Abstract
In this work, we investigate the feasibility to estimating causal interactions between brain regions based on multivariate autoregressive models (MAR models) fitted to magnetoencephalographic (MEG) sensor measurements. We first demonstrate the theoretical feasibility of estimating source level causal interactions after projection of the sensor‐level model coefficients onto the locations of the neural sources. Next, we show with simulated MEG data that causality, as measured by partial directed coherence (PDC), can be correctly reconstructed if the locations of the interacting brain areas are known. We further demonstrate, if a very large number of brain voxels is considered as potential activation sources, that PDC as a measure to reconstruct causal interactions is less accurate. In such case the MAR model coefficients alone contain meaningful causality information. The proposed method overcomes the problems of model nonrobustness and large computation times encountered during causality analysis by existing methods. These methods first project MEG sensor time‐series onto a large number of brain locations after which the MAR model is built on this large number of source‐level time‐series. Instead, through this work, we demonstrate that by building the MAR model on the sensor‐level and then projecting only the MAR coefficients in source space, the true casual pathways are recovered even when a very large number of locations are considered as sources. The main contribution of this work is that by this methodology entire brain causality maps can be efficiently derived without any a priori selection of regions of interest. Hum Brain Mapp, 2013. © 2012 Wiley Periodicals, Inc.
Keywords: MEG, functional connectivity, multivariate autoregressive models, granger causality, partial directed coherence
INTRODUCTION
The importance of studying interactions between specialized areas in the human brain has been increasingly recognized in recent years [Schnitzler and Gross,2005a,b; Schnitzler et al.,2000; Schoffelen et al.,2005,2008]. Magnetoencephalography (MEG) is particularly suited for connectivity studies as it combines a good spatial resolution with high temporal resolution. The high temporal resolution affords the investigation of transient coupling and is a prerequisite to study frequency dependent coupling. A large number of measures for the quantification of neural interactions have been introduced over the years. For these various measures it is custom to distinguish between functional and effective connectivity. Functional connectivity measures assess interactions by means of similarities between time series (e.g., correlation and coherence) or transformations of these time series (e.g., phase synchronization and amplitude correlation). In contrast, effective connectivity methods are used to study the causal effect of one brain area on another brain area.
Besides the distinction between functional and effective connectivity one has to be aware that connectivity analysis can be performed at the sensor‐level or the source‐level. In the first case, connectivity measures are evaluated on the time series recorded by MEG/EEG sensors. In the second case, connectivity measures are evaluated on time series that represent the activity of individual brain areas. Unfortunately, the interpretation of sensor connectivity results is difficult because of the complex and often diffuse sensitivity profiles of MEG/EEG sensors [Schoffelen and Gross,2009]. Significant connectivity between (even distant) sensors cannot be easily assigned to underlying brain areas, may be spurious, and can be affected by power modulations of nearby or distant brain areas [Schoffelen and Gross,2009]. These negative effects can be reduced (though not abolished) by performing connectivity analysis in source space. Most MEG/EEG source connectivity methods are based on functional connectivity measures such as coherence or phase synchronization [Gross et al.,2002; Hoechstetter et al.,2004; Jerbi et al.,2007; Lachaux et al.,1999; Lin et al.,2004; Pollok et al.,2004,2005; Timmermann et al.,2003]. Effective connectivity in source space has been studied with dynamic causal modeling (DCM) [David et al.,2006; Kiebel et al.,2009] or Granger causality [Astolfi et al.,2005; Gómez‐Herrero et al.,2008].
Here, we present and test a new efficient method for Granger causality analysis in source space. Granger causality is a concept from economics that quantifies the causal effect of one time series on another time series. Specifically, if the past of time series x improves the prediction of the future of time series y time series x is said to granger‐cause y. Classically, Granger causality is defined in the time domain, but a frequency domain extension has been proposed [Geweke,1982]. Granger causality has also been extended from its original pairwise form into a multivariate formulation in both the time and frequency domains, known as conditional Granger causality [Chen et al.,2006,2009; Geweke,1984]. This methodology is comparative in the sense that in a multivariate system if one investigates if y is causing x, then a model of x based on every variable including y is compared with a model of x based on every variable excluding y. In simple terms if inclusion of y reduces significantly the variance of the model of x as compared to the variance of the model of x when y is excluded then y is assumed to cause x. Several other multivariate metrics derived from Granger causality have been suggested, such as partial directed coherence (PDC) [Baccalá and Sameshima,2001] and directed transfer function [Kaminski and Blinowska,1991]. These metrics are estimated in the frequency domain and are thus frequency specific. One of their main differences with conditional Granger causality is that they are not comparative methods but they are computed directly from the multivariate model built based on all the variables in the system.
