Abstract
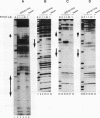
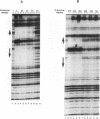
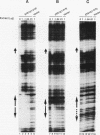
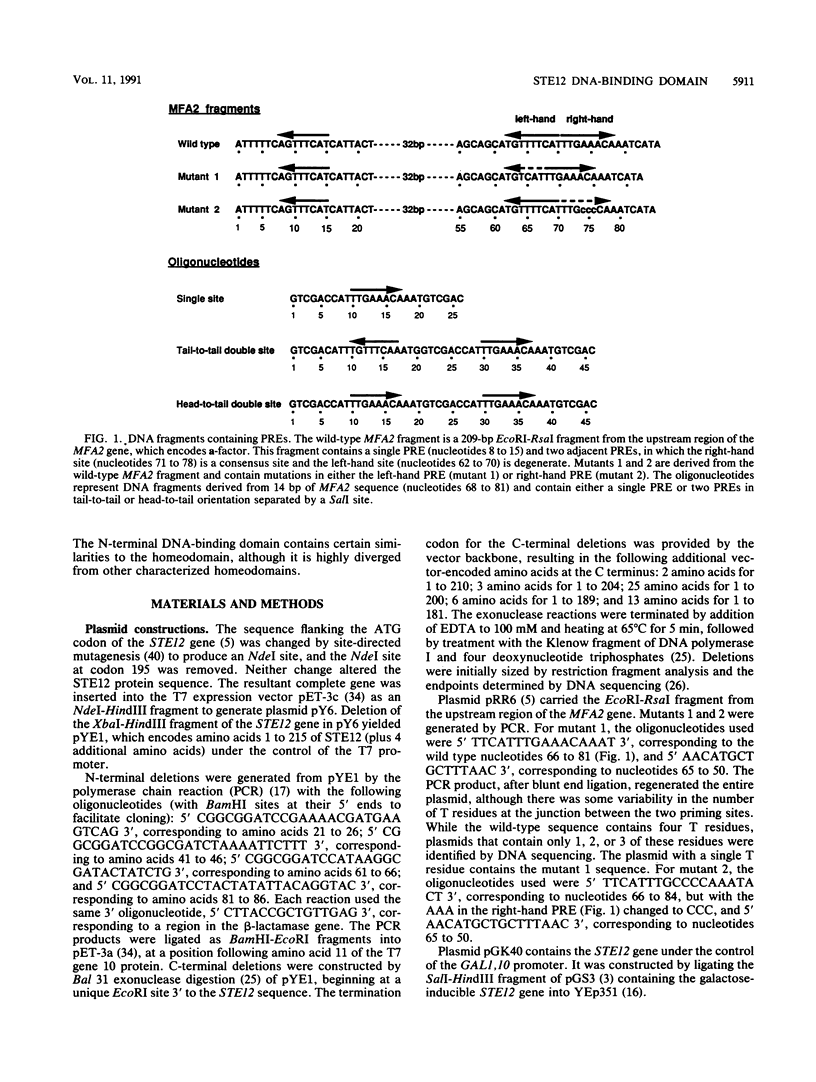
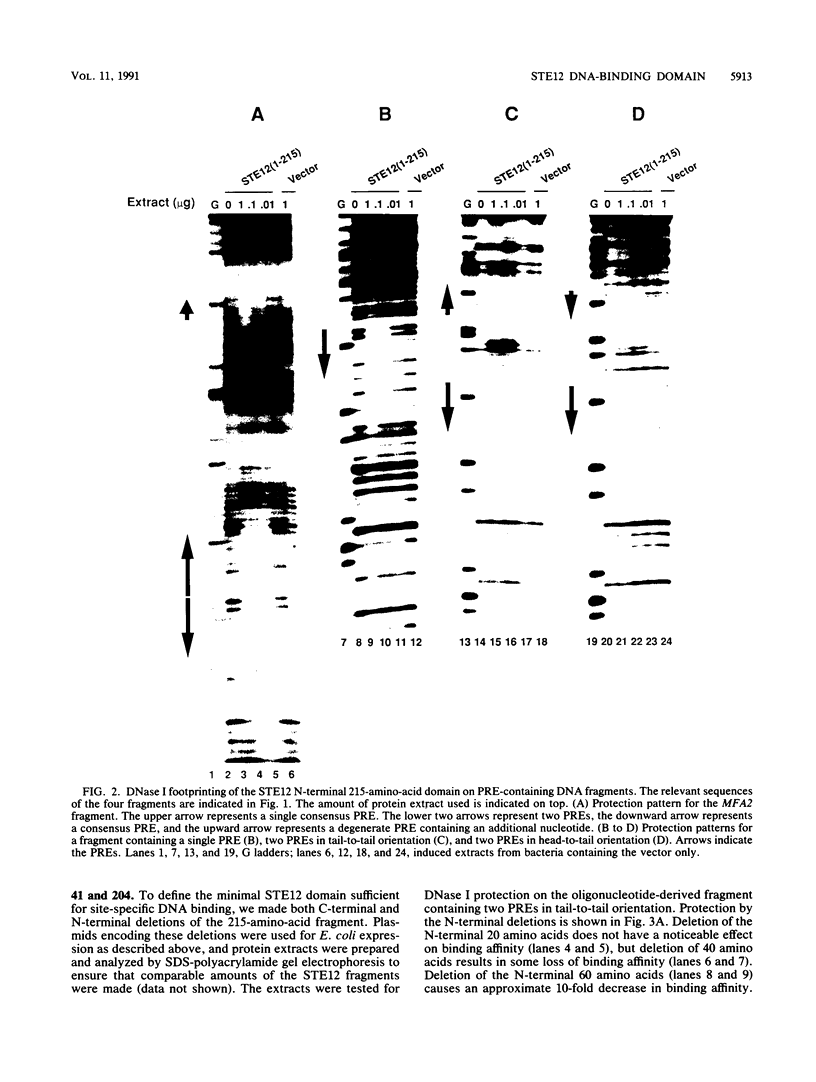
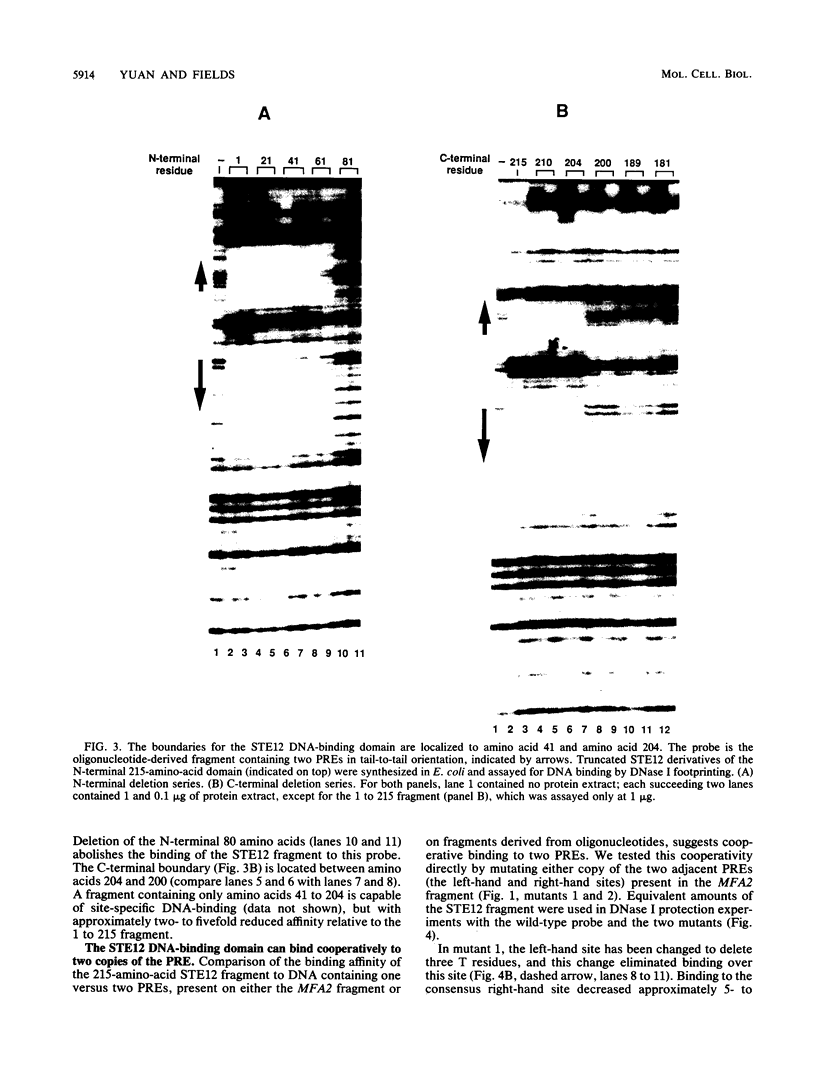
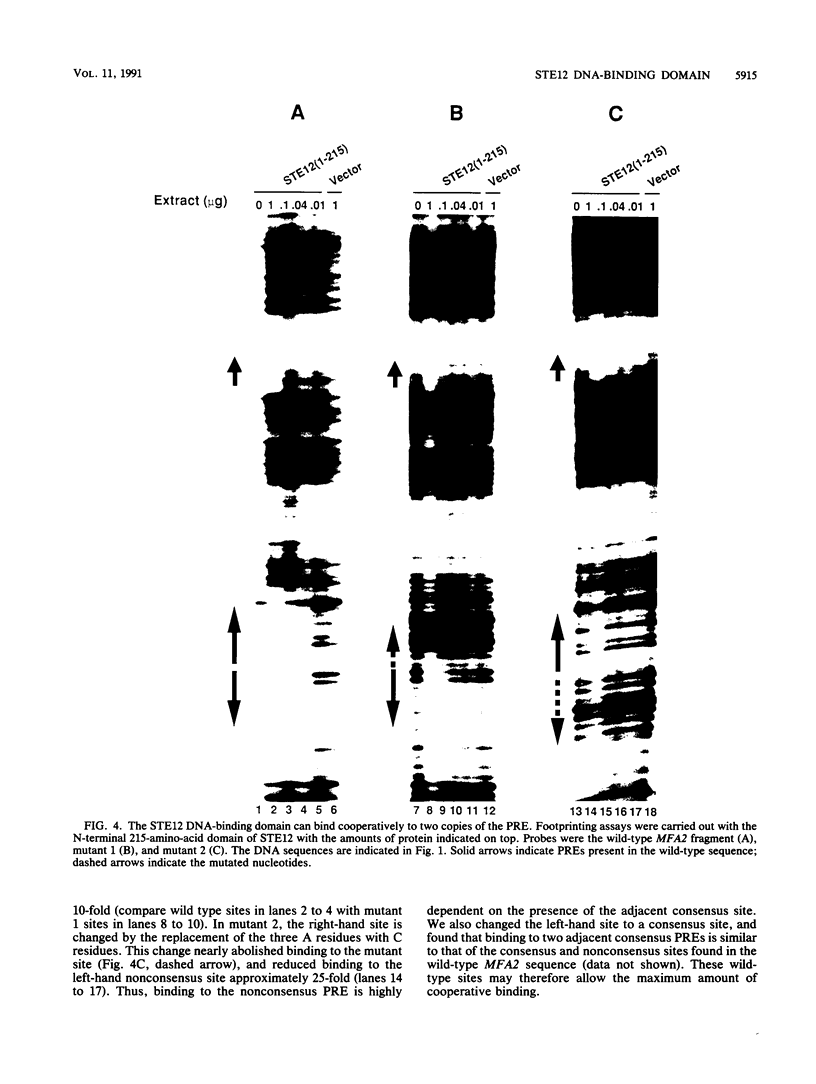
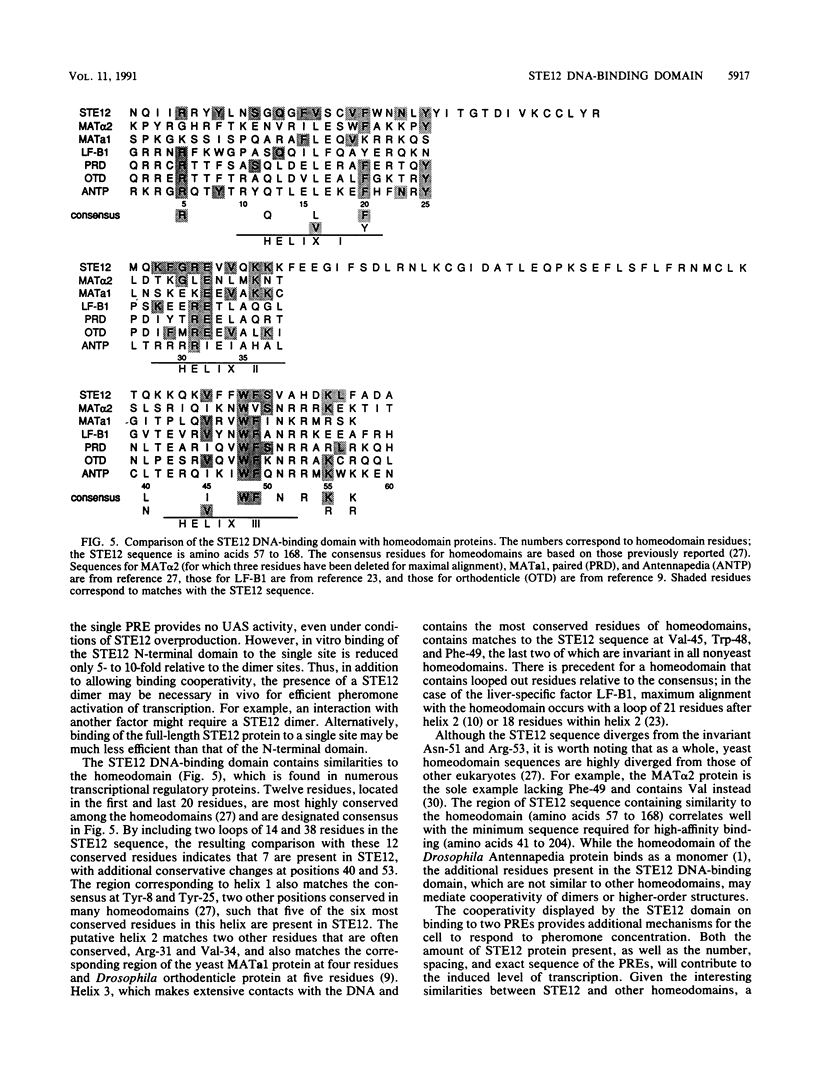
The STE12 protein of the yeast Saccharomyces cerevisiae binds to the pheromone response element (PRE) present in the upstream region of genes whose transcription is induced by pheromone. Using DNase I footprinting assays with bacterially made STE12 fragments, we localized the DNA-binding domain to 164 amino acids near the amino terminus. Footprinting of oligonucleotide-derived sequences containing one PRE, or two PREs in head-to-tail or tail-to-tail orientation, showed that the N-terminal 215 amino acids of STE12 has similar binding affinity to either of the dimer sites and a binding affinity 5- to 10-fold lower for the monomer site. This binding cooperativity was also evident on a fragment from the MFA2 gene, which encodes the a-factor pheromone. On this fragment, the 215-amino-acid STE12 fragment protected both a consensus PRE as well as a degenerate PRE containing an additional residue. Mutation of the degenerate site led to a 5- to 10-fold decrease in binding; mutation of the consensus site led to a 25-fold decrease in binding. The ability of PREs to function as pheromone-inducible upstream activation sequences in yeast correlated with their ability to bind the STE12 domain in vitro. The sequence of the STE12 DNA-binding domain contains similarities to the homeodomain, although it is highly diverged from other known examples of this motif. Moreover, the alignment between STE12 and the homeodomain postulates loops after both the putative helix 1 and helix 2 of the STE12 sequence.