Abstract
From a series of recently published reports, an analytical platform has been proposed for a quantitative and qualitative measure of N- and O-glycosylation, complete with peptide-glycan connectivity and detailed structural understanding. As distant as this may appear, a best methods approach will appear that must move us beyond the cartoon stage of structural understanding. Thus, with this unifying goal in mind, we summarize a series of individually promising first phase protocols of sample preparation (release, purification, and quantification) that remain congruent with a concluding phase (methylation and MSn) for documented structural detail. Sequential enzymatic N-glycan and chemical O-glycan release from glycopeptides with intervening solid phase extraction and derivatization will provide for a comparative quantification measure of glycosylation. The O-glycan release will be nonreductive and coupled with Michael addition to a pyrazolone analog (1-phenyl-3-methyl-5-pyrazolone) with both the peptide and glycan labeled. The product glycans are stable to methylation and appropriate for sequential disassembly (MSn). An application using human serum and cancer samples has been detailed characterizing sLex and comparable valence epitopes. This integrated platform will provide opportunities at variable points to contrast, share, and advance alternative protocols in a collaborative effort that is greatly needed. This integrated platform provides end point opportunities to confirm structural details compiled from synthetic standards and well characterized biologics by MSn.
A UNIFIED GLYCOMICS PLATFORM
The NHLBI, National Institutes of Health, established a Program of Excellence in Glycosciences that supports a study of glycans ubiquitously found on the surfaces of all mammalian cells. Such structures influence a wide variety of cellular and disease-related processes that are governed by the composition and configuration of the interacting partners and the details of structure and conformation within that supramolecular environment. An example of these cohorts are the selectins (E-, L-, and P-) that bind to sialofucosylated ligands displayed on their respective glycans (1, 2). Following an earlier focus by the NIGMS and the NCI of the National Institutes of Health, the NHLBI introduced this Program of Excellence in Glycosciences to bring a more comprehensive glycomics endeavor to support the growing importance of glycan structures in heart, lung, and blood disease research. The Program of Excellence has established three fundamental goals as follows: (i) to develop and expand core facilities across the country; (ii) to facilitate collaborations and distribute research findings, and (iii) to train future generations of scientists to be cognizant in both glycan biology and chemistry. These are laudable goals by any measure, but what blurs the effort and forward momentum must be the considerations of the last goal. With the expanding list of tools contributing to an ever widening array of new applications, the goal for focused training in applications appears disjointed and distant. Thus, we present the underlying motives for this review as follows: first, a platform that would allow a comparative assessment of analytical tools at both a quantitative and qualitative level; second, an opportunity to consider a captivating nonreductive O-glycan release strategy concomitant with peptide site labeling; and finally, a platform where structural details are exposed and can be documented against a spectral library of synthetic standards and well known commercial products (Fig. 1). The lecture aspects of this sequencing application will be reviewed at a Charles Warren Tutorial scheduled at the University of New Hampshire, July 17–20th, 2013.
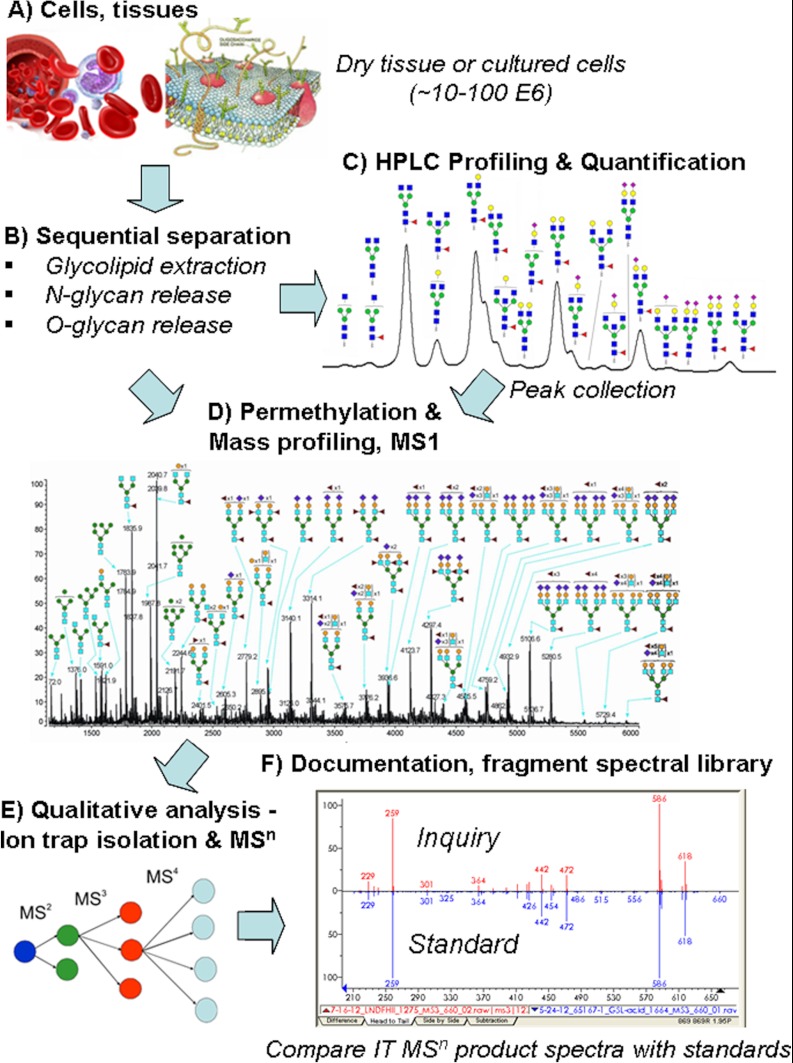
Fig. 1.
