Abstract
The fact that sulfated glycosaminoglycans (GAGs) are necessary for the functioning of all animal physiological systems drives the need to understand their biology. This understanding is limited, however, by the heterogeneous nature of GAG chains and their dynamic spatial and temporal expression patterns. GAGs have a regulated structure overlaid by heterogeneity but lack the detail necessary to build structure/function relationships. In order to provide this information, we need glycomics platforms that are sensitive, robust, high throughput, and information rich. This review summarizes progress on mass-spectrometry-based GAG glycomics methods. The areas covered include disaccharide analysis, oligosaccharide profiling, and tandem mass spectrometric sequencing.
All living cells require a layer of glycoconjugates on their surfaces, sometimes known as a glycocalyx, for survival (1). These glycoconjugates developed in response to evolutionary pressures as a way to maintain organismal fitness. Sulfated glycosaminoglycans (GAGs)1 are found only in the animal kingdom. They appeared with the cnidarians, the earliest animals to have muscles and nervous systems (2). Among eukaryotes, only animal cells have the ability to biosynthesize GAG polysaccharides modified with sulfate groups. Some bacterial pathogens, including Pasturella, Eschericia, and Streptococcus, have capsules containing polysaccharide co-polymers with the same disaccharide structure as nascent animal GAGs, but these arrived through a separate evolutionary path. The apparent functions of these capsules are likely to involve camouflage from host immune systems and adhesion. In animals, the expression of sulfated GAGs appears to be required for the organization of tissue layers, organ compartments, epithelial cells, and basement membranes (3). In mice, the complete elimination of GAG biosynthesis via genetic techniques results in embryonic lethality. Knockout of specific GAG-modifying enzymes results in a variety of organ-specific phenotypes of varying severity (4).
It is compelling to view GAG function through the lens of protein binding. The acidic character of GAGs lends these polysaccharides the tendency to bind basic protein domains, such as are found on numerous classes of growth factors and growth factor receptors. Seen in this way, GAG–protein interactions may be viewed as comparatively nonspecific. We know, however, that there are specific interactions that occur between GAGs and proteins that indicate the presence of both high- and low-affinity interactions in biological systems (5, 6).
According to present understanding, GAGs are expressed on the surfaces of all nucleated cells. Given this wide expression, it has been said that the functioning of every mammalian physiological system depends on the regulated expression of GAGs (3). The expression of GAGs is required for embryogenesis (7), and the knockout of particular GAG biosynthetic enzymes leads to organ-specific phenotypes (4). By extension, nearly every disease has an aspect that impinges upon GAG function. GAGs expressed on cell surfaces serve as co-receptors for growth factor–receptor interactions for downstream receptor tyrosine kinase signaling cascades. GAGs distant from the cell surface participate in the formation of growth factor gradients and sequester growth factors from the cell surface. Thus, there is interplay between the catalysis of growth factor signaling at the surface and sequestration of growth factors away from the surface. Extracellular endosulfatases remove 6-O-sulfate groups from heparan sulfate (HS) chains, altering their interactions with growth factors and growth factor receptors (8, 9). Extracellular heparanase cleavage of HS chains also occurs and has been correlated with cancer phenotypes (10). In addition, heparan sulfate proteoglycans may be endocytosed for degradation or recycling. Thus, the HS structure associated with the cell surface is a dynamic balance of biosynthetic, remodeling, and degradative activities.
Mature chondroitin sulfate (CS)/dermatan sulfate and heparin/HS GAGs are expressed with domains of high and low sulfation, on which substantial heterogeneity is superimposed. Although these general features are widely appreciated, the detailed structures of GAG chains, in the conventional “sequence” sense, are not known. Thus, although it is known that GAG structure varies according to spatial and temporal factors, the dynamic natures of these glycomes are only beginning to emerge. Analytical methods are the primary factors limiting an understanding of GAG structural phenotypes and their roles in physiology.
THE ANALYTICAL CHALLENGE
The analytical challenge is to reduce the heterogeneous GAG populations that exist in a biological context into sequences representative of those that carry out GAG biological activities. Because the biological functions arise from GAG populations in which heterogeneity overlies regulated domain structure, the overall goal is to produce a model representative of the sequences present. An example of this process can be seen from the analytical data used to demonstrate similarity between generic and innovator forms of the low-molecular-weight heparin enoxaparin (11). Under this U.S. Food and Drug Administration ruling, analytical data are an essential part of the totality of evidence used to demonstrate the equivalence of generic and innovator enoxaparins. Disaccharide analysis is used to demonstrate compositional equivalence. Profiling is used to demonstrate the equivalence of oligosaccharide fragments generated via partial depolymerization of the enoxaparin preparation. Sequencing is used to demonstrate the equivalence of functionally relevant enoxaparin structures. The key point is that the analytical data represent a model sufficient to support a conclusion of equivalence between generic and innovator enoxaparin. The heterogeneity of GAG populations is such that it is not possible to sequence every oligosaccharide; however, effort can be focused on the characterization of oligosaccharides that represent the biological function.