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Affolter M., Percival-Smith A., Müller M., Leupin W., Gehring W. J. DNA binding properties of the purified Antennapedia homeodomain. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4093–4097. doi: 10.1073/pnas.87.11.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appeltauer U., Achstetter T. Hormone-induced expression of the CHS1 gene from Saccharomyces cerevisiae. Eur J Biochem. 1989 Apr 15;181(1):243–247. doi: 10.1111/j.1432-1033.1989.tb14718.x. [DOI] [PubMed] [Google Scholar]
- Dolan J. W., Fields S. Cell-type-specific transcription in yeast. Biochim Biophys Acta. 1991 Feb 16;1088(2):155–169. doi: 10.1016/0167-4781(91)90051-m. [DOI] [PubMed] [Google Scholar]
- Dolan J. W., Fields S. Overproduction of the yeast STE12 protein leads to constitutive transcriptional induction. Genes Dev. 1990 Apr;4(4):492–502. doi: 10.1101/gad.4.4.492. [DOI] [PubMed] [Google Scholar]
- Dolan J. W., Kirkman C., Fields S. The yeast STE12 protein binds to the DNA sequence mediating pheromone induction. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5703–5707. doi: 10.1073/pnas.86.15.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B., Ammerer G. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 1989 Sep;3(9):1349–1361. doi: 10.1101/gad.3.9.1349. [DOI] [PubMed] [Google Scholar]
- Fields S., Chaleff D. T., Sprague G. F., Jr Yeast STE7, STE11, and STE12 genes are required for expression of cell-type-specific genes. Mol Cell Biol. 1988 Feb;8(2):551–556. doi: 10.1128/mcb.8.