Quantitative and qualitative sequencing platform with product documentation. Overview of a proposed comprehensive glycan sequencing platform. Dry tissue or cultured cells (A) can be processed. Protocols include processing steps of sequential protein release. B, chromophore/fluorophore labeling of individually released N-linked and nonreductive O-linked glycans for HPLC quantification (data not shown). C, for qualitative understanding and structural interrogation, HPLC peak areas may be collected, methylated, chip-spray infused, and mass profiled (D) for detailed structural understanding by ITMSn. This was designed to provide comprehensive work flow with different points of entry and egress depending on data requirements. Disassembled fragments (E) documented by fragment library match (F) were compiled from known standards. This two-phase approach, sample preparation/purification coupled with second phase structural determination, may provide improved options for comparative collaboration among researchers.
GLYCAN RELEASING
Aberrant glycosylation and disease relationships have been associated with all glycoconjugates, and protocols have been published to separately isolate and partially characterize each glycotype. In the interests of analytical progress and continuity of application, we share and describe how these protocols can be effectively drawn together and equally applied to seek comparative quantification (Fig. 1). Starting with dry ice-pulverized cells, tissues, or liquid samples, initial chloroform/methanol extraction or partition will selectively remove glycolipid classes for independent analysis. These samples can be directly methylated and analyzed by ITMSn.1 Collisional activation and disassembly of precursor ions (ESI) provide conjugate products, permitting detailed structural assignment of each (3). Thus, GSLs will not be considered further in this review.
The diverse solubility and conformational properties of the residual proteins (after lipophilic extraction) make the enzymatic glycan release variable, but these capricious problems can be largely eliminated by the facile chemical steps of reduction, alkylation, and proteolysis. Such well established procedures provide a stable set of glycopeptides for efficient enzymatic (N-linked) and subsequent chemical (O-linked) release. The first enzymatic step utilizes peptide:N-glycosidase F, a very effective peptide:N-glycosidase, the discovery of which must be considered a major milestone in the advancement of mammalian structural glycobiology (4). Hydrazine release (5), although technically more challenging and less sensitive, does provide an orthogonal evaluation that should always be contrasted at least once with new samples, particularly those of nonmammalian origin.
CHROMOPHORIC LABELING, N-GLYCANS
Free glycans (peptide:N-glycosidase F released) can be fluorescently labeled and HPLC-profiled providing quantitative abundance ratios (Fig. 1C) (6). The cartoons annotating the profile suggest structure only within the confines of peptide:N-glycosidase F and ion compositions obtained by mass profiling (7). Peak overlaps, shoulders, glycomers, and even isobars (at higher resolution) can be identified by cutting fractions (Fig. 1D), methylation (Fig. 1E) (8), and direct infusion for a detailed examination of structure as discussed below.
COUPLED CHROMOPHORIC RELEASE AND LABELING OF O-GLYCANS
Strong base release of O-glycans from serine and threonine glycopeptides has been, and remains, a stalwart technique (9). Reduction of the released hemiacetal to the corresponding alditol is an integral part of many studies, but this prevents any opportunity to enhance detection sensitivity or achieve exacting chromophoric quantification. Many variations to this problem have been considered with some success. However, the most intriguing solution may be pyrazolone derivatization, specifically with PMP (10), allowing a direct abundance contrast of N- and O-glycosylation. Early success with mild base catalyzed derivatization of monosaccharides and more recently with larger oligomers; N- and O-glycans (11, 12) support this interest. The synchrony of PMP condensation and glycan elimination prompted Wang et al. (13) (Northwest University, Xian, China) to bring this into a one-pot reaction. The catalyzed β-elimination and rapid Michael addition to the reducing terminus seems to diminish the chance of peeling, an important consideration for select glycans. Similar insight may have been realized (7), and further applications will be exciting to read and apply. There will probably be changes and improvements in the details (14, 15), and preliminary data gathered here indicate that subsequent methylation and sequential disassembly bring together a comparative framework for a quantitative and qualitative assessment for both N- and O-linked glycans of proteins, as outlined in Fig. 1.
GLYCOSYLATION SITE IDENTIFICATION OF O-LINKS
N-Linked glycosylation sites generally have a consensus sequence, whereas O-links do not. To fully understand their biological significance, both glycan and peptide need to detail their specific origins. When applied to glycopeptides, this PMP/DMA release/derivatization scheme now appears to be a reasonable expectation (14). In this application, all groups susceptible to β-elimination become products with only the phospho and sulfo groups not being labeled. The reaction is essentially a combined β-elimination/Michael addition in which a carbon-carbon bond-forming Michael donor rather than a heteroatomic Michael donor is used (complementing earlier studies), but here the released O-glycans are recovered as bis-pyrazolone derivatives, with minimal side reactions (peeling). Using this technique, the O-glycan profiles of model mucin-type glycoproteins were successfully analyzed (14, 15). Carried out in the presence of a thiol (2-mercaptoethanol) provided Michael addition to the peptide (14, 15) analogous to earlier O-GlcN studies (16). With single-site glycopeptides, this specifically ties the O-glycan to a specific sequence, a feature lost with multiple sites.
