Abstract
Dimerization of the transmembrane (TM) adaptor protein DAP12 plays a key role in mediating activation signals through TM-TM association with cell-surface receptors. Herein, we apply the TOXCAT assay and molecular dynamics simulation to analyze dynamics and dimerization of the TM helix of DAP12 in the membrane bilayer. In the TOXCAT assay, we performed site-specific mutagenesis of potential dimerization motifs in the DAP12 TM domain. Instead of the common GxxxG dimerization motif, mutating either of the polar residues Asp-50 and Thr-54 significantly decreased the TOXCAT signal for the dimerization of DAP12 TM domain. Furthermore, through the conformational difference between wild-type and mutant DAP12 TM homodimers, a combined coarse-grained and atomistic molecular dynamics simulation has identified both Asp-50 and Thr-54 at the dimerization interface. The experimental and computational results of the DAP12 TM dimer are in excellent agreement with the previously reported NMR structure obtained in detergent micelles. Such a combination of dynamics simulation and cell-based experiments can be applied to produce insights at the molecular level into the TM-TM association of many other transmembrane proteins.
Introduction
Membrane proteins play a key role in cellular processes involving transmembrane steps, such as signaling, transport, and trafficking. Integral membrane proteins span the membrane bilayer and are thought to assemble via a two-stage folding mechanism (1). In this mechanism, a membrane protein is synthesized and simultaneously integrated into the membrane, then associates into homo- or heterooligomeric complexes. Sequence-specific association between α-helical transmembrane (TM) domains drives correct assembly of many integral membrane proteins (2–4). In some cases, the association involves pairs of α-helices with a right-handed (RH) twist (5). In other cases a leucine zipper type of side-chain packing is evident (6). Determining sequence specificity in the TM-TM association is also crucial to identify potential structural rearrangement involved in TM signaling and channel conduction (7,8). Therefore, to better understand the fundamental principles behind the assembly of membrane proteins, it is important to develop methods for the analysis of the structure, assembly, and dynamics of the TM helix association (9). However, the study of the TM helix association has been a challenging task over the past few decades because high-resolution structural data are difficult to obtain for many membrane proteins. Taking advantage of the regular conformation in α-helical proteins, several biochemical and biophysical methods have been developed to assess the association of TM domain oligomers in detergents and in lipid bilayer membranes (10–12).
TOXCAT assay is a method developed by Russ and Engelman for studying the TM-TM association in the real lipid bilayer membrane system (Escherichia coli inner cell membrane) (13). In this assay, the expression of a reporter gene, chloramphenicol acetyltransferase (CAT) is driven by oligomerization-dependent transcriptional activation by a membrane-inserted fusion protein. The resulting CAT enzymatic activity is largely proportional to the extent of TM helix dimerization. Thus, compared with other in vitro methods, TOXCAT can provide more quantitative measurement to distinguish between strong and weak TM-TM association in a real cell membrane. Furthermore, systematic mutagenesis can be carried out in the TOXCAT system to identify residues critical to the TM-TM association (14). However, little information at the atomic level can be obtained directly from the TOXCAT experiment.
Molecular dynamics (MD) simulation has been used to interrogate the TM-TM association at the atomic level. Recent increases in computing power and improvement in simulation algorithms have made it possible to simulate TM-TM association in a well-defined lipid bilayer environment. Although the simulation typically starts from an available NMR or crystal structure of interacting TM helices, or a homology model, it can be used to explore dynamic configurations in greater detail. Because atomistic MD simulations and docking-based methods were used to investigate the self-assembly of glycophorin A (GpA) and to estimate the energetics of its TM dimerization (15,16), longer effective timescales and larger systems have become accessible with the coarse-grained (CG) approach (17–20), in which small groups of atoms are treated as single particles. A recent CG model has been used to model the self-assembly of GpA monomers in detergent micelles and lipid bilayers (21). In addition to the canonical GxxxG motif, the hydrogen bonding between polar residues play an important role in promoting interhelical association between TM domains (22,23). However, most studies have focused on the GxxxG motif-mediated TM-TM association. There have been no MD simulation studies on polar residue-mediated TM-TM association. Moreover, the accuracy of the MD simulation is uncertain and needs to be revaluated by experimental techniques in some cases. A potential means for overcoming the limitations of the MD simulation approach is to combine it with experimental techniques to arrive at experimentally validated, dynamic, three-dimensional structures of TM associations.
DAP12 (also called KARAP) is a transmembrane adaptor well known for its role in transducing activation signals for an extended array of receptors in the natural killer (NK) cells, granulocytes, monocytes/macrophages, and dendritic cells (24–26). DAP12 is a relatively short polypeptide of 113–114 residues, consisting of a short extracellular tail, a single TM domain, and a cytoplasmic domain containing the immunoreceptor tyrosine-based activation motif sequence motif (27). DAP12 forms a disulfide-bonded signaling homodimer and is associated with a number of activating receptors, such as NK receptors KIR (KIR2DS and KIR3DS) and NKG2C/CD94 (28,29). Recently, the NMR structure of the DAP12 TM homodimer was determined in tetradecylphosphocholine (TDPC)-SDS mixed micelles (30). In the NMR structure, the DAP12 TM domain forms a left-handed (LH) parallel dimer. Two conserved polar residues, D50 and T54, play an important role in homodimerization. In contrast, substitution of T54 by alanine in the full-length DAP12 had no adverse effect on the formation of the DAP12 dimer (30). Moreover, whether the NMR structure of the DAP12 TM homodimer represents the DAP12 TM-TM association in the lipid bilayer is not known.
