Abstract
The Bcl2 pro-survival protein family has long been recognized for its important contributions to cancer. At elevated levels relative to pro-apoptotic effector members, the survival proteins prevent cancer cells from initiating apoptosis in the face of many intrinsic tumour-suppressing pathways and extrinsic therapeutic treatments aimed at controlling tumorigenesis. Recent studies, including genome-wide analyses, have begun to focus attention on a particularly enigmatic member of the family—myeloid cell leukaemia 1 (Mcl1). For reasons that are not clear, Mcl1 in cancer cells is turned over rapidly, eliminated primarily through the ubiquitin–proteasome pathway. Moreover, the mechanistic aspects of this constitutive membrane-associated protein have not been fully elucidated. As the pro-cancer activity of Mcl1 requires elevated expression levels of the protein, the cancer genome adapts to ensure either high levels of synthesis or evasion of degradation, or both. Here, we focus on the complex strategies at play and their therapeutic implications.
Keywords: apoptosis, mitochondria, Bcl2, caspase, proteasome
See Glossary for abbreviations used in this article.
Glossary.
- Abl
Abelson murine leukaemia viral oncogene homologue
- ABT
Abbott
- APC/C
anaphase-promoting complex/cyclosome
- ATF5
activating transcription factor 5
- ATM
ataxia telangiectasia mutated
- Bad
Bcl2-associated agonist of cell death
- Bak
Bcl2 antagonist killer
- Bax
Bcl2-associated protein X
- Bcl2
B-cell lymphoma 2
- Bcl-w
Bcl2-like 2
- Bcl-b
Bcl2-like 10
- Bcl-XL
Bcl2-like protein XL
- Bcr
breakpoint cluster region
- βTRCP
β-transducin-containing protein
- Bfl-1/A1
Bcl2-related protein A1
- Bik
Bcl2-interacting killer
- Bim
Bcl2-like 11
- BimEL
Bim extra long
- Bmf
Bcl2-modifying factor
- Bok
Bcl2-related ovarian killer
- Cdc20
cell division cycle 20 homologue
- Cdk
cyclin-dependent kinase
- c-Jun
jun proto-oncogene
- c-Myc
v-myc myelocytomatosis viral oncogene homologue
- CREB3L2
cAMP response element-binding protein-3-like 2
- DRP1
dynamin-related protein 1
- E2F1
E2F transcription factor 1
- eIF4
eukaryotic translation initiation factor 4
- Erk
extracellular regulated protein kinase
- Fbw7
F-box and WD repeat domain containing 7
- Gsk3
glycogen synthase kinase 3
- HECT
homologous to the E6-AP carboxyl terminus
- Hrk
harakiri, Bcl2-interacting protein
- IL-6/7
interleukin 6/7
- Jak
janus kinase
- Jnk
c-jun N-terminal kinase
- miR29/miR-125b
microRNA 125b
- mTorc1
mammalian target of rapamycin complex 1
- Mule
Mcl1 ubiquitin ligase
- Myc
v-myc myelocytomatosis viral oncogene homologue (avian)
- NFκB
nuclear factor kappa B
- PEST
proline/glutamic acid/serine/threonines
- Pin1
peptidyl-prolyl cis-trans isomerase NIMA interacting protein 1
- Pink1
PTEN-induced putative kinase 1
- Puma
p53 upregulated modulator of apoptosis
- RING
really interesting new gene
- Stat
signal transducers and activators of transcription
- tBid
truncated BH3-interacting domain death agonist
- TIM
translocase of the inner membrane of mitochondria
- TOM
translocase of the outer membrane of mitochondria
- TPA
12-0-tetradecanoylphorbol 13-acetate
- USP9X
ubiquitin-specific peptidase 9, X-linked
Introduction
Evasion of apoptosis continues to be recognized as one of the fundamental characteristics of cancer [1], driven primarily by upregulation from pro-survival members of the Bcl2 family, especially Bcl2, Bcl-XL and myeloid cell leukaemia 1 (Mcl1; [2]). In particular, genome-wide studies have identified Mcl1 as subject to increased gene copy number across many solid and haematological malignancies, and it has emerged as a recognized pathological factor contributing to diverse indications [3]. Moreover, many mutational, epigenetic and signal transduction alterations associated with transformation have the potential to influence Mcl1 expression and function, implying that gene copy number represents only one of many strategies to increase the functional impact of Mcl1. For example, chemical genomics has identified many routes to influence Mcl1 expression through direct or indirect transcriptional regulation [4], arguing that cancer cells have a vast arsenal to exploit changes in Mcl1 levels to escape apoptosis. Moreover, as with other Bcl2 family proteins, Mcl1 has functions that extend beyond apoptosis regulation, yet little is known about their contributions to malignancy or resistance to therapies. In this brief review, we attempt to articulate progress in understanding the complex biology associated with Mcl1, the many mechanisms available for reprogramming Mcl1 expression by cancer cells and how this knowledge can be exploited for potential therapeutic benefit to cancer patients. For an excellent overview of Mcl1 see reference [5].
Myeloid leukaemia cell 1 (MCL1) was discovered by Ruth Craig and colleagues. It is an immediate-early gene, which is activated during TPA-induced myeloid cell differentiation in vitro, and contributes both to cell viability and to regulation of cell proliferation through its linkage to many signal transduction networks [6,7]. Consistent with the expression of Mcl1 across many cancers, the protein is widely distributed in many tissues [8]. In contrast to other Bcl2 survival members, however, germline deletion resulted in peri-implantation lethality during mouse embryogenesis by an as yet undetermined mechanism [9]. Targeted gene deletion in the mouse, on the other hand, indicated Mcl1-dependent survival of many cell lineages including neutrophils [10], lymphocytes [11], haematopoietic stem cells [12], neurons [13], hepatocytes [14] and immunoglobulin-secreting plasma cells [15]. Whether this unique dependence on Mcl1 for survival reflects an unsustainable reduction in the survival threshold conferred by the cohort of redundant Bcl2 proteins expressed in these cells, or reflects a more Mcl1-specific function or cell context-specific regulation of Mcl1, remains to be determined. In the megakaryocyte lineage, for example, loss of both Mcl1 and Bcl-XL has a more profound effect than deletion of either one individually [16,17]. Targeted transgenic expression has confirmed the contribution of Mcl1 to malignancies in several mouse models. For example, overexpression within haematopoietic and lymphoid tissues resulted in B-cell lymphomas over time [18,19,20]. The pathogenesis of acute myeloid leukaemia can also be recapitulated in a murine model, in which Mcl1 expression was required and the disease limited either by Mcl1 deletion [21] or haploinsufficiency [22]. In the solid tumour setting, mouse models of lung adenocarcinoma, in which Myc was targeted to pulmonary alveolar cells, depended on Mcl1 overexpression to circumvent Myc-induced apoptosis [23]. These studies, coupled with numerous examples of correlations between Mcl1 status and cancer cell survival and patient outcomes, drive an intense need to understand—and exploit—Mcl1 biology [24,25,26].