Sequence Documentation, ITMSn General Considerations
Glycans manifest their activity in a multitude of ways, including modulation of a protein's conformation and solubility or serving as conjugate receptors, activators, or blocking agents. It is abundantly clear, however, that a large portion of function resides within properly positioned valence epitopes of very specific structure on the outer surface of lipids and proteins. A listing has been estimated to approximate 3000 (17). This finite number is intriguing with the caveat that much glycomic function might reside on a limited set of epitopes specifically situated on selected conjugates. Focused efforts to characterize such epitopes would be a shortcut to functional glycobiology; however, sensitive procedures to isolate and confirm such details are lacking. Accurate mass compositions and chromatographic separations alone fail to meet the qualitative needs of bioengineers aspiring to design blocking analogs, vaccines, and antibodies to control disease progression. Such conjugates possess a plethora of isomers, both stereo and structural, the details of which exemplify the major obstacles to an understanding of sequence, whether embodied in an epitope or not. Contemporary MS studies can be summarized as efforts to achieve a combination of glycoside and cross-ring fragments to ascertain monomer sequence with branching information. Bond rupture is related to energy deposition (although radical chemistry is altering that feature), where molecular dispersal is immediate cleaving first the weakest bonds (ketosidic, and amido-glycosidic) and finally the cross-ring bonds. Operationally, the pursuit and expectation that all components of structure can be represented in a one- or two-dimensional MS spectrum may be short-sighted, especially when considering the diverse bond strengths and the need to unravel stereo and structural isomeric complexity. Instrumentally, it does appear that careful control of energy deposition could selectively disassemble glycans, but such precision and speed are currently unavailable.
In early pursuit of sequencing goals, we were encouraged with our first venture into collisional activation of carbohydrate structures in 1985 (18), and we realized the opportunities more with the soft ionization techniques of ESI and MALDI in the early 1990s. But the instrument that has captivated our greatest interest has been the unique capabilities of sequential disassembly provided by the ion trap. Data output from these instruments were captivating, especially when contrasted to the transparency of linkage and branched isomers missed in MS/MS analyses. Preparing samples as methyl derivatives, detailed studies with known biologics, and numerous applications were convincing (19–22), and the outgrowth of these and related studies was that glycan isomers have been universally overlooked, even in classic glycoprotein standards, RNase B and ovalbumin (23, 24). In both examples, such findings have recently been confirmed by extensive orthogonal efforts using HPLC (25) and IMMS (26). Motivated by these earlier findings, we initiated a searchable library of fragments compiled from commercially available small oligomers (27), an ongoing effort supported now by major synthetic teams (see Acknowledgments). The unreported isomers (28) in such well studied IgG Fc-glycans brings into question the tremendous bioengineering efforts focused on the design and synthesis of functional vaccines and antibiotics but absent a detailed structural understanding.
Sequential disassembly in the ion trap is intriguing, and its operating principles are most synchronous with glycan disassembly. When supplying energy to orbiting trapped ions, the only defined variable is their size (fewer oscillators) for energy dispersal. Smaller ions accommodate excitation with greater fragmentation (a word description of the quasi-equilibrium theory, QET). In this inverse relationship lies the beauty of the ion trap; the smallest fragments rupture the most stable bonds. This is most obvious in the MS spectra of three sodium adducted ions, a decamer, dimer, and a monomer (Fig. 2). The differences are distinctive and follow QET theory. The patterns of ion masses and neutral losses are characteristic providing a good sense of structure. But, in combination with plotted abundance spectra, the guess work is gone for small oligomers. Linkage points increase ring instability, frequently following a retro-Diels-Alder degradation pattern. Confirmation of these end product structures can be approached by selecting different disassembly pathways and showing a matching product spectrum. Potassium and lithium can be used as alternative cations, although some understanding and experience are important.
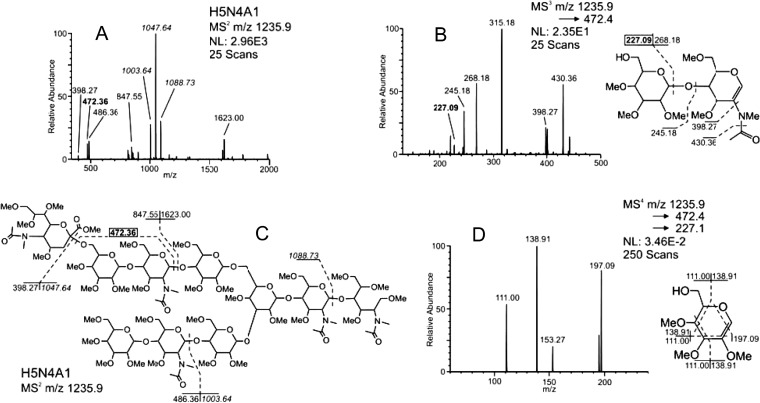
Fig. 2.