A number of biochemical, biophysical, and computational methods have been developed to characterize TM-TM association in detergents and in membrane bilayers. However, it is difficult for each individual method to provide a complete picture of the TM-TM association. In this study, TOXCAT and MD simulation were applied to study the dimerization of the DAP12 TM helix in the membrane bilayer. The computational and experimental results are in excellent agreement with each other. In combination they showed that instead of the GxxxG motif present in the DAP12 TM domain, the polar residues play an important role in the TM-TM association.
Materials and Methods
Materials
E. coli BL21 and DH5 competent cells were purchased from TransGen Biotech (Beijing, China). Anti-MBP monoclonal antibody was purchased from New England Biolabs (Ipswich, MA).
TOXCAT assays
Gene fragments encoding DAP12 TM sequences were amplified by polymerase chain reaction from human DAP12 cDNA (provided by Dr. Lewis L. Lanier), and inserted into the pccKAN plasmid (provided by Dr. Donald M. Engelman)) as described earlier (13). The resulting pccKAN-based plasmids were transformed to E. coli MM39 cells. The topology of the ToxR-TM-MBP fusion protein expressed in MM39 cells was checked with the maltose complementation tests as described (31). Expression of the fusion protein was visualized by western blot against MBP. The activity of CAT expressed in MM39 cells was measured as described (31).
CG-MD simulations
All the CG-MD simulations were performed by GROMACS version 4.5.3 (www.gromacs.org) (32,33) and the MARTINI force field was used for the proteins, lipids, and water (19,34). The model of the wild-type (WT) DAP12-WT TM helices were constructed based on the DAP12 NMR structure (PDB ID 2L34) (30) and other mutants were derived from this NMR structure using Modeler (35). In all structures, the valine in position 48 mutated in the DAP12 NMR structure to avoid secondary cyanogen bromide cleavage of the fusion protein was reverted to WT residue methionine. These atomistic structures and relevant topology files were then converted to corresponding CG counterparts using script from the MARTINI protocol. The assigned backbone particle for Gly residues in the TM segments of the helices changed to atom type Na as the Na-Na pairwise interaction is semiattractive as described in the work by Psachoulia et al. on GpA TM homodimerization (36) and Chng on the integrins TM heterodimerization (37). This change is to mimic the interaction for GxxxG motif in the helices association since the Gly backbones are exposed.
These CG structures were inserted parallelly at a distance of ∼55 Å between the center of masses of the backbone of the two helices in a slab of dipalmitoylphosphatidylcholine (DPPC) lipid bilayer. This bilayer attains equilibrium after hundreds of self-assemblies, containing ∼186 DPPC molecules and 4000 CG waters (each representing four atomistic waters). The N- and C-terminal of the peptide were not acetylated or amidated. Ions were added to electrically neutralize the system. The polar residues D50 and T54 of the two DAP12-WT helices were located in the middle of the bilayer slab as the mutant counterparts.
Lennard-Jones interactions were shifted to zero between 9 and 12 Å, and electrostatics were shifted to zero between 0 and 12 Å, with a relative dielectric constant of 15. The nonbonded neighbor list was updated every 10 steps with a cutoff of 14 Å. A Berendsen thermostat (38) was used for temperature (323 K, coupling constant 1 ps) and pressure (1 bar, coupling constant 5 ps, compressibility 4.5×10−5 bar−1, semiisotropic) coupling. The integration step was 20 fs. These parameters follow from the recommendations of Marrink et al. (39). The energy of each system was again minimized and is then position-restrained in a 3 ns simulation to allow for better packing of the lipid molecules around the TM helices. Seven 3 μs production simulations were carried out for the DAP12-WT and mutants. Each CG system started with the same initial configuration but with different initial velocities.
From CG to atomistic representation
The conversion work from CG to atomistic representation was followed by a fragment-based procedure as described previously (40). In this procedure, the truncated NMR structure was fitted on the protein CG particles and atomistic lipid fragments were aligned with the CG particles of each lipid molecule.
Atomistic MD simulations
Atomistic MD simulations were performed using the GROMOS96 53a6 force field (41) after the conversion to atomistic representation. Each system was performed starting from the same initial position but with different initial velocities. The united-atom lipid models generated for GROMOS96 53a6 force field was used (42). The Berendsen thermostat (323K, coupling constant 0.1 ps) and Parrinello-Rahman barostat (1bar, coupling constant 2 ps, compressibility 4.5 × 10−5 bar−1, semiisotropic) were used for temperature and pressure coupling. LINCS algorithm (43) was used to constrain bond lengths. Long-range electrostatic were modeled up to 9 Å using the Ewald particle mesh. The 14 Å cutoff distance was used for the vander Waals interactions. Every system was energy minimized using the conjugant gradient method and subsequently equilibrated with the protein Cα atoms harmonically restrained for 0.5 ns (force constant = 1000 kJ/mol/A2). Production simulations for 50 ns were performed three times for LH and RH packing dimer.
All trajectory analyses were done with GROMACS tools. Visualization and graphics were performed with VMD software (44).
Results
Analysis of TM sequence specificity of DAP12 dimerization by TOXCAT
Although several reports have suggested that the full-length DAP12 forms a covalent dimer, whether this is mediated by the specific motif of the DAP12 TM domain has not been established. To this end, we first employed the TOXCAT system developed by Russ and Engelman, which measures the association of TM helices in the plasma membrane of E. coli. Table 1 depicts the DAP12 TM sequences that were inserted into the TOXCAT chimera. The length of the inserted TM helix is important in determining the degree of CAT expression levels in the TOXCAT assay (45). Therefore, we identified the optimal length for the DAP12 TM helix (DAP12-WT: from residue V42 to residue Y62) in this assay by incrementally deleting single residues from its N-terminal end (see Table S1 and Fig. S1 in the Supporting Material). Three independent TOXCAT measurements were performed on the DAP12-WT with the average 70.5% dimerization affinity shown in Fig. 1 representing a percent of the CAT activity of GpA. The TOXCAT results indicate there is a relatively strong association in the cytoplasmic membrane of E. coli that is comparable to the signal reported for the TM-TM association of some single-pass receptors. We routinely employ established controls for the TOXCAT assay to measure the expression levels of the chimeras and their correct topology because the location of the junctions between the helix and MBP and ToxR is also the influence factor of CAT activity (14). Specifically, western blots of the TOXCAT chimera containing DAP12-WT showed that their expression levels were comparable to those of GpA-WT and GpA-G38I, and maltose complementation assays indicated the chimeric protein inserted into the membrane with the correct topology (Fig. S2).