The Bcl2 family is defined by the presence of Bcl2 homology (BH) domains and comprises three protein subgroups of which Mcl1, together with Bcl2, Bcl-XL, Bcl-b, Bfl-1/A1 and Bcl-w, confer cell survival. By contrast, Bax and Bak, together with the tissue-restricted Bok, represent the effectors of apoptosis. The group of proteins that contain a single Bcl2 homology domain (BH3) activate Bax and Bak (in the case of tBid, Puma and Bim) and inhibit the pro-survival members (tBid, Puma and Bim together with the sensitizing BH3-only proteins Bad, Bik, Noxa, Hrk and Bmf [27]). With the first discovery of the existence of a Bcl2-related protein, Bax, which interacted with Bcl2 but showed pro-apoptotic rather than anti-apoptotic activity [28], two important principles related to the Bcl2 family have emerged. First, protein–protein interactions that take place between pro-apoptotic and anti-apoptotic members involve docking of the pro-apoptotic amphiphilic BH3 helix into an opposing groove formed by the BH1 and BH2 helices in the pro-survival members, and are exclusively binary interactions; second, it is the functional ratio of the interacting pro-apoptotic and anti-apoptotic members through these interactions that determines whether or not the effector proteins Bax and Bak can initiate apoptosis induction in the face of cell stress stimuli [29]. Cells, including most cancer cells, are considered to be ‘primed’ for cell death if they depend on the Bcl2 family to confer cell survival in the face of extant stress-signalling pathways—for example, oncogenic drivers such as Myc. Otherwise, in unprimed cells, the Bcl2 family status determines cell survival or death only in the face of externally applied stresses. In the case of primed cancer cells, one goal is to overcome both the Bcl2-mediated resistance to intrinsic oncogenic signalling as well as the Bcl2-mediated resistance to therapy. Stress signals ultimately converge on the Bax and Bak effectors through activation of BH3-only proteins, which, because of their multiplicity and differential preferences for binding to individual survival Bcl2 proteins [30], allows differential integration into many signalling pathways.
All three subgroups of the Bcl2 family reside at the endoplasmic reticulum in which, among other functions, they influence calcium homeostasis, calcium transmission to mitochondria, and autophagy [31]. However, by far the best understood roles of this family relate to their regulation of mitochondrial outer membrane permeabilization (MOMP), release of pro-apoptotic factors including cytochrome c from the intermembrane space and activation of caspases [32,33,34]. At least for the initiation of this process—defined as the formation of Bax or Bak oligomers within the outer membrane bilayer, and its prevention by pro-survival members—most of the research has focused on the tBid or Bim BH3-only/Bax/Bcl-XL axis. A combination of high-resolution structure determination of Bax activation intermediates, reconstitution in liposomal systems of defined composition and genetically determined models in cells and animals has led to a fairly well-defined model [35,36,37,38,39]. Although it remains to be determined to what extent the Bcl-XL model is a reflection of Mcl1 activity, similar approaches need to be applied in this case. Activator BH3-only protein is proposed to bind to the trigger site on membrane-free Bax, inducing a conformational change in which the carboxy-terminal transmembrane segment becomes available to mediate membrane insertion. At the same time, the amino-terminus is repositioned, allowing exposure of the BH3 domain of Bax. As the latter can also contact the trigger site on naive Bax, a cascade is set up in which structurally altered Bax monomers within the lipid bilayer can interact through discrete domains (as shown for Bak, [40]) to form oligomers. In reconstituted liposomes, such oligomers breach the lipid bilayer allowing entrapped dyes to be released. Membrane-free Bcl-XL, on the other hand, can interact with the exposed BH3 domains of activator BH3-only proteins or of activated Bax and, as with Bax, it also undergoes conformational alterations and membrane penetration, preventing Bax oligomerization (based on Bcl2 [41]). Intriguingly, Bcl-XL also seems to help cycle membrane-associated Bax back to the cytosol [42]. The model is consistent with the final outcome being determined by the functional ratio of Bcl-XL against the exposed BH3 domains on BH3-only proteins and activated Bax. One way in which this apoptotic rheostat can be adjusted is through inhibition of Bcl-XL by sensitizing BH3-only proteins, such as Bad. Indeed, a combination of Bax activation and Bcl-XL inhibition is probably needed in most circumstances to drive MOMP [43]. In cells, however, many BH3-independent interactions involving all three subgroups of the BH3-only/Bax/Bcl-XL axis have the potential to influence the seeding of intramembrane Bax oligomers. In particular, little is known about the transition of Bax oligomers into pore structures that release the contents of the mitochondrial intermembrane space efficiently. The available evidence however, emphasizes a continued relationship between Bax and the lipid bilayer, potentially involving tethering and hemifusion of membranes induced by DRP1, which provides a platform for formation of the requisite pore functionalities to support cytochrome c release [44]. Moreover, the lipid environment, mediated in part by interactions of mitochondria with heterotypic membranes, seems to have a role [45].
Mcl1—unique or uniquely regulated?
Structural and docking studies have elucidated the binding interactions between BH3 peptides and the BH3 binding groove of Mcl1, which account for the differential specificity of this protein for BH3 ligands compared with other pro-survival family members [46,47]. The Bcl2-related structural core of Mcl1 has a high affinity-binding specificity for the pro-apoptotic BH3 peptides of Bak, Bax, Bid, Puma, Bim and Noxa [30,48]. Additionally, Mcl1 contains a large, flexible approximately 170 amino acid N-terminus, which is dispensable for BH3 binding interactions [46] but is replete with sites governing Mcl1 regulation (see below). A corresponding region is absent from other Bcl2 family proteins. In striking contrast to the tBid or Bim BH3-only/Bax/Bcl-XL MOMP axis described above—in which Bax and Bcl-XL are induced to interact with the membrane—the Bak/Mcl1 MOMP axis comprises Bak and Mcl1, which are constitutively integrated into the MOM by their respective C-terminal transmembrane segments before stimulation by tBid or Bim. Targeting of Bak to the MOM is dependent on its transmembrane segment and does not seem to require obvious members of the mitochondrial protein import machinery [49], whereas constitutive import of Mcl1 into the MOM requires its transmembrane segment and is dependent on the import receptor TOM70 [50]. Although not studied to the same molecular resolution as Bax, tBid-induced activation and oligomerization of MOM-anchored Bak resulting in MOMP and cytochrome c release seem to follow the same cascade of structural transformations [38,40,51,52]. Extrapolating from the detailed understanding of the BH3-only/Bax/Bcl-XL MOMP axis, the corresponding BH3-only/Bak/Mcl1 model is depicted in Fig 1.
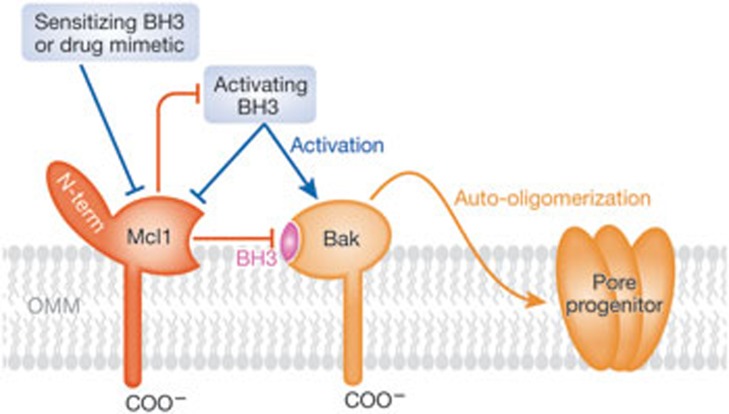
Figure 1.