Oscillator and energy relationships, QET. A presentation of three different precursor ions and their subsequent CID spectrum supports the QET theory of greater instability (fragmentation) with fewer oscillators. MS2, m/z 1235.9+2, of the doubly charged ion provided the spectrum (left top) showing major fragments at neuraminyl and HexN bonds. Smaller fragments, m/z 486 and 472, indicate LacNAc structures, the latter ion 14 Da lower indicating former linkage site and positioning the neuraminyl group. Also apparent in that spectrum was a B/Y-ion linked (nonterminal) monosaccharide fragment, m/z 227. Disassembly of that fragment (lower right) provided a combination of cross-ring cleavages that positioned the hydroxyl group at 6.
In summary, somewhat ironically, a mass measuring instrument, operating in a sequential manner, can be exploited to resolve structural details better than most, if not all, current chromatographic systems, and thereby provides a different type of resolution, based on ion fragments and their relationships. Such details have been compiled in a searchable library, thereby documenting the steps of disassembly and detailing valence epitopes (Figs. 3–6). These evolving results prompted us to move beyond a resolution in time (differing chromatographic media) to a resolution in space (the collision space of the ion trap), and we have for a number of years considered this instrument as having the greatest promise for providing a comprehensive carbohydrate sequence. The many critics are mainly concerned by the concept that a vacuum space could contribute to structural understanding. A further concern has been the need for methylation and an inability to see anomers. However, methylation was only difficult for the untrained, and limited practice usually resolved such problems. Solid phase methylation (8) now makes this an ideal undergraduate exercise. Fragment spectra reflecting anomeric and stereoisomeric differences have been observed and rationalized on the basis of metal ion binding specificity, but much needs to be done to define limits. These questions make good research problems, and C-type ions (29) might be a good place to start. Collisions in space are instantaneous with little chance for equilibration; unlike the anomerization problems in a liquid, so there is no reason to reject such a study outright. Although many details remain to be clarified, we have moved forward assembling a set of supporting tools and techniques that have been summarized (30). A fragment library has been compiled with a limited number of synthetic standards provided by the Consortium for Functional Glycomics (J. Paulson), The Complex Carbohydrate Research Center (G. Boons), and numerous well characterized commercial samples.
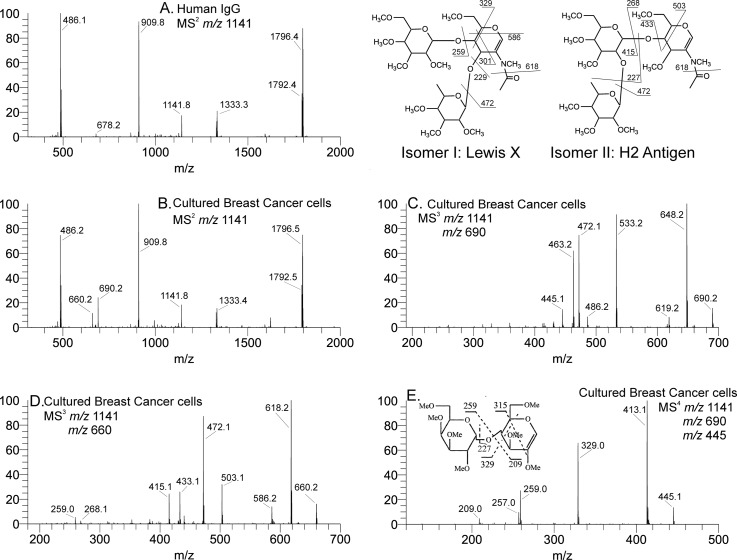
Fig. 3.
Comparative analysis of isomers from two different samples. Released glycans prepared as reduced and methylated analogs and directly infused into a chip-based nanoelectrospray ionization system and analyzed by ITMSn. Comparative analysis of isomers was from two different samples, IgG (A) and breast cancer cells (B). Each sample provided a spectrum, MS2 (A and B), with a composition of Gal2GlcNAc2dHex+Man3GlcNAc suggesting a G2F glycan (expected loss of reducing terminal GlcNAcol) providing the m/z 1141.8 ion. Additional disassembly, MS3–5, of the IgG sample (A) proved the assumption to be correct, G2F (28). Isolation of the same ion from spectrum B provided spectra C–E, which on examination indicated the initial glycan ion to be composed of two different terminal valence epitopes, Lewis x and H2 antigen, isomer I and II, respectively. The message was that analysis limited to MS/MS may not be a comprehensive approach.
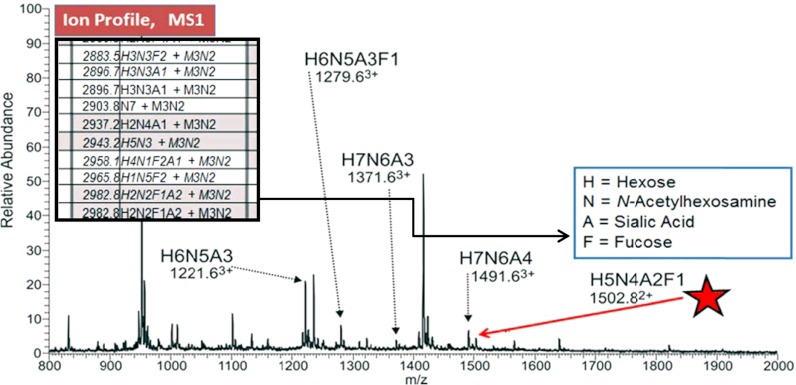
Fig. 4.