Table 1.
Sequences of the DAP12 TM helix and its mutants
| DAP12 helix | TM sequence | TOXCAT | MD |
|---|---|---|---|
| DAP12-WT | VLAGIVMGDLVLTVLIALAVY62 | √ | √ |
| D50N | VLAGIVMGNLVLTVLIALAVY | √ | |
| D50E | VLAGIVMGELVLTVLIALAVY | √ | |
| D50R | VLAGIVMGRLVLTVLIALAVY | √ | |
| D50A | VLAGIVMGALVLTVLIALAVY | √ | √ |
| D50S | VLAGIVMGSLVLTVLIALAVY | √ | |
| D50L | VLAGIVMGLLVLTVLIALAVY | √ | √ |
| T54S | VLAGIVMGDLVLSVLIALAVY | √ | |
| T54A | VLAGIVMGDLVLAVLIALAVY | √ | √ |
| T54V | VLAGIVMGDLVLVVLIALAVY | √ | √ |
| G45I | VLAIIVMGDLVLTVLIALAVY | √ | √ |
| G45F | VLAFIVMGDLVLTVLIALAVY | √ | |
| G49I | VLAGIVMIDLVLTVLIALAVY | √ | √ |
| G49F | VLAGIVMFDLVLTVLIALAVY | √ | |
| D50LT54V | VLAGIVMGLLVLVVLIALAVY | √ | |
| D50AT54A | VLAGIVMGALVLAVLIALAVY | √ | |
| G45IG49I | VLAIIVMIDLVLTVLIALAVY | √ |
Figure 1.
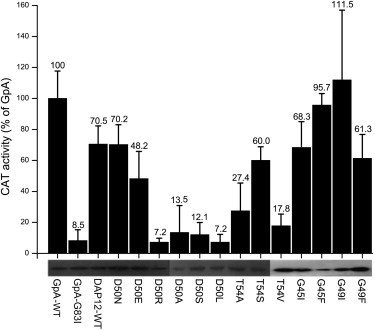
DAP12-WT TM domain self-associated in the Escherichia coli cell membrane and the effect of site-specific mutagenesis in the GxxxG motif and polar residues D50 and T54 of the DAP12-WT TM domain on CAT activity. The enzymatic activity of CAT induced by self-association of the target TM domain and expressed as the percentage of that induced by the GpA-WT TM domain. The GpA-WT and GpA-G83I constructs were used as positive and negative controls, respectively. Black bars represent the CAT activity quantified from cleared lysates, and error bars represent the standard error for three measurements of each lysate. The lower panel shows the expression levels of chimeric ToxR-TM-MBP proteins probed by Western blot.
By inspecting the DAP12-WT and comparing it with previously reported association motifs (46–48), the DAP12 TM domain contains a GxxxG motif and the polar residues D50 and T54. To assess the role of the GxxxG motif and polar residues in the DAP12 TM domain in mediating its dimerization, site-specific mutagenesis was used to alter specific amino acids in the chimera containing the DAP12-WT in TOXCAT. The individual mutational sequences are shown in Table 1. An effective strategy to probe the role of the GxxxG motif in TM association is to introduce steric clashes by changing Gly to amino acids with large side chains. As shown in Fig. 1, mutations G45I, G45F, G49I, and G49F did not disrupt the dimerization of the DAP12 TM domain. On the contrary, the mutations G45F and G49I increased the CAT activity of DAP12-WT by 40%. All these TOXCAT data ruled out the possibility of a role for the GxxxG motif in DAP12 TM homodimerization. Furthermore, mutating polar residues to nonpolar residues (D50L, D50A, T54L, and T54V) caused complete disruption of the DAP12 TM dimer, indicating that the polar residues mediate the dimerization of DAP12 TM domain in membranes. The mutations D50R, D50S, D50N, D50E, and T54S show that the polar residue side chains are also an important determinant of dimerization. Because both the mutations D50R and D50S completely abrogate dimerization of the DAP12 TM and mutations with a similar side chain (D50N, D50E, and T54S) support dimerization of DAP12 TM. These results lead us to the conclusion that it is not the GxxxG motif in the TM domain that mediates the dimerization of DAP12, but the polar residues (D50 and T54) of the DAP12 TM.