A model for Mcl1 regulation of Bak-dependent MOMP. As described in the text, Mcl1 and its binding partner Bak seem to have similarities to Bcl-XL and its binding partner Bax, with the exception that Mcl1 and Bak are constitutively anchored to the outer mitochondrial membrane (OMM) by their respective transmembrane segments. Hybrid models involving Mcl1/Bax and Bcl-XL/Bak are also relevant, as these interactions also exist. As in the Bcl-XL model, antagonism of Mcl1 by a sensitizing BH3-only protein (for example, Noxa) or by a chemical mimetic (such as, obatoclax or sabutoclax) can reinstate Bak-driven MOMP. As in the Bcl-XL model, the latter is probably seeded by Bak oligomers (pore progenitor) but requires additional membrane-dependent events to construct pores capable of releasing the protein content of the mitochondrial intermembrane space, including apoptotic regulators such as cytochrome c and Smac. Bak, Bcl2 antagonist killer; Bcl-XL, Bcl2-like protein XL; BH3, Bcl2 homology domain 3; COO−, carboxyl; Mcl1, myeloid cell leukaemia 1; MOMP, mitochondrial outer membrane permeabilization; Smac, second mitochondrial activator of caspase.
Is the ability of Mcl1 to antagonize pro-apoptotic Bcl2 family members a functionally redundant property of this protein compared with other survival members? A study in the murine model of acute myeloid leukaemia revealed that limiting the expression of Mcl1 alone suppressed the disease [21], whereas enforced expression of Bcl2 rescued Myc-driven disease lethality in Mcl1-haploinsufficient cells [22]. Similarly, the development of pro-B cells in mice harbouring a deletion of the transcriptional factor Stat5 was restored by transgenic expression of the pro-survival protein Bcl2, which compensated for loss of Mcl1 [53]. In the Eμ-Myc transgenic mouse model of lymphoma, the tumour-suppressing activity of knocked-in variants of Bim constructs required that Bim must antagonize all pro-survival members [54]. Conversely, all six pro-survival members could individually accelerate a Myc-induced model of murine myeloid leukaemogenesis [55]. Finally, Mcl1 is a resistance factor for ABT-737, a small molecule chemical antagonist of Bcl2, Bcl-XL and Bcl-w; and yet limiting Mcl1 expression over a wide range of cellular and treatment contexts reinstates lethality to the drug in otherwise resistant cells, indicating that Mcl1 can substitute for inhibited Bcl2, Bcl-XL and Bcl-w to confer survival [56,57]. These results strongly favour the idea that limiting the expression of any Bcl2 survival member, resulting in reduction below a minimal survival threshold, results in lethality in primed cells. In this scenario, the apparent dominant role of Mcl1, in so many circumstances, might arise because Mcl1 expression is crucial to keeping the cell above the Bcl2 survival threshold (Fig 2). Moreover, the fact that Mcl1 expression has evolved to be influenced by so many regulatory networks might facilitate a direct control over the Mcl1 dominated Bcl2 survival threshold. There might be other circumstances, however, reflected for example by the embryonic lethality of the Mcl1-deleted mouse [9] or the induction of a robust autophagic response after targeted deletion of Mcl1 in the cortical neurons of transgenic mice [58], in which additional outcomes exist specifically under Mcl1 control. These ideas are explained in Fig 2.
Figure 2.
The Bcl2 family rheostat. There is substantial evidence for functional redundancy between pro-survival members, especially Bcl2, Mcl1 and Bcl-XL (upper rheostat). In this case, reduction of any one member could adjust the pro-survival rheostat below a crucial threshold, resulting in cell death in the face of cell stress signals. Alternatively, there might be situations in which unique pathways are regulated by the individual pro-survival members (lower rheostats). Although the Bcl2 family is most closely associated with apoptosis, there are or might be many other cellular responses or fates that are controlled by these proteins. Of particular relevance to Mcl1, changes to many cellular pathways can result in changes in the levels of Bcl2 family proteins, thereby regulating cell fate in the face of stress stimuli. Bak, Bcl2 antagonist killer; Bax, Bcl2-associated protein; Bcl2, B-cell lymphoma 2; Bcl-XL, Bcl2-like protein XL; Bim, Bcl2-like 11; Mcl1, myeloid cell leukaemia 1; Puma, p53 upregulated modulator of apoptosis; ROS, reactive oxygen species; tBid, truncated BH3-interacting domain death agonist.
N-terminal regulatory domain of Mcl1
In contrast to other pro-survival members, human and mouse Mcl1 (350 aa and 331 aa, respectively) have an extended N-terminal domain (approximately 170 aa and 140 aa, respectively), connected to a highly conserved Bcl2-like core (containing Bcl2 homology domains 1–3 and the C-terminal transmembrane segment). With the exception of the extreme N-terminus, most of the sequence divergence between the two species occurs within the N-terminal regulatory domain. The overall architecture of the N-terminus is shown in Fig 3, and most importantly is characterized by PEST sequence-associated phosphodegrons. Additionally, the extreme N-terminus is associated with processing by the mitochondrion and contains one of two sites that are required for interaction with the E3 ligase Mule (as discussed below).
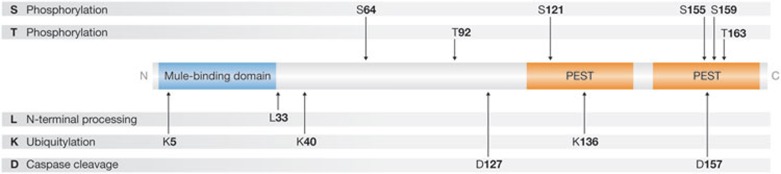
Figure 3.
The amino-terminal regulatory domain of Mcl1. As described in the text, this long (approximately 170 aa), flexible region is unique to Mcl1 among pro-survival Bcl2 proteins and is replete with regulatory sites. In addition to descriptions in the text, discussions about specific sites can be found in reference [5] and below. With the exception of L33 (mouse), amino acid number refers to the human sequence. Ser 64 can be phosphorylated by Cdk1 and Cdk2, and by Jnk1, which does not affect the protein half-life but leads to an enhanced survival function [96]. Erk1 phosphorylates Thr 92 and Thr1 63, which stabilizes Mcl1 through protein association with Pin [97]. Ser 121 in conjunction with Thr 163 is phosphorylated by Jnk, resulting in enhanced stability in one study [98] and unchanged stability but less potency at inhibiting apoptosis in a different study [99]. Ser 155 phosphorylation by Gsk3 decreases Mcl1 expression [69]. Ser 159 phosphorylation leads to Mcl1 destabilization and inhibits interaction with Bim [74], whereas phosphorylation of Thr 163 by Erk increases the half-life of Mcl1 [100]. Bcl2, B-cell lymphoma 2; Bim, Bcl2-like 11; Cdk1/2, cyclin-dependent kinase 1/2; Erk1, extracellular regulated protein kinase 1; Gsk3, glycogen synthase kinase 3; Jnk1, c-jun N-terminal kinase 1; Mcl1, myeloid cell leukaemia 1; PEST, proline/glutamic acid/serine/threonines; Pin, peptidyl-prolyl cis-trans isomerase NIMA interacting protein.