Source space Granger causality analysis is typically performed in the following way. First, regions of interest (ROIs) are selected. Second, the activation time series are computed for all ROIs. Third, a multivariate autoregressive model is computed for these time series and measures of Granger causality are computed. The most significant drawback of this approach is that a large number of potential activation sources correspond to a large number of projected activation time‐series. This is prohibitive for the derivation of numerically robust MAR models without the assumption of sparse connectivity. [Haufe et al.,2010; McQuarrie and Tsai,1998; Valdés‐Sosa et al.,2005] For example, dividing the brain volume into a regular 6 mm grid leads to roughly 10,000 voxels. In addition, Granger causality computation for a different set of ROIs requires time consuming computations because Steps 2 and 3 in the procedure mentioned earlier need to be repeated. The computational complexity precludes a tomographic mapping of Granger causality.
To bypass these limitations, we investigate an alternative approach, which entails the derivation of the MAR model directly on MEG sensor data and its projection into the source space. In this method the modeling process is performed in sensor space, which has moderate dimensionality as compared to the high‐dimensional source space. This leads to greater model robustness as well as significantly reduced computation times. Feasibility of a similar approach for EEG data has already been shown in [Gómez‐Herrero et al.,2008], where the multivariate model was projected onto a small number of locations in source space identified by independent component analysis (ICA) of the residuals of the MAR model and localized by swLORETA [Palmero‐Soler et al.,2007]. Causality was inferred using the directed transfer function metric (DTF). In our work, we demonstrate the feasibility of the methodology when the MAR model is projected in the entire brain volume without any a priori assumption or estimation of the activity locations.
The main advantage of this approach is that all the voxels inside the brain volume can be investigated in terms of causality, something not practical with the traditional approach. This method also offers benefits in terms of data compression as the elements that need to be projected are the coefficients, which are typically significantly less than the data points used to derive them and which would be projected in the traditional case. Another advantage is that the derivation of the MAR model at the sensor space is much more robust, because of the moderate number of variables, than the derivation of the MAR model on projected time‐series in a very large number of brain locations. Additionally, even if different ROIs are recursively selected to examine different network topologies in the brain, the sensor space MAR model is always the same and the only thing that changes is the locations where the model is projected. In the traditional approach, one would have each time to project the sensor time‐series in the new set of brain locations and then build the MAR model again. Finally, due to the computational efficiency of this methodology, application of statistical inference methods on entire brain causality maps from MEG data is feasible.
To infer causality, PDC and the coefficients of the MAR model themselves are used. Although, conditional Granger causality would be, in terms of theory, a more robust choice because of its intrinsic normalization, its computational load for a very large number of considered source locations makes its use problematic. In a traditional approach, if 10,000 voxels are considered, 10,001 multivariate models must be computed. One including all the 10,000 projected voxel time‐series and 10,000 models, each one with one voxel time‐series excluded. In the proposed methodology, where the model is built on the sensor level and only the coefficients are projected, in order to implement conditional Granger causality, again 10,001 models must be built at the sensor level. One on the original sensor data and 10,000 models, each with the effect of one voxel extracted through the derived inverse solution. Then the coefficients of these 10,001 models must be projected in source space. This imposes a heavy computational load.
Also, due to the fact that in each of the 10,000 models the effect of one voxel is extracted through the derived inverse solution, under the condition that the number of sensors is much smaller than the number of voxels, the projected activity will be diffused around the voxels of actual activity. This simply means that even if one voxel's activation effect is excluded from the sensor data, in the context of Geweke's measures computation, the causal pairing will be modeled by the effect of the neighboring voxels.
Another issue regarding the conditional Granger causality in the frequency domain is that is based on the transfer function of the model, which is the inverse of the z‐transform of the MVAR coefficients across model order. For each of the 10,000 models the size of this matrix is 9,999 × 9,999(10,000 × 10,000 for the entire brain model). Inversion of such a large matrix, given also the colinearities because of the projection through the inverse solution, can be very problematic and can lead to singular inverse matrices.