ANALYTICAL METHODS
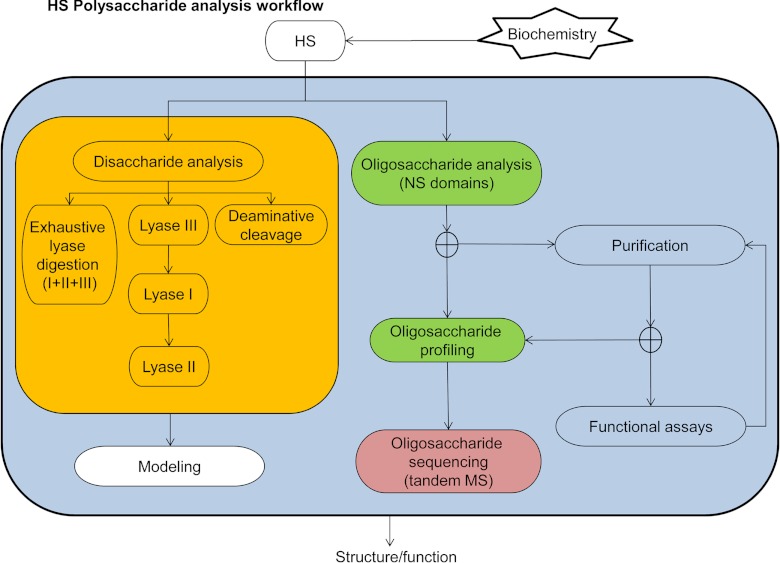
Fig. 1 shows a workflow for the analysis of GAGs using HS/heparin as the target. The goal is to deliver information on GAG structure that is sufficient for an academic or industrial effort. For biomedicine, such a workflow is relevant to the development of structure-function understanding in relation to a particular disease condition. In many such cases today, simple disaccharide analysis after exhaustive digestion using heparin lyases I, II, and III is used to determine the bulk properties of the GAG population in question. Although not yet widely adopted, disaccharide analysis after lyase digestion in series, combined with deaminative cleavage analysis, informs HS structure domain modeling algorithms (12). Oligosaccharide profiling determines the compositions and abundances of GAG oligosaccharides. When combined with purification, functional assays, and/or protein binding experiments, profiling produces useful structural information on functionally relevant GAG domains (13–15). Profiling can also be used to determine oligosaccharide sequences when combined with enzymatic and chemical digestion methods. Mass spectrometric sequencing is made challenging by heterogeneity and the nature of the fragile sulfate groups on GAG oligosaccharides. Nonetheless, recent progress has been made toward these ends.
Fig. 1.
Workflow for analysis of HS/heparin polysaccharides. Biochemically isolated HS chains are subjected to disaccharide analysis. A typical procedure is to digest samples exhaustively using heparin lyases I, II, and III. Modeling of the HS domain structure is most effective when disaccharide analysis is performed on lyase digests performed in series and on deaminative cleavage products.
Disaccharide Analysis
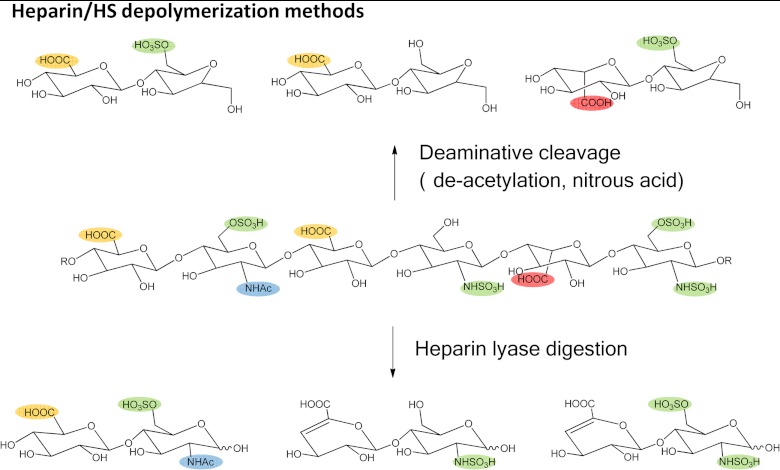
Disaccharide analysis remains the most often used analytical measurement for GAG populations. As shown in Fig. 2, GAGs can be depolymerized to disaccharides through the use of bacterial polysaccharide lyase enzymes (16) or deaminative cleavage by nitrous acid (17). Polysaccharide lyases cleave via an eliminative mechanism, resulting in disaccharides containing a Δ4,5-unsaturated uronic acid residue. As a result, lyase cleavage destroys information concerning the stereochemistry of the C5 position of uronic acid. Deaminative cleavage produces disaccharides containing an anhydromannose (for HS) or an anhydrotalose (for CS). With this method, the stereochemistry of the uronic acid is preserved, but information is lost concerning the substituent on the hexosamine nitrogen. Therefore, the two methods are complementary, and both are necessary in order for complete information on GAG disaccharides to be obtained.
Fig. 2.
Methods for the depolymerization of heparan sulfate. A representative HS hexasaccharide is shown with sulfate groups highlighted in green, glucuronic acid carboxyl groups in yellow, iduronic acid carboxyl groups in red, and acetate groups in blue. Heparin lyase digestion produces disaccharides with Δ4,5-unsaturated uronic acid for which information on the stereochemistry of C5 is lost. Deaminative cleavage produces disaccharides that contain anhydromannose (for CS GAGs, anhydrotalose is produced). Such disaccharides retain information on the stereochemistry of uronic acid C5 but lose information regarding the substitution of the hexosamine amino group.