CG simulation of DAP12 TM dimer
To rationalize the TOXCAT results from a structural perspective, we first performed a CG MD simulation on the DAP12 TM dimer to gain further insight into the dynamic behavior of the TM-TM association within an explicit bilayer-water environment (see Materials and Methods for details). Initially, we used the same DAP12 TM sequences (DAP12-WT) as used in our TOXCAT experiments, which are also predicted by the TM prediction program. The initial structure of the DAP12 TM single chain was obtained from the NMR structure and positioned in a parallel orientation into a preformed DPPC bilayer with the helices separated by a backbone distance of ∼55 Å. This separation between the helices ensures that the initial position does not favor artificial dimer formation. During the simulation, the helices diffuse randomly in the bilayer relative to one another; until an encounter leads to the formation of a stable helix dimer occurs. As observed in earlier self-assembly simulations (Fig. 2 A) the DAP12 TM helices spontaneously form a long-lasting helix dimer within a few hundred nanoseconds. The DAP12 TM dimer reveals a bimodal crossing angle distribution (LH, negative; RH, positive) as shown in Fig. 2 B. Compared with the RH packing mode, the LH packing mode shows a much more narrow distribution with a mean crossing angle of ∼20°, which is in accordance with the observation of the DAP12-TM NMR structure. Furthermore, the trajectories for all of the CG-WT simulations (see Table 1) were concatenated and the backbone particles of the reference helix from each frame was fitted onto a reference structure, and the probability density of the backbone particles of the TM helix at a given point in the bilayer plane was calculated (Fig. 2 C). The DAP12 TM presented only one probability density maxima in the spatial distribution containing both LH and RH structures. This reveals that both the LH packing and RH packing have a similar interface in the CG simulations. The LH structure (Fig. 2 D) includes the same DAP12 dimerization interface from the NMR structure in which the two aspartates move close to each other. Although in the RH structure (Fig. 2 D) with the highest probability of the negative helix crossing angle, the threonine pair are as close as the aspartate pair, and helices did not diverge slightly below T54 as in the NMR structure. Examination of the CG structures of the DAP12-WT LH dimer (Fig. 2 D) suggests that the helices are assembled in a fashion with the D50 and T54 residues forming the helix-helix interface, as indicated by the NMR structures.
Figure 2.
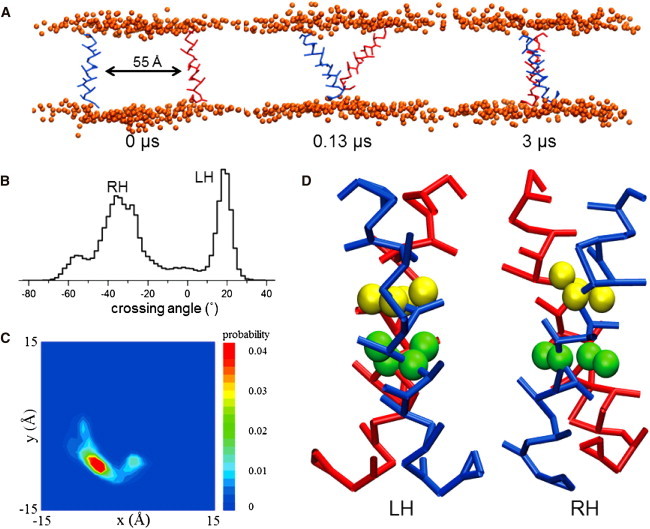
CG simulations to explore DAP12-WT helix homodimer. (A) The DAP12-WT helices were parallel and separated by ∼55 Å in the DPPC bilayer of the initial structure. During the 3 μs CG-WT simulations, they interact with each other and form a homodimer. (B) All the dimer trajectories of CG-WT simulations were converged and crossing angles were analyzed. The positive crossing angle corresponds to LH helix packing and the negative crossing angle corresponds to RH packing. (C) The spatial distribution of helix for the CG-WT simulations. The backbone particles were fitted to a reference structure. This diagram shows the contact probability density between the two TM helix backbone particles in the bilayer plane. The blue and red represents low probability and high probability at that point. The LH and RH dimer have only one maxima area in this distribution. (D) The LH and RH structures were selected to be representative of the modes in the crossing angle distributions. The backbone and side-chain particles of the D50 and T54 particles are in yellow and green.
Atomistic simulation of DAP12 TM dimer
For the purpose of further improving the model of the DAP12 TM helix dimer from the CG simulation self-assembly simulations, as well as to check on the stability of RH and LH conformation, the CG structures of the DAP12 TM dimer were converted to full atomistic models, using the CG2AT fragment-based procedure. This method has been used to refine membrane protein systems by performing subsequent atomistic (AT) simulations of the CG models of the TM helix dimer. We performed simulations of 50 ns duration for two WT dimer structures (LH and RH packing in the CG simulation) generated by the CG2AT procedure, in triplicate (Fig. 3).
Figure 3.
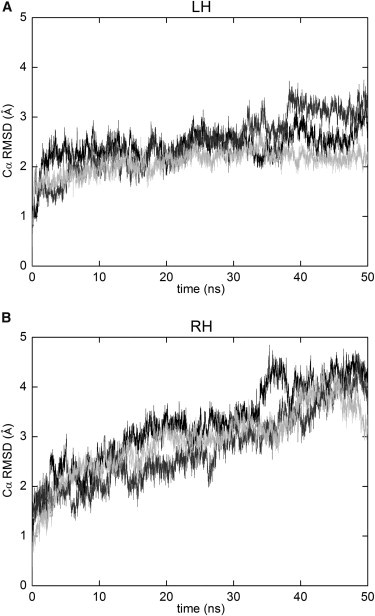
The conformational stability of DAP12-WT helix homodimer in each atomistic simulation is analyzed based on the Cα RMSD from the initial structure as a function of time. The atomistic LH (A) and RH (B) representative initial helix dimer were conversed from the CG structures picked in the crossing angle distribution.
The Cα root mean-squared deviations (RMSD) from the initial structures were monitored over the course of the AT simulations, and the conformational stability of the two WT dimer structures (AT-LH and AT-RH) were assessed. In Fig. 3, the Cα RMSD for the AT-LH dimer rapidly reaches a plateau at 3 Å, indicating the AT-LH dimer is a stable model without conformational change from the initial structures in the AT simulations. In contrast, the Cα RMSD for the AT-RH dimer simulation rises gradually over the 50 ns duration, reaching ∼4 Å. This indicates more conformational drifts occurring in the unstable AT-RH model simulations.