PDC has the implementational advantage that it is computed directly from the coefficients of one MAR model with all the variables included and that it does not require any inversion. Thus, only one model needs to be built at the sensor level, and after the coefficients are projected in source space, PDC can be efficiently computed for a wide range of frequencies. However, due to its semiarbitrary normalization it can only confidently be used to compare causality between voxel pairs that have the same causal voxel [Baccalá and Sameshima,2001].
Because of this drawback of PDC, also the projected MAR model coefficients are examined directly without any normalization. The fact that no normalization is applied means that in this approach causality is not bounded. Also when a continuous linear system with linear coefficients matrix A is periodically sampled with sampling frequency f, the resulting discrete linear coefficients are approximated as
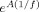 . This means that the discrete coefficients change in amplitude according to the sampling frequency. Nevertheless the aim of examining the MAR model coefficients is to examine if within the same dataset the causal information is correctly represented in the MAR model coefficients when they are derived at the sensor‐level and then projected to a very large number of voxels inside the brain. This examination of the coefficients is only performed in the time‐domain. This approach is used to identify areas inside the entire brain, which are involved in causal interactions. These specific brain areas could then be separately examined with theoretically more robust causality metrics such as the conditional Granger causality.
. This means that the discrete coefficients change in amplitude according to the sampling frequency. Nevertheless the aim of examining the MAR model coefficients is to examine if within the same dataset the causal information is correctly represented in the MAR model coefficients when they are derived at the sensor‐level and then projected to a very large number of voxels inside the brain. This examination of the coefficients is only performed in the time‐domain. This approach is used to identify areas inside the entire brain, which are involved in causal interactions. These specific brain areas could then be separately examined with theoretically more robust causality metrics such as the conditional Granger causality.
First, our proposed approach is investigated theoretically. Subsequently, the method is validated by simulations where pseudo‐MEG data with added noise, uncorrelated, and spatiotemporally correlated, is produced from simulated neural activity in a small number of predefined locations inside the brain with specified causality structure. We show that the PDC reconstructed from the source projection of the MAR model coefficients, is very similar to the PDC, extracted from the simulated source signals directly.
The second part of this work is concerned with the investigation of the causality information that can be derived in the case when a very large number of voxels are considered as potential sources. First, the causality information recovered by PDC is investigated. Then the causality information recovered directly from the MAR model coefficients is investigated. The motivation for the latter comes from the fact that PDC, due to the way it is normalized, is very sensitive to the Signal‐to‐Noise ratio and may not be suitable for applications with very large numbers of voxels [Baccalá and Sameshima,2006; Faes et al.,2010; Schelter et al.,2006,2009]. Here the feasibility of using the model coefficients directly is investigated and it is demonstrated that causality information can be extracted more precisely than with PDC, when a very large number of voxels is considered. Within this context a preliminary evaluation of this methodology is performed with real data from a simple motor planning experiment.
METHODS
Among the many variants of source localization techniques, linear inverse solutions represent an important class of methods because of their efficient computation and numerical stability. These methods are typically used to perform the linear transformation of sensor time series into source space [Baillet et al.,2001; Gross and Ioannides,1999; Hämäläinen,1992]. However, these linear transformations can also be applied to other measures such as the cross‐spectral density to perform tomographic mapping of power or coherence [Gross et al.,2001]. The method suggested here follows a similar logic.
In a first step, a multivariate autoregressive model is computed for the recorded signals of all MEG sensors. The result is a very compact and efficient representation of the data as an NxNxP‐matrix (N: number of channels, P: model order). In a second step, the covariance of all channel combinations is computed and the coefficients of a spatial filter are computed for each volume element in the brain. These coefficients are used in the third step to estimate the MAR‐model for volume elements in the brain. The new MAR‐model (that corresponds to brain areas and not MEG sensor signals) can be used to compute power, coherence, or causality measures (like PDC or DTF) for a given frequency. Interestingly, the computation of these measures given the MAR model is very fast.
In summary, the method is based on the computation of a multivariate AR model using the sensor signals followed by the efficient transformation of the model from the sensor‐level to the brain areas. Once the model (represented by the model coefficients) is defined a large number of measures can be computed that quantify coupling strength and direction of information flow between brain areas. This technique could possibly overcome most of the aforementioned limitations. It is very fast and efficient in terms of memory use and computational load. Because of the fast computation it is ideally suited for randomization techniques that can be used to establish significance levels for the results. Information about directionality is readily available for all volume elements and several partly complementary measures such as PDC and DTF can be easily computed and compared.