The polysaccharide lyase disaccharide composition determines the absolute quantity of GAGs and the relative abundances of each disaccharide. Because the non-reducing chain terminus is not cleaved by the enzyme, the abundances of Δ-unsaturated relative to saturated structures can be used to estimate the average chain length of the GAG chains. The disaccharide composition can also be used to calculate the average number of sulfate and N-acetate groups per 100 disaccharide units (18). An algorithm has been developed to model HS chain structure based on disaccharide analysis data (12). This algorithm requires that the three heparin lyase enzymes be used in series, with disaccharide analysis carried out after each digestion. It also requires analysis of deaminative cleavage disaccharides. This analytical paradigm drives the need for rapid, sensitive, and robust disaccharide analysis methods.
The primary drivers behind the development of new disaccharide analysis methods have been sensitivity and robustness. As described above, the modeling of GAG structure depends on the ability to acquire a series of disaccharide profiles, one after each enzyme digest and after deaminative cleavage. As a result, it is imperative that the analysis system be both sensitive and robust. Sensitivity can be viewed in two ways. The ultimate sensitivity is likely to be highest for the fluorescence-based methods. The practical sensitivity, however, is determined by the quantity of a biological sample (number of cells, weight of tissue, etc.) necessary in order to achieve adequate signal at the end of the workflow, through modification or derivatization if necessary. The practical view takes into account the full workflow and the challenges in working from a biological matrix. Thus, the most appropriate method to use is the one capable of producing results from the biological matrix in question on a routine basis.
There are several methods used for disaccharide analysis. Lyase-generated disaccharides have a UV chromophore that allows detection with modest sensitivity. Efforts have therefore been made to improve sensitivity with fluorescence using reductive amination (19–22) or post-column fluorophore formation (23, 24). Methods that use radio (25) or optical detection systems have the advantage of simplicity but the disadvantage that standards are needed for every peak identified. Standards are commercially available for lyase-generated disaccharides but are not available for 3-O-sulfated disaccharides. In addition, standards are not available for deaminative cleavage disaccharides. MS-based methods have been used increasingly in recent years. Such methods range from the direct infusion of lyase disaccharides (26–28) to a variety of LC/MS systems using chromatography methods including reversed phase (29–31), reversed-phase ion pairing (32–34), size exclusion (18), porous graphitized carbon (51, 52) and hydrophilic interaction. Recently, an LC/MS method for the analysis of deaminative cleavage disaccharides was published (35). MS has the advantage of being able to detect low-abundance products in the digestion mixtures, including saturated disaccharides from the non-reducing polysaccharide termini and rare 3-O-sulfated disaccharides.
Oligosaccharide Profiling
The profiling experiment determines the compositions and abundances of oligosaccharides generated by the partial depolymerization of GAGs. GAG–protein binding requires an oligosaccharide domain of the polysaccharide chain. The use of selective lyase enzymes—for example, heparin lyase III—produces oligosaccharides that correspond to the N-sufated domains of the precursor polysaccharide chain. Profiling determines the compositions and abundances of such oligosaccharides.
MALDI-TOF MS Profiling
The earliest widely applied method for profiling GAG oligosaccharides was developed in the Biemann laboratory in the 1990s (36, 37). Those researchers reported that heparin oligosaccharides with losses of sulfate group equivalents were observed via MALDI-TOF MS. Such sulfate equivalent losses were prevented when the oligosaccharides were paired with a basic peptide of sequence (RG)n. This method was applied for the sequencing of heparin and HS oligosaccharides, with masses determined before and after chemical and enzymatic digestion steps (38). This approach requires a degree of trial and error in order to get the correct ratio of peptides and oligosaccharides for a strong MALDI-TOF MS signal. As a result, several groups have worked to develop matrices that minimize the extent to which losses of sulfate equivalents occur (39–42). In particular, ionic liquids appear to reduce the extent of losses of sulfate equivalents from highly sulfated HS/heparin oligosaccharides. Other authors have used MALDI-TOF MS to determine the masses of oligosaccharides produced by means of limited nitrous acid depolymerization. Bultel et al. minimized the extent of sulfate losses by adding sodium to the matrix solution (43). Although a substantial degree of dissociation occurred, it was possible to observe apparently intact molecular ions. Infrared MALDI is considered softer than UV-MALDI and has been used in the analysis of HS and CS disaccharides (44). The results showed that disaccharides were detected as sodium adducts in negative mode with minimal losses of sulfate equivalents. It was also possible to obtain IR MALDI-TOF mass spectra of a heparin tetramer with five or six sulfate groups without losses of sulfate equivalents; the results were very similar to the profile obtained using electrospray ionization (ESI) MS in terms of the abundance patterns of oligosaccharide ions.
Recently, MALDI-TOF MS has been used to determine the relative quantities of CS from disaccharides enzymatically released from dermal fibroblasts (45–47). This approach has the advantages of simplicity and high throughput. Disaccharides generated from mammalian CS contain predominantly one sulfate group per disaccharide unit. The observation of a singly charged negative ion does allow direct detection of this ion, as well as a variety of salt adducts. It is not clear from the data presented to what extent dissociation of the sulfate groups occurs.
ESI MS
ESI MS results in relatively low vibrational excitation of analytes during the ionization process and is capable of producing ions from highly sulfated GAG oligosaccharides with minimal losses of sulfate (48). Readers wishing to analyze GAGs or other fragile biological molecules using ESI MS are advised to adjust the mass spectrometer source and ion transfer optics appropriately. In many cases, instrument-tuning parameters appropriate for small molecules or peptides will result in the in-source dissociation of fragile ions. It is recommended that a highly sulfated molecule such as Arixtra be used as a performance standard for this purpose. In some cases, it might be necessary to get input from the instrument vendor in order to tune the instrument properly for fragile ions.