Human serum N-glycans; detailing isomers including sLex, m/z 1502.8+2. The released glycans were prepared as reduced and methylated analogs and directly infused into a chip-based nanoelectrospray ionization system and analyzed by ITMSn. The resulting mass profiles (MS1) of IgG-depleted and nondepleted plasma samples were contrasted, and these results were again compared with recent literature reports. Before depletion, ∼50 independent glycan ions were detected; this more than doubled to 106 after depletion. The mass range profiled was 1–5 kDa, which included many doubly and triply charged ions that were resolved by higher MS resolution. Selected ions in the depleted sample were disassembled to define their detailed structure (21). The simplicity of this nonchromatographic, direct infusion, and gas-phase structural characterization compares favorably with the latest reports using alternative instrumentation and adjunct techniques.
Fig. 5.
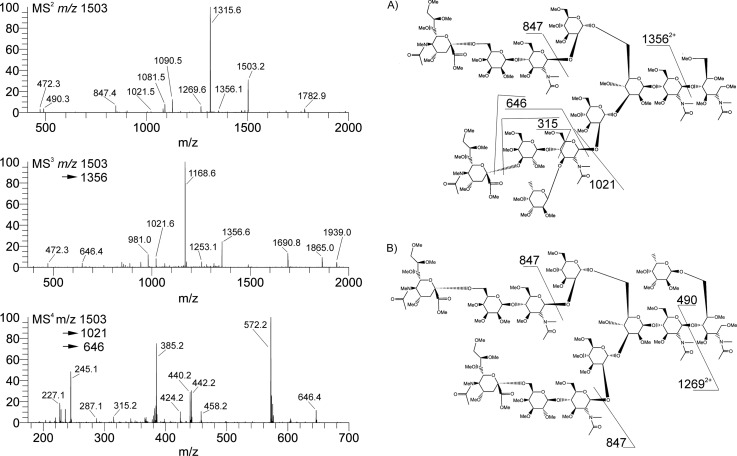
Sequential disassembly, m/z 1502.8+2. The released glycans were prepared as reduced and methylated analogs and directly infused into a chip-based nanoelectrospray ionization system and analyzed by ITMSn (21). A check on one fucosylated ion, m/z 1502.8+2, 2982.6+1 (red star in Fig. 4) was disassembled for structural detail. Two isomers of this composition differed in fucose location (terminal core GlcNAc and antennal). The latter possessed an sLex epitope, detailed in Figs. 5 and 6. Two proposed structures can be considered from the fragment ions in MS3,4,5. Penultimate rupture (MS/MS) defines isomers, m/z 1269.6 and 1356.1, whereas m/z 646.4 and 1021.6 (MS/MS) suggest sLex epitopes. These can only be proven upon further disassembly. Isolation of the m/z 1346.1 for MS3 provides the marker ions m/z 646 and 1021 locating the fucosyl-LacNAc moiety. The proposed galactosyl residue (B) must be isolated to define sialyl linkage. In this blood sample, the ion current was insufficient and that question could not be answered. See Fig. 6.
Fig. 6.
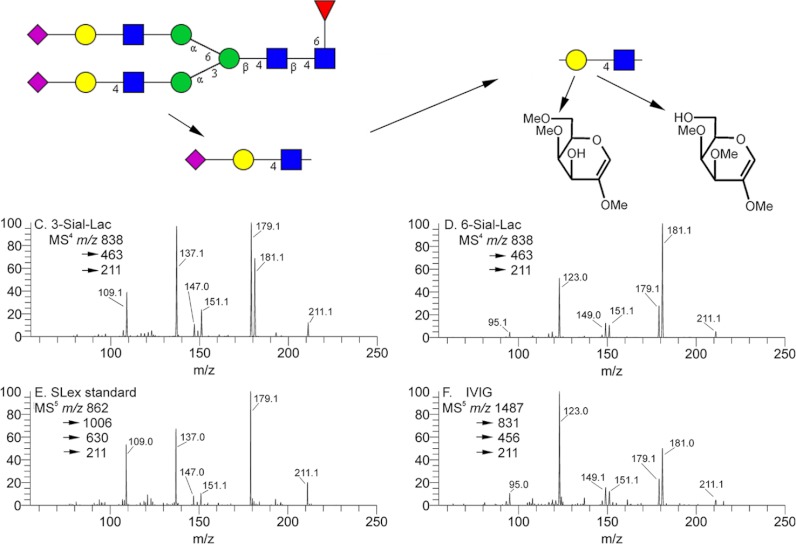
Disassembly of terminal sialyl lactosyl analog. Defining detailed structure on complex structures frequently requires multiple steps of disassembly. Sample purity is the usual problem, i.e. not detection sensitivity. Impure fractions are common, but important problems do get solved (19). Chip infusion (Advion-Triversa) is an essential way to increase duty cycle time, and newer instruments are sure to approach this problem. However, pathways of disassembly exist for full characterization. The aborted sialyl (2–3/6) linkage problem (Fig. 5) can be resolved by the defining cross-ring fragments formed during fragmentation of the B/Y Gal moiety, isolated as the lithiated m/z 211 ion. The two sialyl lactose standards were separately disassembled to expose the Gal residue. The resulting spectra are shown, as well as the same fragment isolated from one sLex standard and an IVIG sample. The localization of the hydroxyl to the 3- or 6-position produces clearly different mass spectra.