Likewise, the helix crossing angle distribution in each of the AT-LH atomistic MD simulations is relatively narrow, changing no more than 20° (Fig. S3 A). In contrast, the crossing angle distribution was slightly wider for the AT-RH dimer simulations compared with the AT-LH dimer, ranging from −43° to 8° for the AT-RH dimer (Fig. S3 B), and from +10° to +45° for the AT-LH dimer. This suggests that the AT-LH dimer model forms a tighter packing interface compared with the AT-RH dimer model.
Furthermore, we compared the most stable structure by the AT simulation to the structure from NMR studies of the DAP12 TM dimer. Both structures have a crossing angle of 20° and a helix-helix interface formed by the same polar residues D50 and T54 (Fig. 4).
Figure 4.
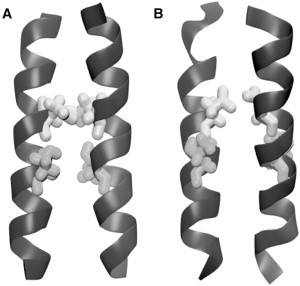
Comparison of the helix dimer interface of the NMR (2L34) structure (A) and a structure from the end of one of the AT-LH simulations (B). The D50 (white) are shown to pack against each other in the interface and the side chains of T54 (gray) stabilize the D50 around the interface. These key interactions are shown in both the NMR and the simulation structure.
Mutational effect on DAP12 TM dimerization
The TOXCAT results suggest that instead of the mutations in the GxxxG motif, the mutations in the polar residues, which mediate the dimerization of DAP12 TM, disrupt the helix dimerization. To replicate the mutational effects on the DAP12-WT dimerization in the E. coli cell membrane, we explored the structure and conformational stability of these mutants via CG simulation. We first compared the distribution of the helix crossing angles for the various mutants observed in the whole CG ensembles (Fig. 5). Thus, all six disruptive mutants (D50L, T54V, D50LT54V, D50A, T54A, and D50AT54A) result in a significant shift, relative to the DAP12-WT, from a LH to a RH distribution. The polar contacts were perturbed and the distribution showed more RH frames than the DAP12-WT dimer, suggesting the weakness of these interactions evidently reduces the strength of the dimerization and perturbs the LH crossing angle bias.
Figure 5.
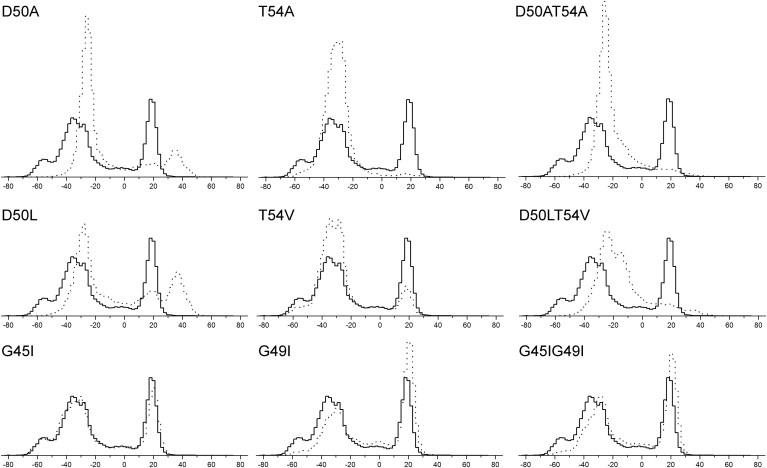
Mutants of DAP12-WT helix homodimer crossing angle histograms. Helix crossing angle histograms. For each set of simulations, all the dimer frames were merged. The helix crossing angles were evaluated from these merged dimer trajectories. The positive crossing angle corresponds to LH helix packing and the negative crossing angle corresponds to RH packing. The WT is drawn in a solid line, and the mutants are drawn in a dashed line.
The crossing angle distribution of the nondisruptive mutants G45I, G49I, and G45IG49I is almost identical to that of the WT simulations. Moreover, these nondisruptive mutations significantly increase the LH dimer frames, implying their dimerization was moderately reinforced.
The polar side-chain residues, D50 and T54, constitute the DAP12-WT dimerization motif and have been shown in previous experiments to be important for dimer stability. In our CG simulations, the side chain of T54 is able to interact with the D50 residues on the same helix of the LH packing dimer, corresponding with the observation in the NMR structures (Fig. 4). However, the side chain of T54 is also able to respectively interact with D50 or T54 on the opposing helix of the RH packing dimer (Fig. S4). These interactions of T54 partially perturb the critical D50-D50 interactions and account for the observation that the amount of the RH packing dimer is greater compared with the LH packing in dimer to some degree, resulting in a bimodal distribution (Fig. 2 B). Comparing the RMSD of all the sets of CG simulation relative to the NMR CG structure as the reference structure (Fig. S5), the WT and nondisruptive mutants have more frames of low RMSD (∼1.8 Å), but the disruptive mutants have a much higher RMSD. This observation suggests the nondisruptive mutants can form dimers similar to the NMR structure, whereas the disruptive mutants seldom form.
The spatial distribution comparison of all the mutants is shown in Fig. 6. For the nondisruptive mutants (G45I, G49I, and G45IG49I) the spatial distribution is similar to the WT and they all favor the same region of the fixed helix. For the disruptive mutants (D50L, T54V, D50LT54V, D50A, T54A, and D50AT54A) the spatial distribution is diverse. In the D50L and D50A mutants, the vital D50-D50 polar interaction is lost, and the favorable contact of DAP12-WT has entirely changed to a different side of the reference helix. A symmetrical distribution is observed in the double mutants D50LT54V and D50AT54A, where the two polar residues are replaced. Consequently, the helices of the double mutants do not have a preferred packing site. Without the Thr polar side-chain interaction in T54V and T54A, the preservation of D50 in this mutant partially maintains some spatial contact relative to DAP12-WT, but it is relatively unstable. This certifies that the D50 residue is more important than the T54 residue in maintaining this strong helix-helix interface.