The most straightforward way to analyze GAGs using ESI is as deprotonated ions in the negative mode. This approach leverages the tendency of these acidic compounds to form negative ions in aqueous solutions, even at acidic pH values. In order to quantify GAG oligosaccharides, however, a separation step is strongly recommended. The separation step removes matrix components such as salts and surfactants that render direct analysis using either MALDI or ESI susceptible to fluctuations in ion abundances for which compensation is difficult to achieve.
LC/MS
The use of a chromatography dimension for GAG analysis removes background from the biological matrix, enabling quantitative comparisons among different samples. Several chromatography systems have been used for this purpose. Among the most widely used, reversed-phase ion pairing allows the use of reversed-phase chromatography columns for the separation of GAG oligosaccharides (49, 50). There is high resolution of the oligosaccharides in the chromatographic dimension using this method. Reversed-phase ion pairing LC/MS is most appropriate for instruments dedicated to GAG analysis owing to the need to infuse mobile phases containing millimolar quantities of amine compounds. Porous graphitized carbon LC/MS has been applied to GAGs (51, 52). This approach appears most suited to the analysis of GAGs, such as CS, containing no more than one sulfate group per disaccharide unit. More highly sulfated oligosaccharides are likely to irreversibly bind to the stationary phase. Hydrophilic interaction chromatography MS has been used to profile both CS and HS/heparin oligosaccharides (14, 53–56). The hydrophilic interaction chromatography dimension separates oligosaccharides according to size and polarity. The mobile phase includes volatile ammonium formate and is compatible with extended, robust LC/MS operation.
Tandem MS
Collisionally Activated Dissociation Tandem MS
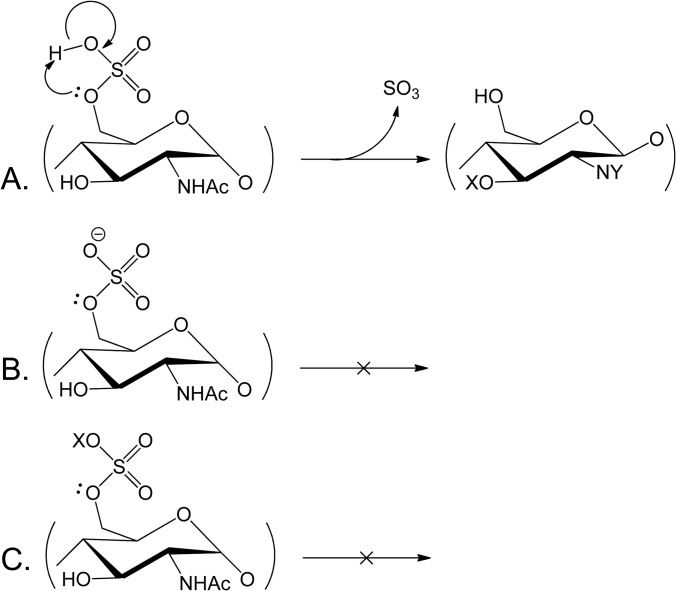
The effort to sequence highly sulfated GAGs using tandem MS is complicated by the tendency of sulfate groups under some conditions to undergo dissociation in response to a relatively low degree of vibrational excitation. As shown in Fig. 3A, protonated sulfate groups dissociate easily to lose SO3, a process that may be observed in the source or during collisional dissociation (48). The extent of such losses is considerably decreased for deprotonated sulfate groups (Fig. 3B) (57). Thus, the extent of losses of sulfate group equivalents diminishes with increased negative charge density. Charge–charge repulsion increases, however, ultimately limit the achievable charge density. Metal cations may be used to pair with sulfate anions, resulting in a stabilizing effect that increases the relative abundances of glycosidic bond cleavages (Fig. 3C) (57–59).
Fig. 3.
The tendency for the loss of sulfate group equivalents during vibrational excitation depends on the charge and the nature of the ion pair. A, protonated sulfate groups are readily lost as SO3 during vibrational excitation. B, deprotonated sulfate groups are much more stable. Thus, the higher the density of negative charge, the lower the extent of SO3 loss, and the greater the abundances of ions from glycosidic bond dissociation. C, pairing of a metal cation (X) also stabilizes sulfate groups and enables glycosidic bond cleavage.
These observations are consistent with the conclusion that the density of free protons in the sulfated oligosaccharide ion determines the extent of deleterious SO3 losses that occur during a collisional excitation experiment (60). This density can conveniently be expressed as a free proton index (FPI):
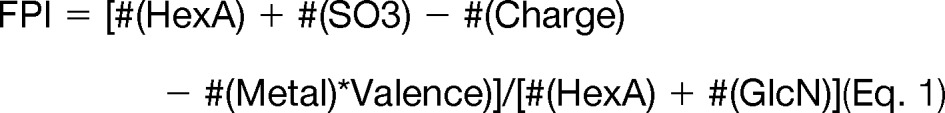 |
Thus, as the charge state increases, the FPI decreases. Generally speaking, an ion with an FPI < 0.5 will produce abundant ions from glycosidic bond and cross-ring cleavage with low abundances of ions from losses of SO3. The extent of SO3 losses increases to the extent that FPI > 0.5.