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Herskowitz I. The yeast STE12 product is required for expression of two sets of cell-type specific genes. Cell. 1985 Oct;42(3):923–930. doi: 10.1016/0092-8674(85)90288-0. [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Smouse D., Capaci T. M., Spradling A. C., Perrimon N. The orthodenticle gene encodes a novel homeo domain protein involved in the development of the Drosophila nervous system and ocellar visual structures. Genes Dev. 1990 Sep;4(9):1516–1527. doi: 10.1101/gad.4.9.1516. [DOI] [PubMed] [Google Scholar]
- Finney M. The homeodomain of the transcription factor LF-B1 has a 21 amino acid loop between helix 2 and helix 3. Cell. 1990 Jan 12;60(1):5–6. doi: 10.1016/0092-8674(90)90708-m. [DOI] [PubMed] [Google Scholar]
- Hagen D. C., McCaffrey G., Sprague G. F., Jr Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):2952–2961. doi: 10.1128/mcb.11.6.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen D. C., Sprague G. F., Jr Induction of the yeast alpha-specific STE3 gene by the peptide pheromone a-factor. J Mol Biol. 1984 Oct 5;178(4):835–852. doi: 10.1016/0022-2836(84)90314-0. [DOI] [PubMed] [Google Scholar]
- Hartig A., Holly J., Saari G., MacKay V. L. Multiple regulation of STE2, a mating-type-specific gene of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2106–2114. doi: 10.1128/mcb.6.6.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. A regulatory hierarchy for cell specialization in yeast. Nature. 1989 Dec 14;342(6251):749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Jarvis E. E., Hagen D. C., Sprague G. F., Jr Identification of a DNA segment that is necessary and sufficient for alpha-specific gene control in Saccharomyces cerevisiae: implications for regulation of alpha-specific and a-specific genes. Mol Cell Biol. 1988 Jan;8(1):309–320. doi: 10.1128/mcb.8.