STRUCTURAL DETAIL BEYOND MS/MS
Mass spectrometry remains the instrument of choice for glycoprotein analyses. The sensitivity, flexibility, and moderate cost warrant all claims. In the previous section, we summarized several factors that make sequential disassembly a more effective strategy to expose carbohydrate details. In this section, we extend that understanding to isomeric glycan structures isolated from human serum IgG and from cultured breast cancer cells. N-Glycans were released, methylated, and mass profiled. A common m/z 2260.3 ion was observed having a composition of Gal2GlcNAc2dHex+Man3GlcNAc2 and a doubly charged ion, m/z 1141.8+2, an expected IgG-G2F glycan. MS/MS analysis of that ion is shown in Fig. 3A, top left panel. The same ion was isolated from cultured breast cancer cells and worked up in a similar manner as shown in Fig. 3B. We have previously shown that upon further analyses (MS3,4) the IgG sample was composed mostly of a G2 structure, with two smaller isomers (28). The MS2 spectrum from the cultured breast cancer cells shows additional fragments at m/z 660 and 690 (Fig. 3B), which are absent from the IgG spectrum (Fig. 3A). Isolation and CID of the m/z 660 ion provided the MS3 spectrum (Fig. 3D). These spectral results indicate a mixture of Lex and the H2 antigen, and the B-type fragment (29) structures are shown in Fig. 3C, top right panel. The m/z 690 fragment is indicative of a Gal-Gal-GlcNAc B-type fragment. Disassembly produced the MS3 and MS4 spectra shown. Importantly, the MS4 spectrum clearly identifies a 4 linkage between the galactose residues; this would be difficult to determine at earlier MSn stages. These fragments were confirmed by spectral matches (27).
Colorectal Cancer Cell Lines and sLex Epitope Structural Details
The high expression of sLea appears to be an prognostic measure for predicting recurrence in colorectal cancer patients. It has been suggested that a clinical evaluation of sLea CA-19-9 antigen could be useful in the planning of coadjuvant therapies after surgical resection, especially in patients with high sLea expression. ITMSn has the ability to structurally differentiate the isomeric sLea/sLex antigens and, equally important, to specify their setting on different glycoconjugate platforms (lipids, N-, or O-linked glycoproteins). Following the platform outline (Fig. 1), dried samples were first extracted with lipophilic solvents (Fig. 1B), and soluble GSLs were removed and residues solubilized for the separate release of N- and O-glycans. We have not evaluated carefully PMP nonreductive release, so classical base-reductive elimination was used to prepare these O-glycans. Released N-glycans were solid phase extraction-purified, reduced, and directly methylated for ITMSn analyses. An ion composition H5N4A2F1 (Fig. 4) fits a fucosylated, disialyl, and biantennary glycan that was resolved into two isomers by MS2 (Fig. 5). The product ion, m/z 1356+2, H5N3A2F1, formed by loss of the reducing-end GlcNAc, retains the fucose and was thus selected for MS3 analyses (Fig. 5, middle spectrum), which provided fragment compositions suggesting sLex, is at m/z 646.4 and 1021.6. But proof could only be achieved by an additional disassembly step, MS4/5. This spectrum provided a most informative product ion, m/z 315.2. This fragment usually provides a cross-ring 3,5X-ion fixing three structural components, confirming a sLex structure. Unfortunately, the ion current from this 50-μl serum sample was insufficient for evaluation at the monomer level. These sialyl linkage details are illustrated in Fig. 6.
Details of Sialyl Linkage in Smaller Oligomers
Disassembly to small oligomers and a spectral match are imperative for a comprehensive understanding of the structure. Sample impurities are a fact of biological research, but important problems do get solved by including additional purification steps (19). Chip infusion (Advion-Triversa) is a way to increase duty cycle time, and newer instruments are sure to develop comparable sensitivity-enhancing features. But pathways of disassembly do exist for full characterization with purer samples and smaller oligomers. The sialyl (2–3/6) linkage question (Fig. 5) in the serum sample may well be resolved with one additional step. As shown in Fig. 6, the sialyl linkage can be resolved as demonstrated in Fig. 6 by exposing the B/Y-type galactose moiety. In this strategy, lithium adduction proved advantageous in terms of superior ion current and spectral differentiation. The two sialyl-lactose standards were separately disassembled to expose the linking Gal residue by selecting the B/Y-type ion, m/z 211, which is structurally the same as the sodiated m/z 227 ion in Fig. 5. The bottom two spectra are the same fragment isolated from an sLex standard and an IVIG sample.