Figure 6.
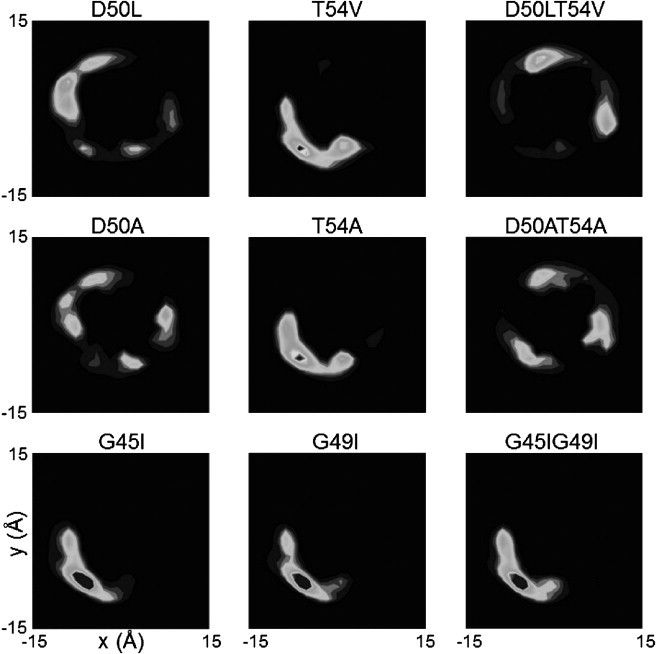
Spatial distributions of all the mutants of DAP12-WT helix. The backbone particles were fitted to a unique reference structure used in Fig. 2C. This diagram shows the contact probability density between the two TM helix backbone particles in the bilayer plane; white and black represent low and high probability.
Discussion
Understanding the dimerization structure of the DAP12 TM is likely to provide relevant insight into the assembly of a variety of activating immune receptors with adaptor proteins. In our studies, the importance of characterizing the nature of the interactions of membrane proteins with their lipid bilayer environment is taken into consideration. A combination of TOXCAT and MD simulation has revealed the dynamics and dimerization of DAP12 TM helix in the lipid bilayer membrane. First, we apply the TOXCAT assay to experimentally study the effects of mutations of polar residues D50 and T54 on the molecular association of DAP12 TM-TM helix in a natural lipid bilayer system (E. coli inner cell membrane). Second, we performed a simulation strategy that characterizes the structural differences between the mutant and the WT DAP12 homodimers, as well as identifying the amino acids located at the helical interface that may promote the DAP12 homodimerization. Overall, the computational and experimental results are in excellent agreement. Combining the TOXCAT with molecular simulation, our results show that instead of the GxxxG motif, which is also present in the DAP12 TM domain, the polar residues play an important role in the DAP12 TM-TM association.
With the development of progress in simulation algorithms, MD simulation has been used to study the TM-TM association in a well-defined lipid bilayer environment (16,17). However, most MD simulations focus on the classical GxxxG dimerization motif within the TM-TM association. Our current studies give the first, to our knowledge, examples to study polar residues’ role on the TM-TM association using MD simulation. Alignment of the TM domain of DAP12 from a variety of different species demonstrated that the DAP12 TM domain contains the conserved polar residues D50 and T54 (29). Notably, the DxxxT motif is also present in the CD3ε and CD3δ chains associated with TCR (49), whereas the ExxS in CD3γ remind us of the dimerization motif QxxS reported by Sal-Man and Shai (50). Consistent with the NMR results, the DAP12 TM helix dimer was prone to adopt an ∼20° LH conformation by crossing angle distribution analysis, and the association interface was apparently dominated by the key polar residues D50 and T54, according to the examination of helix-helix contacts (Fig. S6). Polar residues D50 and T54 are forming hydrogen bonds to drive DAP12 TM dimerization, but this polar-polar interaction is not as strong and tight as the docking and packing of the small residues GxxxG motif and GxxxG-like motifs, which was observed from previous TOXCAT results. Moreover, the threonine pair is not as close as the aspartate pair; and as a consequence of the relative loose conformation, helices of DAP12 TM dimer are prone to rotate. These speculations could postulate why there is also a RH population in the CG simulation. Although the RH conformation accounts for some proportion of all the dimers, the span of it was so wide (>40°) there was not a single dominated peak to compare to the LH conformation. To gain greater accuracy of the DAP12 TM helix dimer, we further employed AT-MD simulation with the initial structures, which we converted from our previous representative CG models. As predicted, the LH dimer was stable with little conformational change from the initial structures using Cα RSMD calculation throughout the simulations, in contrast with the RH dimer. In all three AT-RH simulations, we found the RH dimer trend to rotate to the LH conformation as the crossing angle of the dimer increases from negative to positive (Fig. S3 B). This interesting variation gives a manifest evidence of the dominant LH conformation of DAP12 TM, indicating the stable LH dimer is more probable than the RH dimer. These key polar residues in our DAP12-WT AT-LH structure had the same spatial positions in comparison with the NMR structure, providing solid interaction interface confirmation of D50 and T54.