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Herskowitz I. A repressor (MAT alpha 2 Product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985 Aug;42(1):237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- Kronstad J. W., Holly J. A., MacKay V. L. A yeast operator overlaps an upstream activation site. Cell. 1987 Jul 31;50(3):369–377. doi: 10.1016/0092-8674(87)90491-0. [DOI] [PubMed] [Google Scholar]
- Martinez E., Wahli W. Cooperative binding of estrogen receptor to imperfect estrogen-responsive DNA elements correlates with their synergistic hormone-dependent enhancer activity. EMBO J. 1989 Dec 1;8(12):3781–3791. doi: 10.1002/j.1460-2075.1989.tb08555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey G., Clay F. J., Kelsay K., Sprague G. F., Jr Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987 Aug;7(8):2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicosia A., Monaci P., Tomei L., De Francesco R., Nuzzo M., Stunnenberg H., Cortese R. A myosin-like dimerization helix and an extra-large homeodomain are essential elements of the tripartite DNA binding structure of LFB1. Cell. 1990 Jun 29;61(7):1225–1236. doi: 10.1016/0092-8674(90)90687-a. [DOI] [PubMed] [Google Scholar]
- Otting G., Qian Y. Q., Müller M., Affolter M., Gehring W., Wüthrich K. Secondary structure determination for the Antennapedia homeodomain by nuclear magnetic resonance and evidence for a helix-turn-helix motif. EMBO J. 1988 Dec 20;7(13):4305–4309. doi: 10.1002/j.1460-2075.1988.tb03329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. P., Tamkun J. W., Hartzell G. W., 3rd The structure and function of the homeodomain. Biochim Biophys Acta. 1989 Jul 28;989(1):25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4115–4119. doi: 10.1073/pnas.81.13.