Multivariate Modeling of MEG Data
If s(t) is the column vector of all the activation signals in brain space at time t and x(t) is the column vector of the sensor measurements in the sensor space at the same time t, then the following relationship holds:
| (1) |
where Λ is the leadfield matrix or forward operator.
In the same fashion the inverse projection can be described with the use of an inverse operator. The source signals inside the brain can be described as projections of the sensor signals as:
| (2) |
where ϕ is the inverse operator.
Assuming that one could measure the activation time‐series of an arbitrarily large number of potential activation sources symbolized by s(t), a multivariate model built on them would have the form:
| (3) |
where P is the model order across time lags, τ is the time lag, B(τ) is the model coefficients matrix for lag τ, and ε is the model residual column vector.
Combining Eqs. (1), (2), and (3) gives the following expression for the multivariate model in the sensor space:
| (4) |
In the above derivation it is assumed that ϕΛ = I. This is evident if Eq. (1) is used in Eq. (2) to derive
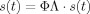 . This means that if a source activation signal is projected through its leadfield to sensor space and then back to source space through the inverse solution, the recovered signal should be the same as the original. Because of the nonuniqueness of the inverse solution the product ϕΛ deviates from the identity matrix. This form of deviation depends on the inverse method used. For beamformers ϕΛ = I is satisfied for each voxel individually, as this is the constraint used for the inverse solution for each brain location. When the leadfields and spatial filters for all voxels are entered in product ϕΛ, then the off‐diagonal components deviate from 0. Similar deviation from the identity matrix is also occurring with minimum norm solutions. However, as there is no unique solution to the inverse problem by these methods, it is assumed that by projecting the sensor data through the inverse solution, the original activation signals are recovered and not a distorted version of them (as there is no way to eliminate this distortion due to the nonuniqueness). This assumption is described by Eq. (2). By combining again Eqs. (1) and (2) to give
. This means that if a source activation signal is projected through its leadfield to sensor space and then back to source space through the inverse solution, the recovered signal should be the same as the original. Because of the nonuniqueness of the inverse solution the product ϕΛ deviates from the identity matrix. This form of deviation depends on the inverse method used. For beamformers ϕΛ = I is satisfied for each voxel individually, as this is the constraint used for the inverse solution for each brain location. When the leadfields and spatial filters for all voxels are entered in product ϕΛ, then the off‐diagonal components deviate from 0. Similar deviation from the identity matrix is also occurring with minimum norm solutions. However, as there is no unique solution to the inverse problem by these methods, it is assumed that by projecting the sensor data through the inverse solution, the original activation signals are recovered and not a distorted version of them (as there is no way to eliminate this distortion due to the nonuniqueness). This assumption is described by Eq. (2). By combining again Eqs. (1) and (2) to give
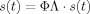 , it is seen that this assumption is translated in the assumption ϕΛ = I.
, it is seen that this assumption is translated in the assumption ϕΛ = I.
Because of the typical spatial proximity of MEG sensors and to the structure in the leadfield operator, it is expected that there will be colinearity between different sensor time‐series and a model derived directly on these time‐series would provide a poor solution. This can be avoided by applying principal component analysis (PCA) [Jolliffe,2002] to the time‐series, and additionally selecting only the principal components corresponding to the largest eigenvalues (typically those that explain 99% of the variance). Then the MAR model is built on these components. In this way colinearity is largely reduced and components representing mainly noise are omitted. The projection from sensor space to the principal component space is performed through the matrix of selected feature vectors [Jolliffe,2002] for the principal components as:
| (5) |
where V is the matrix of feature vectors mapping from the original recorded time‐series to the reduced PCA‐space.
The number of significant components, explaining the 99% of data variance, is lower than the number of sensors due to the presence of noise. Assuming that the excluded components represent noise, and assuming that the Moore‐Penrose pseudo‐inverse of V exists, the following assumptions can be made:
| (6) |
| (7) |
where + denotes the Moore‐Penrose pseudo‐inverse.Consequently:
| (8) |
Then combining the model in Eqs. (4) and (5) gives the MAR model in principal component space:
| (9) |
If a MAR model of the form:
| (10) |
is developed directly on the principal components of the MEG sensor measured time‐series, it is evident from Eq. (9) that for each time lag τ up to the model order, the MAR coefficients B(τ) in source space can be estimated through:
| (11) |
and the MAR model residual time‐series in source space can be estimated through:
| (12) |
For completeness, the data and noise covariance in source space can be derived from the ones in principal component space through:
| (13) |
| (14) |
where C s,N s are the data and noise covariance in source space, respectively and C PCA,N PCA in principal component space, respectively.