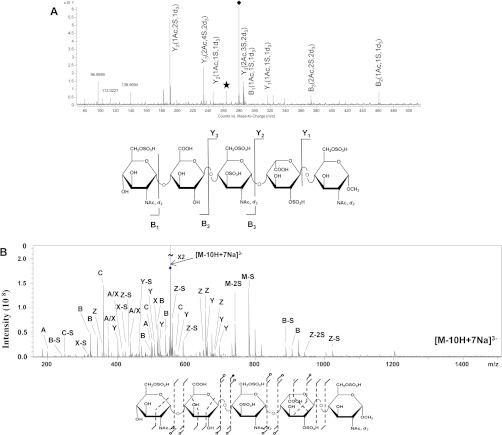
Two approaches for manipulating the FPI for highly sulfated HS/heparin oligosaccharides in order to enable effective collisionally activated dissociation (CAD) tandem MS are summarized in Fig. 4. The target compound is a synthetic octasulfated pentasaccharide that readily undergoes losses of SO3 when subjected to collisional dissociation as a deprotonated ion. In the first approach (Fig. 4A), N-sulfate groups were chemically replaced by N-d3-acetate, and the density of charge on the precursor ions was maximized using sulfolane (60). When using on-line LC-tandem MS with collisional dissociation, the abundances of losses of SO3 from the precursor ion were low relative to those from backbone dissociation. Cleavage was observed to every glycosidic bond, enabling a determination of the numbers of sulfate and acetate groups on every monosaccharide. The second approach (Fig. 4B) involved static infusion of the synthetic octasulfated pentasaccharide in the presence of sodium salts (59). Tandem MS of the 3− ion with 7 sodium adducts resulted in a greater variety of product ions than in Fig. 4A, including a complete series of glycosidic bond cleavages. Cross-ring cleavage ions were observed across three of the monosaccharide residues. The increased structural information relative to Fig. 4A is offset by the need to add salts to the spray solution. A third option, that of electron detachment dissociation, has been demonstrated for GAG oligosaccharides (61–63), but not those having the same density of sulfate groups as the octasulfated pentasaccharide shown in Fig. 4. Permethylation of GAGs followed by chemical de-sulfation and re-acetylation has been explored as a means of replacing sulfate groups with chemically stable reporter groups (64). This approach has been demonstrated for CS saccharides and the resulting derivatives amenable to reversed-phase LC-tandem MS.
Fig. 4.
Comparison of methods for CAD tandem MS of highly sulfated HS/heparin oligosaccharides. A, CAD tandem MS of the octasulfated pentasaccharide Arixtra, subjected to the replacement of N-sulfate groups with d3-N-acetate groups as a 4− ion. Ions were supercharged with sulfolane (60). The data were acquired using an Agilent 6520 QTOF instrument. The ion labeled with a diamond is the precursor, and that with a star resulted from the loss of SO3 from the precursor. Product ions are labeled using the widely accepted Domon–Costello nomenclature (65). B, CAD tandem MS of a synthetic octasulfated hexasaccharide adducted with 7 equivalents of Na as a 3− ion (59). Data were acquired using a Fourier transform ion cyclotron resonance mass spectrometer. The CAD product ion is indicated with a slash (/), open circles indicate SO3 loss, and solid circles indicate the loss of two or more equivalents of SO3.
CONCLUSIONS
The field is moving toward analytical platforms for GAG analysis that inform bioinformatics modeling of structure–function relationships and chemo/enzymatic synthesis efforts. As of this writing, there are effective methods for MS-based disaccharide analysis that are being adopted by groups around the world. We can expect to see analytical platforms capable of the throughput necessary for the effective use of combinations of disaccharide analyses from polysaccharide lyase and deaminative cleavage in conjunction with algorithmic modeling of chain structure in the near future. The profiling of GAG oligosaccharides has progressed to the point that robust and sensitive LC/MS methods are available. As with any glycomics project, glycomics methods are most likely to be successful when a suitable block of instrument time is set aside for that purpose. For tandem MS, it is necessary to decrease the FPI of highly sulfated GAG saccharides in order to produce backbone dissociation with a minimum of sulfate losses. A decrease in the FPI can be accomplished via a combination of chemical derivatization, supercharging, and metal cation adduction.
Footnotes
* The author acknowledges NIH Grant Nos. P41GM104603 and R01HL098950.
1 The abbreviations used are:
- CAD
- collisionally activated dissociation
- CS
- chondroitin sulfate
- ESI
- electrospray ionization
- FPI
- free proton index
- GAG
- glycosaminoglycan
- HS
- heparan sulfate
- MALDI-TOF
- matrix-assisted laser desorption/ionization time-of-flight
- MS
- mass spectrometry.