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P., Cochran B. H. The PRE and PQ box are functionally distinct yeast pheromone response elements. Mol Cell Biol. 1990 Dec;10(12):6809–6812. doi: 10.1128/mcb.10.12.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J. C., McGinnis W., Carrasco A. E., De Robertis E. M., Gehring W. J. Fly and frog homoeo domains show homologies with yeast mating type regulatory proteins. Nature. 1984 Jul 5;310(5972):70–71. doi: 10.1038/310070a0. [DOI] [PubMed] [Google Scholar]
- Song D., Dolan J. W., Yuan Y. L., Fields S. Pheromone-dependent phosphorylation of the yeast STE12 protein correlates with transcriptional activation. Genes Dev. 1991 May;5(5):741–750. doi: 10.1101/gad.5.5.741. [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Jr Combinatorial associations of regulatory proteins and the control of cell type in yeast. Adv Genet. 1990;27:33–62. doi: 10.1016/s0065-2660(08)60023-1. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989 Feb 24;56(4):619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Trueheart J., Boeke J. D., Fink G. R. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987 Jul;7(7):2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. Y., Tsai M. J., O'Malley B. W. Cooperative binding of steroid hormone receptors contributes to transcriptional synergism at target enhancer elements. Cell. 1989 May 5;57(3):443–448. doi: 10.1016/0092-8674(89)90919-7. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. Methods Enzymol. 1987;154:329–350. doi: 10.1016/0076-6879(87)54083-6. [DOI] [PubMed] [Google Scholar]