As the projection of the multivariate coefficients is performed through the use of the spatial filters and the leadfield matrices, it is important to mention how the inverse solution affects the projection.
If we denote the model coefficients matrix at the sensor level for lag τ as Γ(τ) then from Eq. (11) it can be seen that:
| (15) |
and
| (16) |
If we consider just the coefficient from source j to source k inside brain then:
| (17) |
where φkq is the spatial filter weight from sensor q to source k, λrj is the leadfield weight from source j to sensor r, γqr(τ) is the multivariate model coefficient from sensor r to sensor q for lag τ, N denotes number of sensors, and r, q denotes sensor index with values 1 to N.
In this representation it can be seen that the terms that dominate the sum are the ones in which all three components
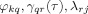 have significant values. These three terms can be depicted in Figure 1 where three activated sources i, j, and k are shown
have significant values. These three terms can be depicted in Figure 1 where three activated sources i, j, and k are shown
Figure 1.
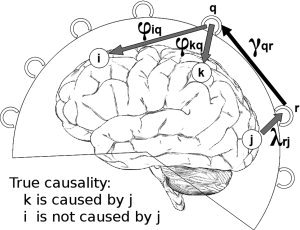
Visualization of the effect of mixing during the projection of coefficients from sensor space to source space. i, j, k: activated sources, r, q: MEG sensors, φkq:spatial filter weight from sensor q to source k, λrj: leadfield weight from source j to sensor r, γqr(τ): multivariate model coefficient from sensor r to sensor q. Source j is causing source k but not source i. If the spatial filter weight φiq has a value other than zero then true causality between sources j and k can be misrepresented as causality between sources j and i.
γqris the MAR coefficient from sensor r to sensor q. According to the dipoles' orientation of the actual activated brain sources that have a causal relationship, causality should be also present between the sets of sensors where the magnetic fields from the activated sources converge to local‐maxima and ‐minima.
φkq depends only on the caused source k and λrj depends only on the causal source j. The double sum in Eq. (17) iterates through all the possible sensor pairs. In Figure 2 are shown the histograms of the leadfield and spatial filter weights to and from all sensors for six simulated active sources and six inactive sources inside the brain. The leadfields and the spatial filters have been combined with the estimated dipole orientation from the inverse solution, which was derived through an LCMV beamformer.
Figure 2.
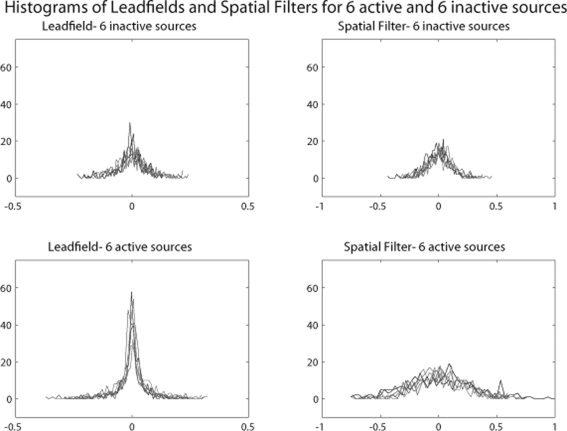
Histograms of leadfield and spatial filter weights to and from all the MEG sensors respectively, for six activated and six nonactivated sources. For the activated sources the leadfield and especially the spatial filter weights have a much wider distribution than the nonactivated sources. The values at the tales correspond to sensors in the vicinity of the local maxima and minima of the magnetic field generated by the activated dipole.
From Figure 2 it can be seen that the leadfield and the spatial filter weights for active sources have distributions with heavier tails than these for the inactive sources. The significantly higher positive and negative values at the tails correspond to the sensors that are located in the vicinity of the local‐maxima and ‐minima of the magnetic fields produced by the activated sources.