REFERENCES
- 1. Varki A. (2011) Evolutionary forces shaping the Golgi glycosylation machinery: why cell surface glycans are universal to living cells. Cold Spring Harb. Perspect. Biol. 3:a005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeAngelis P. L. (2002) Evolution of glycosaminoglycans and their glycosyltransferases: implications for the extracellular matrices of animals and the capsules of pathogenic bacteria. Anat. Rec. 268, 317–326 [DOI] [PubMed] [Google Scholar]
- 3. Bishop J. R., Schuksz M., Esko J. D. (2007) Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 4. Bulow H. E., Hobert O. (2006) The molecular diversity of glycosaminoglycans shapes animal development. Annu. Rev. Cell Dev. Biol. 22, 375–407 [DOI] [PubMed] [Google Scholar]
- 5. Chu C. L., Goerges A. L., Nugent M. A. (2005) Identification of common and specific growth factor binding sites in heparan sulfate proteoglycans. Biochemistry 44, 12203–12213 [DOI] [PubMed] [Google Scholar]
- 6. Lindahl U. (2007) Heparan sulfate-protein interactions—a concept for drug design? Thromb. Haemost. 98, 109–115 [PubMed] [Google Scholar]
- 7. Perrimon N., Bernfield M. (2000) Specificities of heparan sulphate proteoglycans in developmental processes. Nature 404, 725–728 [DOI] [PubMed] [Google Scholar]
- 8. Dhoot G. K., Gustafsson M. K., Ai X., Sun W., Standiford D. M., Emerson C. P., Jr. (2001) Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science 293, 1663–1666 [DOI] [PubMed] [Google Scholar]
- 9. Uchimura K., Morimoto-Tomita M., Bistrup A., Li J., Lyon M., Gallagher J., Werb Z., Rosen S. D. (2006) HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 7, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vlodavsky I., Beckhove P., Lerner I., Pisano C., Meirovitz A., Ilan N., Elkin M. (2011) Significance of Heparanase in Cancer and Inflammation. Cancer Microenviron. 5, 115–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Docket No. FDA-2003-P-0273, July 23, 2010 [Google Scholar]
- 12. Spencer J. L., Bernanke J. A., Buczek-Thomas J. A., Nugent M. A. (2010) A computational approach for deciphering the organization of glycosaminoglycans. PLoS ONE 5, e9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Naimy H., Buczek-Thomas J. A., Nugent M. A., Leymarie N., Zaia J. (2011) Highly sulfated non reducing end-derived heparan sulfate domains bind fibroblast growth factor-2 with high affinity and are enriched in biologically active fractions. J. Biol. Chem. 286, 19311–19319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Naimy H., Leymarie N., Bowman M., Zaia J. (2008) Characterization of heparin oligosaccharides binding specifically to antithrombin III using mass spectrometry. Biochemistry 47, 3155–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Naimy H., Leymarie N., Zaia J. (2010) Screening for anticoagulant heparan sulfate octasaccharides and fine structure characterization using tandem mass spectrometry. Biochemistry 49, 3743–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ernst S., Langer R., Cooney C. L., Sasisekharan R. (1995) Enzymatic degradation of glycosaminoglycans. Crit. Rev. Biochem. Mol. Biol. 30, 387–444 [DOI] [PubMed] [Google Scholar]
- 17. Conrad H. E. (2001) Degradation of heparan sulfate by nitrous acid. Methods Mol. Biol. 171, 347–351 [DOI] [PubMed] [Google Scholar]
- 18. Shi X., Zaia J. (2009) Organ-specific heparan sulfate structural phenotypes. J. Biol. Chem. 284, 11806–11814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Skidmore M. A., Guimond S. E., Dumax-Vorzet A. F., Atrih A., Yates E. A., Turnbull J. E. (2006) High sensitivity separation and detection of heparan sulfate disaccharides. J. Chromatogr. A 1135, 52–56 [DOI] [PubMed] [Google Scholar]
- 20. Kinoshita A., Sugahara K. (1999) Microanalysis of glycosaminoglycan-derived oligosaccharides labeled with a fluorophore 2-aminobenzamide by high-performance liquid chromatography: application to disaccharide composition analysis and exosequencing of oligosaccharides. Anal. Biochem. 269, 367–378 [DOI] [PubMed] [Google Scholar]
- 21. Calabro A., Midura R., Wang A., West L., Plaas A., Hascall V. C. (2001) Fluorophore-assisted carbohydrate electrophoresis (FACE) of glycosaminoglycans. Osteoarthritis Cartilage 9 Suppl A, S16–S22 [DOI] [PubMed] [Google Scholar]
- 22. Calabro A., Hascall V. C., Midura R. J. (2000) Adaptation of FACE methodology for microanalysis of total hyaluronan and chondroitin sulfate composition from cartilage. Glycobiology 10, 283–293 [DOI] [PubMed] [Google Scholar]
- 23. Toyoda H., Motoki K., Tanikawa M., Shinomiya K., Akiyama H., Imanari T. (1991) Determination of human urinary hyaluronic acid, chondroitin sulphate and dermatan sulphate as their unsaturated disaccharides by high-performance liquid chromatography. J. Chromatogr. 565, 141–148 [DOI] [PubMed] [Google Scholar]
- 24. Toyoda H., Kinoshita-Toyoda A., Fox B., Selleck S. B. (2000) Structural analysis of glycosaminoglycans in animals bearing mutations in sugarless, sulfateless, and tout-velu. Drosophila homologues of vertebrate genes encoding glycosaminoglycan biosynthetic enzymes. J. Biol. Chem. 275, 21856–21861 [DOI] [PubMed] [Google Scholar]