N-terminal processing
Craig and colleagues observed that an N-terminally truncated (approximately 30 aa) form of Mcl1 (Mcl1ΔN) was the predominant species present in lymphomas from transgenic mice and human tumour lines of B-lymphoid origin, and was characterized by enhanced stability and survival features compared with full-length Mcl1 [59]. Remarkably, the truncated region has characteristics of a mitochondrial matrix targeting sequence and requires the mitochondrial electrochemical potential (ΔΨ) across the inner membrane for processing [48,60,61]. On the basis of murine models, one study concluded that Mcl1ΔN resides in the outer membrane and evades interaction with the E3 protein ubiquitin ligase Mule, resulting in enhanced stability [48]. ΔΨ-dependent processing could arise either because the long flexible N-terminal region of outer membrane-anchored Mcl1 reversibly accesses—through the TOM–TIM complexes—processing enzyme(s) at the inner membrane or matrix, or because ΔΨ controls the fate of proteins at the outer membrane. For example, loss of Δψ rescues Pink1 from degradation at the inner membrane by allowing it to relocalize to the outer membrane, at which it can influence mitophagy [62]. By contrast, alternative findings concluded that Mcl1ΔN arises because Mcl1 can be entirely translocated to the inner membrane and/or matrix, in which it is processed after Leu33 [60,61]. Intriguingly, such intra-mitochondrial Mcl1ΔN was proposed to regulate mitochondrial dynamics, ATP production and respiration [61]. It is not known if the proposed locations of the Mcl1ΔN isoform at the outer membrane and inner membrane and/or matrix represent distinct models, or if both can exist dependent on cell context. In either case, strategies that enhance Mcl1 stability or Mcl1-dependent mitochondrial fitness could provide a selective survival advantage for cancer cells.
Strategies to evade Mcl1 degradation
Physiological turnover of full-length Mcl1 is typically rapid and executed by the ubiquitin–proteasome system. Thus, strategies to evade or dampen the rate of degradation and increase the levels of Mcl1 include suppression or loss of Mcl1 E3 ubiquitin ligases, modification of the Mcl1 substrate—for example, processing or phosphodegron status—so that it is no longer targeted by cognate E3 ligases, and enhancement of the levels of Mcl1 deubiquitinases.
The HECT domain E3 ligase Mule requires its BH3 domain to target Mcl1 for rapid degradation [63,64]. Overexpression of a competing BH3-containing protein such as Bim, on the other hand, can sequester and stabilize Mcl1 [63]. Under normal conditions, however, Mule might have a competitive advantage over other BH3-dependent Mcl1-binding partners because the interaction between Mcl1 and Mule is cooperatively bimodal due to the existence of the second essential site of interaction between the two proteins, located within the N-terminal 30 aa of Mcl1 [48]. Thus, stimuli that interfere with global protein synthesis, such as radiation damage, typically lead to rapid loss of Mcl1 and, in cells in which Mcl1 is required for survival, result in cell death [64]. Mule targets many protein substrates and so far its association with malignancy—for example, as a tumour suppressor—has not been established, presumably because of other essential housekeeping functions. Targeted gene deletion in the mouse, however, indicated that Mule regulates the ATM–p53 axis to maintain B-cell homeostasis [65], loss of which could ultimately be deleterious. As noted earlier, however, evasion from Mule might be accomplished through N-terminal processing of Mcl1. In cultured cell lines, Mule seems to be responsible for constitutive Mcl1 degradation [63,64], yet in the knockout mouse Mcl1 turnover was largely unaffected [65]. This suggests that Mcl1 turnover in a physiological setting is linked to regulatory networks involving other ligases or that a compensatory mechanism was co-selected.
The cognate RING-type Mcl1 E3 ligase Fbw7, is the substrate recognition subunit of the multi-component Skp1–Cullin–F box (SCF) complex and a tumour suppressor, which is frequently lost in diverse cancer types including breast and colon cancer and T-cell acute lymphocytic leukaemia. In addition to Mcl1, the SCFFbw7 complex targets several other oncoproteins for polyubiquitylation and degradation, including c-Jun and c-Myc. Loss of Fbw7 in both human and mouse can result in elevation of Mcl1, providing resistance both to the apoptotic stimuli conferred by the corresponding upregulation of oncoproteins such Myc, as well as conferring resistance to certain therapeutics [66,67]. Moreover, recent evidence indicates that Fbw7 itself is negatively regulated by the peptidyl-prolyl cis/trans isomerase Pin1, which is frequently overexpressed in many tumour settings. Pin1 binds to and disrupts phosphorylated Fbw7 dimers, resulting in its auto-ubiquitylation and degradation [68]. Thus, there exist many avenues for Mcl1 to escape polyubiquitylation by the SCFFbw7 complex. Finally, another E3 ligase component of the SCF complex, βTRCP, targets both Mcl1 [69] and BimEL [70], emphasizing the complexity and fine-tuning that exists within the cohort of apoptosis regulators, which are targets of the ubiquitin–proteasome system [71].
SCFFbw7, as well as another multi-component RING E3 ligase APC/Ccdc20, is important in ubiquitylating and degrading Mcl1 during mitotic arrest as part of a cell cycle quality control mechanism controlled by apoptosis [66,67,72]. Escape from mitotic arrest is a characteristic of proliferating cancer cells, and the failure of Mcl1 to be degraded in response to loss or negative regulation of these ligases might help to keep the cells viable long enough to achieve this [73].
As outlined in Fig 3, the long unstructured N-terminal region of Mcl1 contains PEST-associated phosphodegrons, and these control the access of the Fbw7, βTRCP and Cdc20 E3 ligases described above and potentially others to Mcl1. This, of course, affords the control of Mcl1 degradation potentially by many signalling pathways. In addition to Cdk1 and Cdk2, Jnk and Erk [5], a predominant kinase controlling Mcl1 degradation is Gsk3 [74]. Of note, however, Jnk-dependent phosphorylation at Thr 163 provides the priming site for phosphorylation of Ser 159 by Gsk3, creating a phosphodegron targeting Mcl1 for ubiquitylation-dependent degradation [75]. Given that upstream signalling to control Jnk and Gsk3 can either support or promote cellular survival, the Mcl1 Ser 159–Thr 163 phosphodegron axis might integrate complex signalling information to determine cell fate. Moreover, evidence indicates a distinct difference in outcomes in cancer cells compared with normal cells. Erk-mediated phosphorylation at Thr 163 resulted in Mcl1 stabilization in BL41-3 lymphoma cells, due to loss of the Gsk3–Mcl1 phosphorylation pathway, and resulted in significant resistance to many chemotherapeutic agents [76]. Finally, and as outlined in Fig 3, phosphorylation of different residues can have an opposing function.