In the case of two sources with actual causal relationship like sources j and k in Figure 1, if it is assumed that sensor r is located at the local maximum of the magnetic field produced by source j and sensor q is located at the local maximum of the magnetic field produced by source k, then the coefficient γqr(τ) of the MAR model at the sensor level will be significant representing the underlying causal relationship. In this case, the product
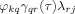 will attain a high value. The further away sensors r and q are from the local maxima (or minima) of the actual activation dipoles, the lower will be the above product. Consequently, the sum in Eq. (17) is dominated by the factors that correspond to the sensors that are located in the vicinity of the local maxima and minima of the actual activated dipoles, which represent the true causality at sensor level.
will attain a high value. The further away sensors r and q are from the local maxima (or minima) of the actual activation dipoles, the lower will be the above product. Consequently, the sum in Eq. (17) is dominated by the factors that correspond to the sensors that are located in the vicinity of the local maxima and minima of the actual activated dipoles, which represent the true causality at sensor level.
From Figure 1 it is also evident that the spatial filter weights have much wider distribution away from 0 for the active sources as compared to inactive sources, than in the case of the leadfield weights. This means that for sensors away from the local maxima and minima. While the leadfield weights decrease sharply, the spatial filter weights decrease more smoothly. This can result in mixing of causality information.
In the case that there is another activated source with no causal interaction with the other activated sources, like source i in Figure 1, then mixing of causal information can result if the spatial filter weights for source i have a wide distribution. This can be seen if in the computation of the coefficient b
kj(τ) from Eq. (17), the term
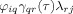 is examined. As discussed earlier, the product
is examined. As discussed earlier, the product
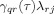 will have a high value due to the underlying true causality between sources j and k. If φiq has also a value different from zero then it is evident that a portion of the true causal relationship between sources j and k will be erroneously projected also between sources j and i.
will have a high value due to the underlying true causality between sources j and k. If φiq has also a value different from zero then it is evident that a portion of the true causal relationship between sources j and k will be erroneously projected also between sources j and i.
This mixing affects mostly activated sources for which the spatial filter weights have wider distributions. As nonactivated sources have a much narrower distribution of spatial filter weights, causality information is less likely to be represented in nonactivated areas.
As mixing depends mostly on the distribution of the spatial filter weights, the inverse solution used for deriving them plays a central role on the extend and pattern of mixing. Beamformers (LCMV, DICS) cannot separate highly correlated sources (i.e., auditory activations) and tend to represent two such sources with one source located in‐between. In this case, causality information would be projected in erroneous nonactivated locations. Another factor that affects beamformers is the use of a regularization parameter. This regularization parameter is used to make the inverse solution wider so that actual activated sources situated between grid points will not be missed. The use of high regularization values creates spatial filter weights with wide distributions and thus the level of causality information mixing will be higher. In the case if minimum norm solutions the main disadvantage is that they tend to assign observed activity to cortical areas because they are closer to the sensors and thus they fit better to the minimum norm constraint. This has the consequence that even inactive cortical voxels attain spatial filter weights with wider distributions than in the case of beamformers.
Consequently, in order to minimize the effect of mixing in this work, an LCMV beamformer is used with no regularization. When the entire brain is considered with no a priori selection of brain locations, a fine grid of 6 mm resolution is used (8,942 voxels).
Using the above formulation, in this work we investigate the feasibility of building the MAR model in the Principal Component space of sensor data and projecting it into source space, as described earlier, where causality is estimated from the model coefficients. Here we quantify causality by means of PDC.
Partial Directed Coherence
Partial directed coherence (PDC) is a metric aimed to identify causal relationships between signals at different frequencies in a multivariate system [Baccalá and Sameshima,2001]. It belongs to a family of methods that analyze the coefficients from MAR models. Another widely used metric is the DTF [Kaminski et al.,2001] but it has been shown [Astolfi et al.,2005] that PDC is superior over DTF in correctly identifying both direct and indirect causal pathways. The PDC from voxel j to voxel i at frequency f is given by the following equation:
| \font\abc=cmmib10\def\bi#1{\hbox{\abc #1}} $$ \pi _{ij} = {{\bar A_{ij} (f)} \over {\sqrt {{\bar {\bi a}}_j^H (f) \cdot {\bar {\bi a}}_j (f)} }} $$ | (18) |
where
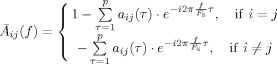 |
(19) |
are the elements of matrix :
| \font\abc=cmmib10\def\bi#1{\hbox{\abc #1}} $$ {{\bar {\bi A}}} = I ‐ \left. {\sum\limits_{\tau = 1}^p {A(\tau ) \cdot z^{ ‐ \tau } } } \right|_{z = e^{ ‐ i2\pi {f \over {Fs}}\tau } } = \left[ {{{\bar {\bi a}}}_1 ,{\bar {\bi a}}_2 , \cdots ,{\bar {\bi a}}_N } \right] $$ | (20) |
where f is the frequency, F s is the sampling frequency, τ is the model time lag, H is the Hermitian transpose.