MSn SPECTRA HANDLING AND LIBRARY SEARCH
Library confirmation of spectral data is a fundamental component underpinning this beginning technology. The library has been largely compiled from numerous small, commercially available oligomers and standard glycoproteins, but synthetic contributions are arriving periodically. These results have also been substantiated when following alternative pathways of disassembly and when contrasted with synthetic standards provided by collaborators (James Paulson, Consortium for Functional Glycomics; Geert-Jan Boons, The Complex Carbohydrate Research Center). A software set of tools (FragLib tool kit) has been designed for constructing MSn libraries and facilitating the management of raw spectral files (27, 30). The tool captures raw data files, the pathway, and a peak/abundance list of each spectrum, converting them into a single spectral library component having a smaller disk footprint. Various community-accepted spectral formats are supported by the tool. Therefore, the NIST MS search tool, and other commonly used software can be chosen as the library searching engine to match unknowns to the accumulated glycan standards. With small fragments and enhanced fragmentation, an MSn library provides detailed spectral evidence of a glycan structure.
CONCLUSIONS
In contemporary glycoproteome reports, carbohydrate isomers, linkage, and branching details are usually unresolved and instrumentally transparent, even when coupled with a host of hyphenated techniques. This inability to detect often projects to an absence, but an absence of evidence is not evidence of absence. Alternative information, such as high resolution (mass or chromatographic), biological inference, statistics, tandem MS, or linkage analysis does provide more data, but conclusive and reproducible structural information remains elusive. Terminal valence epitopes (17) are common and may drive a large component of glycobiology, and a more focused accounting of such structures is sorely needed. However, the gaps to achieve such goals are considerable, especially when trying to match the details needed by bioengineers and the promises envisioned from personalized medicine. When considering the obstacles to such details, divergent sample preparation must be high on the list and, as the sample amounts get smaller, the problems approach the impossible. Here is where collaborative efforts remain critical and maybe a start would be around unified ways of sample preparation. From such samples, an attack on structural detail can progress most effectively, and we would like to suggest ITMSn is clearly the technology of choice. This approach, although qualitatively very high, has limits in that it requires somewhat larger amounts of sample, a significant investment in time per sample, a particular MS instrumentation, and a degree of expertise and database querying. In the short history of MS applications, these problems will clearly get resolved. However, with wider adoption of such approaches, improvements in sample handling, instrumentation, and software automation, such hurdles can eventually be overcome.
Acknowledgments
We greatly appreciate the synthetic standards supplied by Jim Paulson (Consortium for Functional Glycomics), Geert-Jan Boons (The Complex Carbohydrate Research Center), and Dr. Khushi Matta.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grants P01 HL107146-01 and RO1A1090866. This work was also supported by Glycan Connections, LLC.
1 The abbreviations used are:
- ITMSn
- ion trap mass spectrometry
- PMP
- 1-phenyl-3-methyl-5-pyrazolone
- DMA
- dimethylamine
- IMMS
- ion mobility mass spectrometry.
REFERENCES
- 1. Lühn K., Wild M. K. (2012) Human deficiencies of fucosylation and sialylation affecting selectin ligands. Semin. Immunopathol. 34, 383–399 [DOI] [PubMed] [Google Scholar]
- 2. Sackstein R. (2009) Glycosyltransferase-programmed stereosubstitution (GPS) to create HCELL: engineering a roadmap for cell migration. Immunol. Rev. 230, 51–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Levery S. B. (2005) Glycosphingolipid structural analysis and glycosphingolipidomics. Methods Enzymol. 405, 300–369 [DOI] [PubMed] [Google Scholar]
- 4. Tarentino A. L., Gómez C. M., Plummer T. H., Jr. (1985) Deglycosylation of asparagine-linked glycans by peptide:N-glycosidase F. Biochemistry 24, 4665–4671 [DOI] [PubMed] [Google Scholar]
- 5. Patel T. P., Parekh R. B. (1994) Release of oligosaccharides from glycoproteins by hydrazinolysis. Methods Enzymol. 230, 57–66 [DOI] [PubMed] [Google Scholar]
- 6. Anumula K. R. (2012) Quantitative glycan profiling of normal human plasma derived immunoglobulin and its fragments Fab and Fc. J. Immunol. Methods 382, 167–176 [DOI] [PubMed] [Google Scholar]
- 7. Zauner G., Kozak R. P., Gardner R. A., Fernandes D. L., Deelder A. M., Wuhrer M. (2012) Protein O-glycosylation analysis. Biol. Chem. 393, 687–708 [DOI] [PubMed] [Google Scholar]
- 8. Mechref Y., Kang P., Novotny M. V. (2009) Solid-phase permethylation for glycomic analysis. Methods Mol. Biol. 534, 53–64 [DOI] [PubMed] [Google Scholar]