- 25. Kariya Y., Yoshida K., Morikawa K., Tawada A., Miyazono H., Kikuchi H., Tokuyasu K. (1992) Preparation of unsaturated disaccharides by eliminative cleavage of heparin and heparan sulfate with heparitinases. Comp. Biochem. Physiol. B 103, 473–479 [DOI] [PubMed] [Google Scholar]
- 26. Desaire H., Leary J. (2000) Detection and quantification of the sulfated disaccharides in chondroitin sulfate by electrospray tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 11, 916–920 [DOI] [PubMed] [Google Scholar]
- 27. Saad O. M., Leary J. A. (2003) Compositional analysis and quantification of heparin and heparan sulfate by electrospray ionization ion trap mass spectrometry. Anal. Chem. 75, 2985–2995 [DOI] [PubMed] [Google Scholar]
- 28. Behr J. R., Matsumoto Y., White F. M., Sasisekharan R. (2005) Quantification of isomers from a mixture of twelve heparin and heparan sulfate disaccharides using tandem mass spectrometry. Rapid Commun. Mass Spectrom. 19, 2553–2562 [DOI] [PubMed] [Google Scholar]
- 29. Mason K. E., Meikle P. J., Hopwood J. J., Fuller M. (2006) Characterization of sulfated oligosaccharides in mucopolysaccharidosis type IIIA by electrospray ionization mass spectrometry. Anal. Chem. 78, 4534–4542 [DOI] [PubMed] [Google Scholar]
- 30. Ramsay S. L., Meikle P. J., Hopwood J. J., Clements P. R. (2005) Profiling oligosaccharidurias by electrospray tandem mass spectrometry: quantifying reducing oligosaccharides. Anal. Biochem. 345, 30–46 [DOI] [PubMed] [Google Scholar]
- 31. Ramsay S. L., Meikle P. J., Hopwood J. J. (2003) Determination of monosaccharides and disaccharides in mucopolysaccharidoses patients by electrospray ionisation mass spectrometry. Mol. Genet. Metab. 78, 193–204 [DOI] [PubMed] [Google Scholar]
- 32. Lawrence R., Olson S. K., Steele R. E., Wang L., Warrior R., Cummings R. D., Esko J. D. (2008) Evolutionary differences in glycosaminoglycan fine structure detected by quantitative glycan reductive isotope labeling. J. Biol. Chem. 283, 33674–33684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xue B., Alves S., Desbans C., Souchaud M., Filali-Ansary A., Soubayrol P., Tabet J. C. (2011) Heparin-like glycosaminoglycan/amine salt-bridge interactions: a new potential tool for HLGAGs analysis using mass spectrometry. J. Mass Spectrom. 46, 689–695 [DOI] [PubMed] [Google Scholar]
- 34. Zhang Z., Xie J., Liu H., Liu J., Linhardt R. J. (2009) Quantification of heparan sulfate disaccharides using ion-pairing reversed-phase microflow high-performance liquid chromatography with electrospray ionization trap mass spectrometry. Anal. Chem. 81, 4349–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gill V. L., Wang Q., Shi X., Zaia J. (2012) Mass spectrometric method for determining the uronic acid epimerization in heparan sulfate disaccharides generated using nitrous acid. Anal. Chem. 84, 7539–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Juhasz P., Biemann K. (1994) Mass spectrometric molecular-weight determination of highly acidic compounds of biological significance via their complexes with basic polypeptides. Proc. Natl. Acad. Sci. U.S.A. 91, 4333–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Juhasz P., Biemann K. (1995) Utility of non-covalent complexes in the matrix-assisted laser desorption ionization mass spectrometry of heparin-derived oligosaccharides. Carbohydr. Res. 270, 131–147 [DOI] [PubMed] [Google Scholar]
- 38. Venkataraman G., Shriver Z., Raman R., Sasisekharan R. (1999) Sequencing complex polysaccharides. Science 286, 537–542 [DOI] [PubMed] [Google Scholar]
- 39. Laremore T. N., Zhang F., Linhardt R. J. (2007) Ionic liquid matrix for direct UV-MALDI-TOF-MS analysis of dermatan sulfate and chondroitin sulfate oligosaccharides. Anal. Chem. 79, 1604–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Laremore T. N., Linhardt R. J. (2007) Improved matrix-assisted laser desorption/ionization mass spectrometric detection of glycosaminoglycan disaccharides as cesium salts. Rapid Commun. Mass Spectrom. 21, 1315–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tissot B., Gasiunas N., Powell A. K., Ahmed Y., Zhi Z. L., Haslam S. M., Morris H. R., Turnbull J. E., Gallagher J. T., Dell A. (2007) Towards GAG glycomics: analysis of highly sulfated heparins by MALDI-TOF mass spectrometry. Glycobiology 17, 972–982 [DOI] [PubMed] [Google Scholar]
- 42. Przybylski C., Gonnet F., Bonnaffe D., Hersant Y., Lortat-Jacob H., Daniel R. (2010) HABA-based ionic liquid matrices for UV-MALDI-MS analysis of heparin and heparan sulfate oligosaccharides. Glycobiology 20, 224–234 [DOI] [PubMed] [Google Scholar]
- 43. Bultel L., Landoni M., Grand E., Couto A. S., Kovensky J. (2010) UV-MALDI-TOF mass spectrometry analysis of heparin oligosaccharides obtained by nitrous acid controlled degradation and high performance anion exchange chromatography. J. Am. Soc. Mass Spectrom. 21, 178–190 [DOI] [PubMed] [Google Scholar]
- 44. Tajiri M., Wada Y. (2011) Infrared matrix-assisted laser desorption/ionization mass spectrometry for quantification of glycosaminoglycans and gangliosides. Int. J. Mass Spectrom. 305, 164–169 [Google Scholar]