The same CG simulation approach was used to characterize the effect of the mutations on the stability and structure. For the nondisruptive mutants G45I, G49I, and G45IG49I, the crossing angle distribution and spatial distribution is almost identical to that of the WT simulations. As with the six disruptive mutants, the crossing angle had shifted significantly from LH to RH, thus completely disturbing the DAP12-WT TM helix interaction into an unstable state. The spatial distribution of the disruptive mutants revealed diverse results that all mutants that involved Asp would largely shift the interaction interface, whereas T54A and T54V showed similarity to the WT. Based on a recent study of NMR structure refinement of DAP12-NKG2C, the T54 residues in the DAP12 TM dimer could form intra- and interhelical H-bonds to keep the DAP12 TM dimer compact, further stabilizing the TM physical contact (51). Given that the polar interaction is not as strong as the GxxxG motif, which could dock and pack tightly, the DAP12 TM dimer without the disulfide bond in the ectodomain would be able to rotate and switch between RH and LH formations. Notably in our AT-MD simulations we found a similar effect, giving a reasonable explanation for the importance of T54 in DAP12-TM dimer. All these simulation results are not only well matched to the experimental data but also shed light on the association mechanism of DAP12-TM helix.
As our knowledge of membrane protein folding evolved, searching for sequence motifs to mediate dimerization cannot solve this problem alone. Because the presence of known sequence motifs alone does not guarantee interactions, TM sequences that do not contain any recognizable motifs can interact, and lipid bilayer properties may significantly modulate the strength of a sequence-specific transmembrane helix-helix interaction as well (47,48). In this study, we adopt a desirable multiscale approach, whereby CG-MD simulations are used to explore TM-TM interactions efficiently, yielding system configuration that could then be converted to atomistic resolution for accurate analysis, and our approach of employing both experimental and computational methods and comparing mutational effects in both can be potentially applicable for the ternary interaction between DAP12 dimer and the associating receptors TM domain and many other proteins.
Acknowledgments
We thank Prof. Renhao Li, Saburo Aimoto, and Baeyens Jan for helpful discussions.
This work was supported by the National Basic Research Program of China (973 program) (2013CB910700) and the National Natural Science Foundation of China (No. 20932003, No. 21002007). This work was performed under the International Collaborative Research Program of the Institute for Protein Research, Osaka University.
Supporting Material
References
- 1.Popot J.L., Engelman D.M. Membrane protein folding and oligomerization: the two-stage model. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 2.Melnyk R.A., Partridge A.W., Deber C.M. Transmembrane domain mediated self-assembly of major coat protein subunits from Ff bacteriophage. J. Mol. Biol. 2002;315:63–72. doi: 10.1006/jmbi.2001.5214. [DOI] [PubMed] [Google Scholar]
- 3.Gaddie K.J., Kirley T.L. Conserved polar residues stabilize transmembrane domains and promote oligomerization in human nucleoside triphosphate diphosphohydrolase 3. Biochemistry. 2009;48:9437–9447. doi: 10.1021/bi900909g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escher C., Cymer F., Schneider D. Two GxxxG-like motifs facilitate promiscuous interactions of the human ErbB transmembrane domains. J. Mol. Biol. 2009;389:10–16. doi: 10.1016/j.jmb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Treutlein H.R., Lemmon M.A., Brünger A.T. The glycophorin A transmembrane domain dimer: sequence-specific propensity for a right-handed supercoil of helices. Biochemistry. 1992;31:12726–12732. doi: 10.1021/bi00166a003. [DOI] [PubMed] [Google Scholar]
- 6.Simmerman H.K.B., Kobayashi Y.M., Jones L.R. A leucine zipper stabilizes the pentameric membrane domain of phospholamban and forms a coiled-coil pore structure. J. Biol. Chem. 1996;271:5941–5946. doi: 10.1074/jbc.271.10.5941. [DOI] [PubMed] [Google Scholar]
- 7.Moore D.T., Berger B.W., DeGrado W.F. Protein-protein interactions in the membrane: sequence, structural, and biological motifs. Structure. 2008;16:991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bormann B.J., Engelman D.M. Intramembrane helix-helix association in oligomerization and transmembrane signaling. Annu. Rev. Biophys. Biomol. Struct. 1992;21:223–242. doi: 10.1146/annurev.bb.21.060192.001255. [DOI] [PubMed] [Google Scholar]
- 9.Wimley W.C. Protein folding in membranes. Biochim. Biophys. Acta. 2012;1818:925–926. doi: 10.1016/j.bbamem.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Chen L., Merzlyakov M., Hristova K. Energetics of ErbB1 transmembrane domain dimerization in lipid bilayers. Biophys. J. 2009;96:4622–4630. doi: 10.1016/j.bpj.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anbazhagan V., Cymer F., Schneider D. Unfolding a transmembrane helix dimer: a FRET study in mixed micelles. Arch. Biochem. Biophys. 2010;495:159–164. doi: 10.1016/j.abb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Berger B.W., Kulp D.W., DeGrado W.F. Consensus motif for integrin transmembrane helix association. Proc. Natl. Acad. Sci. USA. 2010;107:703–708. doi: 10.1073/pnas.0910873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russ W.P., Engelman D.M. TOXCAT: a measure of transmembrane helix association in a biological membrane. Proc. Natl. Acad. Sci. USA. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei P., Liu X., Luo S.Z. The dimerization interface of the glycoprotein Ibβ transmembrane domain corresponds to polar residues within a leucine zipper motif. Protein Sci. 2011;20:1814–1823. doi: 10.1002/pro.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braun R., Engelman D.M., Schulten K. Molecular dynamics simulations of micelle formation around dimeric Glycophorin A transmembrane helices. Biophys. J. 2004;87:754–763. doi: 10.1529/biophysj.104.040279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuthbertson J.M., Bond P.J., Sansom M.S.P. Transmembrane helix-helix interactions: comparative simulations of the glycophorin a dimer. Biochemistry. 2006;45:14298–14310. doi: 10.1021/bi0610911. [DOI] [PubMed] [Google Scholar]