Finally, deubiquitination is another means of sparing Mcl1 from degradation. For example, the deubiquitinase USP9X removes polyubiquitin chains that normally mark Mcl1 for proteasomal degradation, resulting in enhanced Mcl1 stability. Increased USP9X expression and elevated Mcl1 protein levels are found in human follicular lymphomas and diffuse large B-cell lymphomas, and many myeloma patients with elevated USP9X have poor outcomes [77].
Strategies to enhance Mcl1 biosynthesis
The arsenal available to cancer cells to maintain a high level of Mcl1 biosynthesis include chromosomal changes favouring enhanced transcription of the Mcl1 gene—factors targeting the Mcl1 promoter; epigenetic changes to enhance promoter accessibility; gene dosage; chromosomal translocations; and splicing—enhanced access of Mcl1 messenger RNA (mRNA) to the translation initiation machinery and stabilization of Mcl1 mRNA through regulation of microRNAs (miRNAs).
Mcl1 transcriptional regulation, especially in response to growth factors, has been extensively studied [5]. For example, the transcription factor ATF5 is a direct activator of the Mcl1 promoter; ATF5 itself is upregulated in malignant glioma and other solid tumours from complex upstream signalling pathways converging on the ATF5-activating transcription factor CREB3L2 [78]. In normal pro-B-cells, Mcl1 transcription is required for survival, and is maintained by the Mcl1 transcription factor Stat5 in response to IL-7 signalling within the normal bone marrow niche. B-cell acute lymphoblastic leukaemia driven by Bcr-Abl or by activating mutations in Jak1 or Jak2, on the other hand, has escaped dependence on cytokines to maintain the Stat5–Mcl1-mediated survival pathway, which supports the transformation to malignancy [53]. A similar process seems to be operating in chronic lymphocytic leukaemia, in which Stat3 rather than Stat5 has a crucial role in maintaining high constitutive Mcl1 transcription [79]. Conversely, the tumour suppressor E2F1 is a transcriptional repressor of the Mcl1 promoter [80], yet it can also switch from a mediator of cell death towards an accelerator of tumour progression [81]. How such repression of Mcl1 transcription is lost during the progression to tumour invasion remains to be determined.
Opportunities to maintain elevated Mcl1 expression also exist at the level of Mcl1 RNA splicing, translation and stability. A complex balance might exist between Mcl1 RNA splicing that generates either pro-survival or pro-death Mcl1 transcripts [5], especially coupled to cell cycle regulation [82]. Such quality control mechanisms might become subverted in cancer through loss of splicing factors that promote the pro-apoptotic isoforms. Not surprisingly, global changes in protein synthesis capacity are required to maintain the demands of proliferating tumour cells and this can be accomplished, in part, by upregulation of key components of the translational machinery, such as eIF4A, members of the eIF4F complex and eIF4E. Indeed, overexpression of 4E has transforming activity and can cooperate with Myc to induce lymphoma [83]. As with the near global influences that Myc has on gene expression, the increase in protein synthesis capacity that accompanies tumorigenesis favours many rapidly turned over proteins associated with cell proliferation and survival. Accelerated oncogenesis caused by mTorc1 activation in a murine model of lymphomagenesis, for example, was accompanied by translational maintenance of Mcl1 [84]. An important question, however, is whether or not the structure of Mcl1 mRNA confers a competitive edge to take advantage of the increased capacity of the translation machinery.
Finally, several miRNAs have been implicated in the turnover of Mcl1 mRNA. One well-documented example is miR-29, which is lost in several cancer settings including cholangiocarcinoma [85], acute myeloid leukaemia [86] and anaplastic large cell lymphoma [87], and for which there is a direct correlation with the maintenance of Mcl1 expression. Another example is miR-125b expression that diminishes Mcl1 and Bcl-w expression indirectly by affecting IL-6/STAT3 signalling [88]. A challenge related to the identity of the cohort of miRNAs that are associated with any given gene is the many genes that each miRNA can regulate and the many miRNAs that can regulate a single gene [89]. In the case of Mcl1, this challenge was addressed by conducting a genome-wide miRNA screen to identify genes that otherwise confer resistance to the lethality associated with ABT-737 treatment, which is primarily determined by Mcl1 [90]. Ten miRNAs were found to regulate Mcl1 mRNA directly and several more regulated Mcl1 indirectly by targeting other genes. Moreover, ABT-sensitizing miRNAs were lower in tumour tissue compared with normal tissues. Thus, loss of specific miRNAs represents yet another strategy for cancer cells to maintain an adequate supply of Mcl1 mRNA.
The cancer genome favours Mcl1
As documented above, the potential to influence indirectly the expression level and/or function of the Mcl1 protein is extraordinarily extensive. Many of these strategies could well be missed in genome-wide screens focusing on chromosomal DNA or mRNA expression signatures. Nevertheless, genome-wide somatic copy number (SCN) and association studies (GWAS) of clinical samples argue that important drivers of malignancy should be represented, given the selective mutational pressure in most cancer indications. Importantly, Mcl1 is a widely represented signature. An extensive genomic analysis of SCN amplification (SCNA) in more than 3,000 cancer specimens representing 26 histological types, for example, identified several gene families that are enriched among regions of focal SCNA, including the Bcl2 family (especially MCL1 and BCL2L1) and NFκB pathway [3]. Focal amplifications for MCL1 were found in more than 10% of cancers, but were much higher in lung and breast cancers. At least in the few cell lines tested, amplifications in MCL1 and BCL2L1 correlated with dependency on these proteins for survival.
Therapeutic opportunities
Two general strategies can be contemplated for therapeutic manipulation of the Bcl2 survival set point in oncology, both to overcome intrinsic Bcl2-mediated dependence for cell survival and to overcome resistance to external therapies. One strategy—particularly relevant to Mcl1—is to exploit drug regimens targeting pathways that ultimately downregulate Mcl1 protein expression. The literature pertaining to Mcl1-mediated resistance to ABT-737 is replete with such examples. The second strategy involves direct chemical antagonism of pro-survival Bcl2 family members [91]. Intensive efforts over the past 15 years have resulted in several such clinical candidates targeting the Bcl2 family in oncology, including obatoclax, navitoclax and sabutoclax. The United States Accepted Names/International Nonproprietary Names index for these includes the stem ‘toclax’, reflecting their activity targeting Bcl–Bax interactions. As Bcl2 family proteins contribute extensively to normal development and cellular physiology, however, the question of adequate therapeutic index requires a rational strategy.