- 9. Carlson D. M. (1966) Oligosaccharides isolated from pig submaxillary mucin. J. Biol. Chem. 241, 2984–2986 [PubMed] [Google Scholar]
- 10. Honda S., Akao E., Suzuki S., Okuda M., Kakehi K., Nakamura J. (1989) High-performance liquid chromatography of reducing carbohydrates as strongly ultraviolet-absorbing and electrochemically sensitive 1-phenyl-3-methyl-5-pyrazolone derivatives. Anal. Biochem. 180, 351–357 [DOI] [PubMed] [Google Scholar]
- 11. Kakehi K., Suzuki S., Honda S., Lee Y. C. (1991) Precolumn labeling of reducing carbohydrates with 1-(p-methoxy)phenyl-3-methyl-5-pyrazolone: analysis of neutral and sialic acid-containing oligosaccharides found in glycoproteins. Anal. Biochem. 199, 256–268 [DOI] [PubMed] [Google Scholar]
- 12. Shen X., Perreault H. (1998) Characterization of carbohydrates using a combination of derivatization, high-performance liquid chromatography and mass spectrometry. J. Chromatogr. A 811, 47–59 [DOI] [PubMed] [Google Scholar]
- 13. Wang C., Fan W., Zhang P., Wang Z., Huang L. (2011) One-pot nonreductive O-glycan release and labeling with 1-phenyl-3-methyl-5-pyrazolone followed by ESI-MS analysis. Proteomics 11, 4229–4242 [DOI] [PubMed] [Google Scholar]
- 14. Furukawa J., Fujitani N., Araki K., Takegawa Y., Kodama K., Shinohara Y. (2011) A versatile method for analysis of serine/threonine post-translational modifications by β-elimination in the presence of pyrazolone analogues. Anal Chem. 83, 9060–9067 [DOI] [PubMed] [Google Scholar]
- 15. Zauner G., Koeleman C. A., Deelder A. M., Wuhrer M. (2012) Mass spectrometric O-glycan analysis after combined O-glycan release by β-elimination and 1-phenyl-3-methyl-5-pyrazolone labeling. Biochim. Biophys. Acta 1820, 1420–1428 [DOI] [PubMed] [Google Scholar]
- 16. Vosseller K., Hansen K. C., Chalkley R. J., Trinidad J. C., Wells L., Hart G. W., Burlingame A. L. (2005) Quantitative analysis of both protein expression and serine/threonine post-translational modifications through stable isotope labeling with dithiothreitol. Proteomics 5, 388–398 [DOI] [PubMed] [Google Scholar]
- 17. Cummings R. D. (2009) The repertoire of glycan determinants in the human glycome. Mol. Biosyst. 5, 1087–1104 [DOI] [PubMed] [Google Scholar]
- 18. Carr S. A., Reinhold V. N., Green B. N., Hass J. R. (1985) Enhancement of structural information in FAB-ionized carbohydrate samples by neutral gas collision. Biomed. Mass Spectrom. 12, 288–295 [DOI] [PubMed] [Google Scholar]
- 19. Anthony R. M., Nimmerjahn F., Ashline D. J., Reinhold V. N., Paulson J. C., Ravetch J. V. (2008). Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG Fc. Science 320, 373–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sheeley D. M., Reinhold V. N. (1998) Structural characterization of carbohydrate sequence, linkage, and branching in a quadrupole ion trap mass spectrometer: neutral oligosaccharides and N-linked glycans. Anal. Chem. 70, 3053–3059 [DOI] [PubMed] [Google Scholar]
- 21. Stumpo K. A., Reinhold V. N. (2010) The N-glycome of human plasma. J. Proteome Res. 9, 4823–4830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spaink H. P., Sheeley D. M., van Brussel A. A., Glushka J., York W. S., Tak T., Geiger O., Kennedy E. P., Reinhold V. N., Lugtenberg B. J. (1991) A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature 354, 125–130 [DOI] [PubMed] [Google Scholar]
- 23. Prien J. M., Ashline D. J., Lapadula A. J., Zhang H., Reinhold V. N. (2009) The high mannose glycans from bovine ribonuclease B isomer characterization by ion trap MS. J. Am. Soc. Mass Spectrom. 20, 539–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiao J., Zhang H., Reinhold V. N. (2011) High performance IT-MS sequencing of glycans (spatial resolution of ovalbumin isomers). Int. J. Mass Spectrom. 303, 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Prien J. M., Prater B. D., Cockrill S. L. (2010) A multi-method approach toward de novo glycan characterization: a Man-5 case study. Glycobiology 20, 629–647 [DOI] [PubMed] [Google Scholar]
- 26. Zhu F., Lee S., Valentine S. J., Reilly J. P., Clemmer D. E. (2012) Mannose7 glycan isomer characterization by IMS-MS/MS analysis. J. Am. Soc. Mass Spectrom. 23, 2158–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang H., Singh S., Reinhold V. N. (2005) Congruent strategies for carbohydrate sequencing. 2. FragLib: an MSn spectral library. Anal. Chem. 77, 6263–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ashline D. J., Lapadula A. J., Liu Y. H., Lin M., Grace M., Pramanik B., Reinhold V. N. (2007) Carbohydrate structural isomers analyzed by sequential mass spectrometry. Anal. Chem. 79, 3830–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Domon B., Costello C. E. (1988) Structure elucidation of glycosphingolipids and gangliosides using high-performance tandem mass spectrometry. Biochemistry 27, 1534–1543 [DOI] [PubMed] [Google Scholar]
- 30. Reinhold V. N., Ashline D. J., Zhang H. (2010) in Practical Aspects of Trapped Ion Mass Spectrometry (March R. E., Todd J. F., eds) pp. 705–736, CRC Press, Inc., Boca Raton, FL [Google Scholar]