
- 112. Abe K., McKibbin J. M., Hakomori S. (1983) The monoclonal antibody directed to difucosylated type 2 chain (Fuc alpha 1 leads to 2Gal beta 1 leads to 4[Fuc alpha 1 leads to 3]GlcNAc, Y Determinant, J. Biol. Chem. 258 (19), 11793–11797 [PubMed] [Google Scholar]
- 113. Lloyd K. O., Larson G., Stromberg N., Thurin J., Karlsson K. A. (1983) Mouse monoclonal antibody F-3 recognizes the difucosyl type-2 blood group structure, Immunogenet. 17 (5), 537–541 [DOI] [PubMed] [Google Scholar]