Navitoclax (the clinical analogue of ABT-737) is the most family-specific of these antagonists, but its specificity is also restricted to Bcl2, Bcl-XL and Bcl-w. Moreover, its activity in cells might be further restricted due to its apparent inability to antagonize binding of Bim to Bcl2 and Bcl-XL in this context [92]. Nevertheless, there are obvious Bcl2 family signatures that predict the response to navitoclax/ABT-737 in various indications, and susceptibility to the most downstream line of Bcl2 defence—such as, Bcl2–activated Bax interactions—might be relevant (for example, see [93]). In contrast to navitoclax, obatoclax and sabutoclax are pan-Bcl2 antagonists (including Mcl1) but are widely considered to have additional off-target activities especially when used at clinically irrelevant high doses. Obatoclax inhibits Mcl1 both in isolated mitochondria at low doses (10 nM) and in cells, and can overcome Mcl1-mediated resistance to ABT-737 [94]. It is an insoluble compound in aqueous solutions, and therefore inactive, but readily partitions into lipid membranes. In synthetic liposomes reconstituted with the tBid or Bim BH3/Mcl1/Bak axis, obatoclax is a potent inhibitor of Mcl1 but only when Mcl1 is constitutively anchored to the bilayer (Nguyen M, Shore GC, unpublished work). Furthermore, concluding that off-target activity exists for a compound that antagonizes Mcl1 is valid only if such activity does not arise in response to effective Mcl1 knockdown by RNA interference.
Given the concerns described above and the prominent contribution of Mcl1 to cancer biology, second generation Mcl1 antagonists are actively being sought. The combined computational, structural, biochemical and genetic approaches that are available almost guarantee that these efforts will prove successful [95]. Nevertheless, pushing the pendulum too far in favour of Mcl1 will also introduce risk, as Bcl2 and Bcl-XL are proven to substitute for Mcl1 in many cancer models, and will be under strong selective pressure to do so. On the other hand, if the efficacy and safety profile can be best achieved by combining optimal agents with complementary Bcl2 specificity, then the regulatory hurdle of combining two (or more) experimental agents in patients will need to be addressed in trial designs. Perhaps the most attractive attribute of an effective Mcl1 antagonist in the clinic resides in the many opportunities to exploit synthetic lethality through rational drug combinations and patient stratification. These opportunities for second generation Bcl2 family antagonists are particularly relevant as the likelihood for regulatory approval of the first-generation agents is diminishing.
Future directions
It seems that Mcl1 has the continuing capacity to surprise and in particular engage pathways that were not anticipated. As reported here, many laboratories have made spectacular discoveries over the past few years on the role of Mcl1 in tumorigenesis, but one is left with the strong suspicion that research thus far has only grazed the surface. There is no question of the central role of Mcl1 in apoptosis and cancer, which has been the focus of our report. It is what is not known about this protein, however, that might well turn out to be the most intriguing; and as illustrated in Fig 2, this might well include activities other than the canonical role of Mcl1 in apoptosis. As outlined in part in Sidebar A, we eagerly await the many new surprises that are surely to come.
Sidebar A | In need of answers.
The sum of many publications clearly links Mcl1 expression and function to a large array of regulators, including transcriptional and translational regulators, kinases, phosphatases, E3 ligases, deubiquitinases and processing enzymes. What we lack—and what is crucial for therapeutic modulation—is a detailed understanding of how these many regulators are integrated into a network. Are there master regulators of the network that could represent an Achilles heel?
Bcl2 proteins including Mcl1 are validated targets in oncology but there remains the challenge of their pervasive role in normal development and physiology as a potential barrier to a manageable therapeutic index. Articulating a rational treatment strategy continues to be a challenge. Will the underlying research that identifies synthetic lethal opportunities—disease selection, patient stratification and rational drug combinations—provide the requisite therapeutic window?
Can the Bax–Bak gateway to cell lethality be breached and, if so, what, if any, is the involvement of the pro-survival Bcl2 proteins and what pathways do they target? As emphasized in Fig 2, Bcl2 proteins might be involved in many non-apoptotic pathways influencing cell survival. In support of this, an intriguing finding is our unpublished observation that epithelial cells derived for the Bax/Bak double-knockout mouse and transformed by co-expression of E1A and dominant-negative p53 resist lethality induced by ABT-737, as expected; however, knockdown of Mcl1, although on its own not lethal, strongly sensitized the cells to ABT-737 lethality, as determined by colony formation assays.

Gordon C Shore, Mai Nguyen, Franziska Ertel & Anne Roulston
Acknowledgments
We thank the reviewers of this article for their critical input. Research from the authors' laboratory was supported by grants from the Canadian Institutes of Health Research (FRN6192), Consortium québécois sur la découverte du médicament and Génome Québec. F.E. is supported by a grant from the German Research Foundation (Forschungsstipendium GZ: ER 678/1-1).
Footnotes
The authors declare that they have no conflict of interest.
References
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Yip KW, Reed JC (2008) Bcl-2 family proteins and cancer. Oncogene 27: 6398–6406 [DOI] [PubMed] [Google Scholar]
- Beroukhim R et al. (2010) The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G et al. (2012) Chemical genomics identifies small-molecule MCL1 repressors and BCL-xL as a predictor of MCL1 dependency. Cancer Cell 21: 547–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LW, Lam C, Edwards SW (2010) Mcl-1; the molecular regulation of protein function. FEBS Lett 584: 2981–2989 [DOI] [PubMed] [Google Scholar]
- Craig RW (2002) MCL1 provides a window on the role of the BCL2 family in cell proliferation, differentiation and tumorigenesis. Leukemia 16: 444–454 [DOI] [PubMed] [Google Scholar]
- Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW (1993) MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci USA 90: 3516–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski S, Bodrug S, Krajewska M, Shabaik A, Gascoyne R, Berean K, Reed JC (1995) Immunohistochemical analysis of Mcl-1 protein in human tissues. Differential regulation of Mcl-1 and Bcl-2 protein production suggests a unique role for Mcl-1 in control of programmed cell death in vivo. Am J Pathol 146: 1309–1319 [PMC free article] [PubMed] [Google Scholar]
- Rinkenberger JL, Horning S, Klocke B, Roth K, Korsmeyer SJ (2000) Mcl-1 deficiency results in peri-implantation embryonic lethality. Genes Dev 14: 23–27 [PMC free article] [PubMed] [Google Scholar]
- Dzhagalov I, St John A, He YW (2007) The antiapoptotic protein Mcl-1 is essential for the survival of neutrophils but not macrophages. Blood 109: 1620–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ (2003) Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature 426: 671–676 [DOI] [PubMed] [Google Scholar]
- Opferman JT, Iwasaki H, Ong CC, Suh H, Mizuno S, Akashi K, Korsmeyer SJ (2005) Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science 307: 1101–1104 [DOI] [PubMed] [Google Scholar]