- 45. Bohme J., Anderegg U., Nimptsch A., Nimptsch K., Hacker M., Schulz-Siegmund M., Huster D., Schiller J. (2012) De novo biosynthesis of glycosaminoglycans in the extracellular matrix of skin studied by matrix-assisted laser desorption/ionization mass spectrometry. Anal. Biochem. 421, 791–793 [DOI] [PubMed] [Google Scholar]
- 46. Nimptsch A., Schibur S., Schnabelrauch M., Fuchs B., Huster D., Schiller J. (2009) Characterization of the quantitative relationship between signal-to-noise (S/N) ratio and sample amount on-target by MALDI-TOF MS: determination of chondroitin sulfate subsequent to enzymatic digestion. Anal. Chim. Acta 635, 175–182 [DOI] [PubMed] [Google Scholar]
- 47. Schiller J., Arnhold J., Benard S., Reichl S., Arnold K. (1999) Cartilage degradation by hyaluronate lyase and chondroitin ABC lyase: a MALDI-TOF mass spectrometric study. Carbohydr. Res. 318, 116–122 [DOI] [PubMed] [Google Scholar]
- 48. Naggar E. F., Costello C. E., Zaia J. (2004) Competing fragmentation processes in tandem mass spectra of heparin-like glycosaminoglycans. J. Am. Soc. Mass Spectrom. 15, 1534–1544 [DOI] [PubMed] [Google Scholar]
- 49. Kuberan B., Lech M., Zhang L., Wu Z. L., Beeler D. L., Rosenberg R. (2002) Analysis of heparan sulfate oligosaccharides with ion pair-reverse phase capillary high performance liquid chromatography-microelectrospray ionization time-of-flight mass spectrometry. J. Am. Chem. Soc. 124, 8707–8718 [DOI] [PubMed] [Google Scholar]
- 50. Thanawiroon C., Linhardt R. J. (2003) Separation of a complex mixture of heparin-derived oligosaccharides using reversed-phase high-performance liquid chromatography. J. Chromatogr. A 1014, 215–223 [DOI] [PubMed] [Google Scholar]
- 51. Estrella R. P., Whitelock J. M., Packer N. H., Karlsson N. G. (2007) Graphitized carbon LC-MS characterization of the chondroitin sulfate oligosaccharides of aggrecan. Anal. Chem. 79, 3597–3606 [DOI] [PubMed] [Google Scholar]
- 52. Karlsson N. G., Schulz B. L., Packer N. H., Whitelock J. M. (2005) Use of graphitised carbon negative ion LC-MS to analyse enzymatically digested glycosaminoglycans. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 824, 139–147 [DOI] [PubMed] [Google Scholar]
- 53. Staples G. O., Naimy H., Yin H., Kileen K., Kraiczek K., Costello C. E., Zaia J. (2010) Improved hydrophilic interaction chromatography LC/MS of heparinoids using a chip with postcolumn makeup flow. Anal. Chem. 82, 516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Staples G., Bowman M., Costello C. E., Hitchcock A., Lau J., Leymarie N., Miller C., Naimy H., Shi X., Zaia J. (2009) A chip-based amide-HILIC LC/MS platform for glycosaminoglycan glycomics. Proteomics 9, 686–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hitchcock A., Yates K. E., Costello C., Zaia J. (2008) Comparative glycomics of connective tissue glycosaminoglycans. Proteomics 8, 1384–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li L., Zhang F., Zaia J., Linhardt R. J. (2012) Top-down approach for the direct characterization of low molecular weight heparins using LC-FT-MS. Anal. Chem. 84, 8822–8829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zaia J., Costello C. E. (2003) Tandem mass spectrometry of sulfated heparin-like glycosaminoglycan oligosaccharides. Anal. Chem. 75, 2445–2455 [DOI] [PubMed] [Google Scholar]
- 58. Wolff J. J., Laremore T. N., Busch A. M., Linhardt R. J., Amster I. J. (2008) Influence of charge state and sodium cationization on the electron detachment dissociation and infrared multiphoton dissociation of glycosaminoglycan oligosaccharides. J. Am. Soc. Mass Spectrom. 19, 790–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kailemia M. J., Li L., Ly M., Linhardt R. J., Amster I. J. (2012) Complete mass spectral characterization of a synthetic ultralow-molecular-weight heparin using collision-induced dissociation. Anal. Chem. 84, 5475–5478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shi X., Huang Y., Mao Y., Naimy H., Zaia J. (2012) Tandem mass spectrometry of heparan sulfate negative ions: sulfate loss patterns and chemical modification methods for improvement of product ion profiles. J. Am. Soc. Mass Spectrom. 23, 1498–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wolff J. J., Laremore T. N., Busch A. M., Linhardt R. J., Amster I. J. (2008) Electron detachment dissociation of dermatan sulfate oligosaccharides. J. Am. Soc. Mass Spectrom. 19, 294–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wolff J. J., Chi L., Linhardt R. J., Amster I. J. (2007) Distinguishing glucuronic from iduronic acid in glycosaminoglycan tetrasaccharides by using electron detachment dissociation. Anal. Chem. 79, 2015–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wolff J. J., Amster I. J., Chi L., Linhardt R. J. (2007) Electron detachment dissociation of glycosaminoglycan tetrasaccharides. J. Am. Soc. Mass Spectrom. 18, 234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Huang R., Pomin V., Sharp J. (2011) LC/MSn Analysis of Isomeric Chondroitin Sulfate Oligosaccharides Using a Chemical Derivatization Strategy. J. Am. Soc. Mass Spectrom. 22, 1577–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Domon B., Costello C. E. (1988) A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate J. 5, 397–409 [Google Scholar]