- 17.Marrink S.J., de Vries A.H., Mark A.E. Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B. 2004;108:750–760. [Google Scholar]
- 18.Sansom M.S.P., Scott K.A., Bond P.J. Coarse-grained simulation: a high-throughput computational approach to membrane proteins. Biochem. Soc. Trans. 2008;36:27–32. doi: 10.1042/BST0360027. [DOI] [PubMed] [Google Scholar]
- 19.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen S.O., Lopez C.F., Srinivas G., Klein M.L. Coarse grain models and the computer simulation of soft materials. J. Phys.: Condens. Matter. 2004;16:R481–R512. [Google Scholar]
- 21.Bond P.J., Sansom M.S.P. Insertion and assembly of membrane proteins via simulation. J. Am. Chem. Soc. 2006;128:2697–2704. doi: 10.1021/ja0569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevens T.J., Arkin I.T. Are membrane proteins “inside-out” proteins? Proteins: Struct. Funct. Genet. 1999;36:135–143. doi: 10.1002/(sici)1097-0134(19990701)36:1<135::aid-prot11>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 23.Bañó-Polo M., Baeza-Delgado C., Mingarro I. Polar/Ionizable residues in transmembrane segments: effects on helix-helix packing. PLoS ONE. 2012;7:e44263. doi: 10.1371/journal.pone.0044263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanier L.L., Corliss B.C., Phillips J.H. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 25.Lanier L.L., Corliss B.C., Phillips J.H. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998;8:693–701. doi: 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 26.Lanier L.L. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Humphrey M.B., Lanier L.L., Nakamura M.C. Role of ITAM-containing adapter proteins and their receptors in the immune system and bone. Immunol. Rev. 2005;208:50–65. doi: 10.1111/j.0105-2896.2005.00325.x. [DOI] [PubMed] [Google Scholar]
- 28.Lanier L.L. DAP10- and DAP12-associated receptors in innate immunity. Immunol. Rev. 2009;227:150–160. doi: 10.1111/j.1600-065X.2008.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Call M.E., Wucherpfennig K.W. Common themes in the assembly and architecture of activating immune receptors. Nat. Rev. Immunol. 2007;7:841–850. doi: 10.1038/nri2186. [DOI] [PubMed] [Google Scholar]
- 30.Call M.E., Wucherpfennig K.W., Chou J.J. The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nat. Immunol. 2010;11:1023–1029. doi: 10.1038/ni.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo S.Z., Li R. Specific heteromeric association of four transmembrane peptides derived from platelet glycoprotein Ib-IX complex. J. Mol. Biol. 2008;382:448–457. doi: 10.1016/j.jmb.2008.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 33.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 34.Monticelli L., Kandasamy S.K., Marrink S.J. The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 35.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 36.Psachoulia E., Fowler P.W., Sansom M.S. Helix-helix interactions in membrane proteins: coarse-grained simulations of glycophorin a helix dimerization. Biochemistry. 2008;47:10503–10512. doi: 10.1021/bi800678t. [DOI] [PubMed] [Google Scholar]
- 37.Chng C.P., Tan S.M. Leukocyte integrin αLβ2 transmembrane association dynamics revealed by coarse-grained molecular dynamics simulations. Proteins. 2011;79:2203–2213. doi: 10.1002/prot.23044. [DOI] [PubMed] [Google Scholar]
- 38.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 39.Marrink S.J., Periole X., de Vries A.H. Comment on “On using a too large integration time step in molecular dynamics simulations of coarse-grained molecular models” by M. Winger, D. Trzesniak, R. Baron and W. F. van Gunsteren, Phys. Chem. Chem. Phys., 2009, 11, 1934. Phys. Chem. Chem. Phys. 2010;12:2254–2256. doi: 10.1039/b915293h. 2257–2258. [DOI] [PubMed] [Google Scholar]
- 40.Stansfeld P.J., Sansom M.S.P. From coarse-grained to atomistic: a serial multi-scale approach to membrane protein simulations. J. Chem. Theory Comput. 2011;7:1157–1166. doi: 10.1021/ct100569y. [DOI] [PubMed] [Google Scholar]
- 41.Oostenbrink C., Villa A., van Gunsteren W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 42.Kukol A. Lipid models for united-atom molecular dynamics simulations of proteins. J. Chem. Theory Comput. 2009;5:615–626. doi: 10.1021/ct8003468. [DOI] [PubMed] [Google Scholar]
- 43.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 44.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 45.Li R., Gorelik R., Bennett J.S. Dimerization of the transmembrane domain of Integrin alphaIIb subunit in cell membranes. J. Biol. Chem. 2004;279:26666–26673. doi: 10.1074/jbc.M314168200. [DOI] [PubMed] [Google Scholar]
- 46.Fink A., Sal-Man N., Shai Y. Transmembrane domains interactions within the membrane milieu: principles, advances and challenges. Biochim. Biophys. Acta. 2012;1818:974–983. doi: 10.1016/j.bbamem.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 47.Cymer F., Veerappan A., Schneider D. Transmembrane helix-helix interactions are modulated by the sequence context and by lipid bilayer properties. Biochim. Biophys. Acta. 2012;1818:963–973. doi: 10.1016/j.bbamem.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 48.Li E., Wimley W.C., Hristova K. Transmembrane helix dimerization: beyond the search for sequence motifs. Biochim. Biophys. Acta. 2012;1818:183–193. doi: 10.1016/j.bbamem.2011.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Call M.E., Schnell J.R., Wucherpfennig K.W. The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127:355–368. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sal-Man N., Gerber D., Shai Y. The identification of a minimal dimerization motif QXXS that enables homo- and hetero-association of transmembrane helices in vivo. J. Biol. Chem. 2005;280:27449–27457. doi: 10.1074/jbc.M503095200. [DOI] [PubMed] [Google Scholar]
- 51.Cheng X., Im W. NMR observable-based structure refinement of DAP12-NKG2C activating immunoreceptor complex in explicit membranes. Biophys. J. 2012;102:L27–L29. doi: 10.1016/j.bpj.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