- Arbour N et al. (2008) Mcl-1 is a key regulator of apoptosis during CNS development and after DNA damage. J Neurosci 28: 6068–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick B et al. (2009) Knockout of myeloid cell leukemia-1 induces liver damage and increases apoptosis susceptibility of murine hepatocytes. Hepatology 49: 627–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peperzak V et al. (2013) Mcl-1 is essential for the survival of plasma cells. Nat Immunol 14: 290–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debrincat MA et al. (2012) Mcl-1 and Bcl-x(L) coordinately regulate megakaryocyte survival. Blood 119: 5850–5858 [DOI] [PubMed] [Google Scholar]
- Kodama T et al. (2012) Mcl-1 and Bcl-xL regulate Bak/Bax-dependent apoptosis of the megakaryocytic lineage at multistages. Cell Death Differ 19: 1856–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KJ, Bath ML, Turner ML, Vandenberg CJ, Bouillet P, Metcalf D, Scott CL, Cory S (2010) Elevated Mcl-1 perturbs lymphopoiesis, promotes transformation of hematopoietic stem/progenitor cells, and enhances drug resistance. Blood 116: 3197–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Levy NB, Xie H, Qian L, Lee CY, Gascoyne RD, Craig RW (2001) MCL1 transgenic mice exhibit a high incidence of B-cell lymphoma manifested as a spectrum of histologic subtypes. Blood 97: 3902–3909 [DOI] [PubMed] [Google Scholar]
- Zhou P, Qian L, Bieszczad CK, Noelle R, Binder M, Levy NB, Craig RW (1998) Mcl-1 in transgenic mice promotes survival in a spectrum of hematopoietic cell types and immortalization in the myeloid lineage. Blood 92: 3226–3239 [PubMed] [Google Scholar]
- Glaser SP et al. (2012) Anti-apoptotic Mcl-1 is essential for the development and sustained growth of acute myeloid leukemia. Genes Dev 26: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Luo H, Payton JE, Cain J, Ley TJ, Opferman JT, Tomasson MH (2010) Mcl1 haploinsufficiency protects mice from Myc-induced acute myeloid leukemia. J Clin Invest 120: 2109–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TD, Zhu CQ, Jone KD, Yanagawa N, Tsao MS, Bishop JM (2011) Interaction between MYC and MCL1 in the genesis and outcome of non-small-cell lung cancer. Cancer Res 71: 2212–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awan FT et al. (2009) Mcl-1 expression predicts progression-free survival in chronic lymphocytic leukemia patients treated with pentostatin, cyclophosphamide, and rituximab. Blood 113: 535–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann SH, Karp JE, Svingen PA, Krajewski S, Burke PJ, Gore SD, Reed JC (1998) Elevated expression of the apoptotic regulator Mcl-1 at the time of leukemic relapse. Blood 91: 991–1000 [PubMed] [Google Scholar]
- Warr MR, Shore GC (2008) Unique biology of Mcl-1: therapeutic opportunities in cancer. Curr Mol Med 8: 138–147 [DOI] [PubMed] [Google Scholar]
- Happo L, Strasser A, Cory S (2012) BH3-only proteins in apoptosis at a glance. J Cell Sci 125: 1081–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltvai ZN, Milliman CL, Korsmeyer SJ (1993) Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 74: 609–619 [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ (2004) Cell death: critical control points. Cell 116: 205–219 [DOI] [PubMed] [Google Scholar]
- Chen L et al. (2005) Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 17: 393–403 [DOI] [PubMed] [Google Scholar]
- Heath-Engel HM, Chang NC, Shore GC (2008) The endoplasmic reticulum in apoptosis and autophagy: role of the BCL-2 protein family. Oncogene 27: 6419–6433 [DOI] [PubMed] [Google Scholar]
- Martinou JC, Youle RJ (2011) Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell 21: 92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamas-Din, Brahmbhatt H, Leber B, Andrews DW (2011) BH3-only proteins: orchestrators of apoptosis. Biochim Biophys Acta 1813: 508–520 [DOI] [PubMed] [Google Scholar]
- Tait SW, Green DR (2010) Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 11: 621–632 [DOI] [PubMed] [Google Scholar]
- Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD (2010) BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol Cell 40: 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH (2006) Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol 8: 1348–1358 [DOI] [PubMed] [Google Scholar]
- Kushnareva Y, Andreyev AY, Kuwana T, Newmeyer DD (2012) Bax activation initiates the assembly of a multimeric catalyst that facilitates Bax pore formation in mitochondrial outer membranes. PLoS Biol 10: e1001394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL, Dillon CP, Green DR (2011) A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell 44: 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, Andrews DW (2008) Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell 135: 1074–1084 [DOI] [PubMed] [Google Scholar]
- Dewson G, Kratina T, Czabotar P, Day CL, Adams JM, Kluck RM (2009) Bak activation for apoptosis involves oligomerization of dimers via their α6 helices. Mol Cell 36: 696–703 [DOI] [PubMed] [Google Scholar]
- Bogner C, Leber B, Andrews DW (2010) Apoptosis: embedded in membranes. Curr Opin Cell Biol 22: 845–851 [DOI] [PubMed] [Google Scholar]
- Edlich F, Banerjee S, Suzuki M, Cleland MM, Arnoult D, Wang C, Neutzer A, Tiandra N, Youle RJ (2011) Bcl-x(L) retrotranslocates Bax from the mitochondria into the cytosol. Cell 145: 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino D et al. (2009) The role of BH3-only protein Bim extends beyond inhibiting Bcl-2-like prosurvival proteins. J Cell Biol 186: 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montessuit S et al. (2010) Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell 142: 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, McStay GP, Bharti A, Kuwana T, Clarke CJ, Siskind LJ, Obeid LM, Green DR (2012) Sphingolipid metabolism cooperates with BAK and BAX to promote the mitochondrial pathway of apoptosis. Cell 148: 988–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabotar PE, Lee EF, van Delft MF, Day CL, Smith BJ, Huang DC, Fairlie WD, Hinds MG, Colman PM (2007) Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc Natl Acad Sci USA 104: 6217–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S et al. (2010) Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL. J Mol Biol 398: 747–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr MR et al. (2011) Mitochondrion-dependent N-terminal processing of outer membrane Mcl-1 protein removes an essential Mule/Lasu1 protein-binding site. J Biol Chem 286: 25098–25107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross K, Rudel T, Kozjak-Pavlovic V (2009) TOM-independent complex formation of Bax and Bak in mammalian mitochondria during TNFα-induced apoptosis. Cell Death Differ 16: 697–707 [DOI] [PubMed] [Google Scholar]
- Chou CH, Lee RS, Yang-Yen HF (2006) An internal EELD domain facilitates mitochondrial targeting of Mcl-1 via a Tom70-dependent pathway. Mol Biol Cell 17: 3952–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffolo SC, Shore GC (2003) BCL-2 selectively interacts with the BID-induced open conformer of BAK, inhibiting BAK auto-oligomerization. J Biol Chem 278: 25039–25045 [DOI] [PubMed] [Google Scholar]
- Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M, Thompson CB, Korsmeyer SJ (2000) tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev 14: 2060–2071 [PMC free article] [PubMed] [Google Scholar]
- Malin S, McManus S, Cobaleda C, Novatchkova M, Deloqu A, Bouillet P, Strasser A, Busslinger M (2010) Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol 11: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérino D, Strasser A, Bouillet P (2012) Bim must be able to engage all pro-survival Bcl-2 family members for efficient tumor suppression. Oncogene 31: 3392–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverly LJ, Varmus HE (2009) MYC-induced myeloid leukemogenesis is accelerated by all six members of the antiapoptotic BCL family. Oncogene 28: 1274–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopleva M et al. (2006) Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell 10: 375–388 [DOI] [PubMed] [Google Scholar]
- van Delft MF et al. (2006) The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 10: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain M, Nguyen AP, Le Grand JN, Arbour N, Vanderluit JL, Park DS, Opferman JT, Slack RS (2011) MCL-1 is a stress sensor that regulates autophagy in a developmentally regulated manner. EMBO J 30: 395–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biasio A et al. (2007) N-terminal truncation of antiapoptotic MCL1, but not G2/M-induced phosphorylation, is associated with stabilization and abundant expression in tumor cells. J Biol Chem 282: 23919–23936 [DOI] [PubMed] [Google Scholar]
- Huang CR, Yang-Yen HF (2010) The fast-mobility isoform of mouse Mcl-1 is a mitochondrial matrix-localized protein with attenuated anti-apoptotic activity. FEBS Lett 584: 3323–3330 [DOI] [PubMed] [Google Scholar]
- Perciavalle RM et al. (2012) Anti-apoptotic MCL-1 localizes to the mitochondrial matrix and couples mitochondrial fusion to respiration. Nat Cell Biol 14: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N et al. (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 189: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr MR, Acoca S, Liu Z, Germain M, Watson M, Blanchette M, Wing SS, Shore GC (2005) BH3-ligand regulates access of MCL-1 to its E3 ligase. FEBS Lett 579: 5603–5608 [DOI] [PubMed] [Google Scholar]
- Zhong Q, Gao W, Du F, Wang X (2005) Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 121: 1085–1095 [DOI] [PubMed] [Google Scholar]
- Hao Z et al. (2012) The E3 ubiquitin ligase Mule acts through the ATM-p53 axis to maintain B lymphocyte homeostasis. J Exp Med 209: 173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H et al. (2011) SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 471: 104–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE et al. (2011) Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 471: 110–114 [DOI] [PubMed] [Google Scholar]
- Min SH et al. (2012) Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Mol Cell 46: 771–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q et al. (2007) Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol 27: 4006–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehan E et al. (2009) betaTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol Cell 33: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic D, Dixit VM, Wertz IE (2011) Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol 12: 439–452 [DOI] [PubMed] [Google Scholar]
- Harley ME, Allan LA, Sanderson HS, Clarke PR (2010) Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. EMBO J 29: 2407–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman SE, Pagano M (2011) MCL1 meets its end during mitotic arrest. EMBO Rep 12: 384–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR (2006) Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell 21: 749–760 [DOI] [PubMed] [Google Scholar]
- Morel C, Carlson SM, White FM, Davis RJ (2009) Mcl-1 integrates the opposing actions of signaling pathways that mediate survival and apoptosis. Mol Cell Biol 29: 3845–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nifoussi SK, Vrana JA, Domina AM, De Biasio A, Gui J, Gregory MA, Hann SR, Craig RW (2012) Thr 163 phosphorylation causes Mcl-1 stabilization when degradation is independent of the adjacent GSK3-targeted phosphodegron, promoting drug resistance in cancer. PLoS ONE 7: e47060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickart M et al. (2010) Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 463: 103–107 [DOI] [PubMed] [Google Scholar]
- Sheng Z, Li L, Zhu LJ, Smith TW, Demers A, Ross AH, Moser RP, Green MR (2010) A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nat Med 16: 671–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JC, Talab F, Zuzel M, Lin K, Slupsky JR (2011) c-Abl regulates Mcl-1 gene expression in chronic lymphocytic leukemia cells. Blood 117: 2414–2422 [DOI] [PubMed] [Google Scholar]
- Croxton R, Ma Y, Song L, Haura EB, Cress WD (2002) Direct repression of the Mcl-1 promoter by E2F1. Oncogene 21: 1359–1369 [DOI] [PubMed] [Google Scholar]
- Engelmann D, Pützer BM (2012) The dark side of E2F1: in transit beyond apoptosis. Cancer Res 72: 571–575 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA (2010) An alternative splicing network links cell-cycle control to apoptosis. Cell 142: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist L, Pelletier J (2009) Inhibitors of translation initiation as cancer therapeutics. Future Med Chem 1: 1709–1722 [DOI] [PubMed] [Google Scholar]
- Mills JR et al. (2008) mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci USA 105: 10853–10858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott JL et al. (2007) mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 26: 6133–6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R et al. (2009) MicroRNA 29b functions in acute myeloid leukemia. Blood 114: 5331–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjobert C et al. (2011) MiR-29a down-regulation in ALK-positive anaplastic large cell lymphomas contributes to apoptosis blockade through MCL-1 overexpression. Blood 117: 6627–6637 [DOI] [PubMed] [Google Scholar]
- Gong J, Zhang JP, Li B, Zeng C, You K, Chen MX, Yuan Y, Zhuang SM (2012) MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene [Epub ahead of print] doi:; DOI: 10.1038/onc.2012.318 [DOI] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP (2005) The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 310: 1817–1821 [DOI] [PubMed] [Google Scholar]
- Lam LT, Lu X, Zhang H, Lesniewski R, Rosenberg S, Semizarov D (2010) A microRNA screen to identify modulators of sensitivity to BCL2 inhibitor ABT-263 (navitoclax). Mol Cancer Ther 9: 2943–2950 [DOI] [PubMed] [Google Scholar]
- Thomas S et al. (2012) Targeting the Bcl-2 family for cancer therapy. Expert Opin Ther Targets 17: 61–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranovich A, Liu Q, Collins T, Geng F, Dixit S, Leber B, Andrews DW (2012) Differences in the mechanisms of proapoptotic BH3 proteins binding to Bcl-XL and Bcl-2 quantified in live MCF-7 cells. Mol Cell 45: 754–763 [DOI] [PubMed] [Google Scholar]
- Bodet L et al. (2011) ABT-737 is highly effective against molecular subgroups of multiple myeloma. Blood 118: 3901–3910 [DOI] [PubMed] [Google Scholar]
- Nguyen M et al. (2007) Small molecule obatoclax (GX15–070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc Natl Acad Sci USA 104: 19512–19517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NA, Stewart ML, Gavathiotis E, Tepper JL, Bruekner SR, Koss B, Opferman JT, Walensky LD (2012) A competitive stapled peptide screen identifies a selective small molecule that overcomes MCL-1-dependent leukemia cell survival. Chem Biol 19: 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S et al. (2007) Serine 64 phosphorylation enhances the antiapoptotic function of Mcl-1. J Biol Chem 282: 18407–18417 [DOI] [PubMed] [Google Scholar]
- Ding Q et al. (2008) Down-regulation of myeloid cell leukemia-1 through inhibiting Erk/Pin 1 pathway by sorafenib facilitates chemosensitization in breast cancer. Cancer Res 68: 6109–6117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y et al. (2009) Antiapoptotic effect of c-Jun N-terminal kinase-1 through Mcl-1 stabilization in TNF-induced hepatocyte apoptosis. Gastroenterology 136: 1423–1434 [DOI] [PubMed] [Google Scholar]
- Inoshita S, Takeda K, Hatai T, Terada Y, Sano M, Hata J, Umezawa A, Ichijo H (2002) Phosphorylation and inactivation of myeloid cell leukemia 1 by JNK in response to oxidative stress. J Biol Chem 277: 43730–43734 [DOI] [PubMed] [Google Scholar]
- Domina AM, Vrana JA, Gregory MA, Hann SR, Craig RW (2004) MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene 23: 5301–5315 [DOI] [PubMed] [Google Scholar]