As it can be seen from Eq. (19) for a given voxel pair i−j and frequency, the element
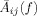 is in effect the z‐transform of the MAR coefficients series across time lags a
ij(τ), modeling the effect of voxel j on voxel i. The vectors
\font\abc=cmmib10\def\bi#1{\hbox{\abc #1}} $ {\bar {\bi a}}_j $, contain the elements of the jth column of matrix
\font\abc=cmmib10\def\bi#1{\hbox{\abc #1}} $ {\bar {\bi A}} $
and they contain all elements
is in effect the z‐transform of the MAR coefficients series across time lags a
ij(τ), modeling the effect of voxel j on voxel i. The vectors
\font\abc=cmmib10\def\bi#1{\hbox{\abc #1}} $ {\bar {\bi a}}_j $, contain the elements of the jth column of matrix
\font\abc=cmmib10\def\bi#1{\hbox{\abc #1}} $ {\bar {\bi A}} $
and they contain all elements
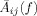 from voxel j to all voxels
from voxel j to all voxels
 . Consequently, it is straightforward to see that the denominator in Eq. (18) is the norm of vector
\font\abc=cmmib10\def\bi#1{\hbox{\abc #1}} $ {\bar {\bi a}}_j $. Thus, PDC is normalized with respect to the transmitting voxel. This means that PDC values between different voxel pairs are comparable only for the same transmitting voxel and comparison between pairs of different transmitting voxels is not feasible. It is also evident that if the activity between two voxels is highly correlated, then the norm will be dominated by the PDC of this pair, and the PDC of all other pairs will be down‐weighted. It is also natural to infer that if, because of poor signal‐to‐noise ratio, the MAR coefficients contain modeled noise, then it is highly probable that this will dominate the PDC and real causal pairs will be down‐weighted. Finally, the norm depends on the number of voxels, and for a very large number of voxels, the normalization factor increases and consequently the PDC values decrease to low‐levels and become more sensitive to erroneous or random correlations.
. Consequently, it is straightforward to see that the denominator in Eq. (18) is the norm of vector
\font\abc=cmmib10\def\bi#1{\hbox{\abc #1}} $ {\bar {\bi a}}_j $. Thus, PDC is normalized with respect to the transmitting voxel. This means that PDC values between different voxel pairs are comparable only for the same transmitting voxel and comparison between pairs of different transmitting voxels is not feasible. It is also evident that if the activity between two voxels is highly correlated, then the norm will be dominated by the PDC of this pair, and the PDC of all other pairs will be down‐weighted. It is also natural to infer that if, because of poor signal‐to‐noise ratio, the MAR coefficients contain modeled noise, then it is highly probable that this will dominate the PDC and real causal pairs will be down‐weighted. Finally, the norm depends on the number of voxels, and for a very large number of voxels, the normalization factor increases and consequently the PDC values decrease to low‐levels and become more sensitive to erroneous or random correlations.
RESULTS
Investigation 1: Small Number of Voxels with Known Causality Structure
As a first step to evaluate the feasibility of building the MAR model on the principal component space of the sensor data and then projecting it into source space where causality is inferred, the following process has been used.
Six simulated signals with predefined causality were generated to represent activity of six sources inside a typical brain volume segmented in a 6 mm grid (8,942 voxels in brain volume).The ‘Ideal’ PDC was computed directly from the simulated signals. Through, the forward solution pseudo‐MEG sensor time‐series were derived. Noise was added to the sensor data. A MAR model was built on the principal component space of the pseudo‐MEG sensor data. Spatial Filters and dipole orientations were estimated by a Linearly‐Constrained Minimum Variance beamformer for the six known locations of dipole activity. The MAR model coefficients were then projected onto the six known locations. PDC was computed from the coefficients of the projected MAR model and compared to the ‘Ideal’ PDC. Tolerance of the method with respect to white Gaussian sensor noise, model order and number of samples is investigated. Additionally, tolerance of the method with respect to spatiotemporally background noise is investigated. Confidence intervals of PDC are examined by a jackknife method.
Simulated brain dipole
MEG sensor data was simulated. Six activation signals inside the brain volume were generated from MAR equations approximating damped oscillators [Baccalá and Sameshima,2001]:
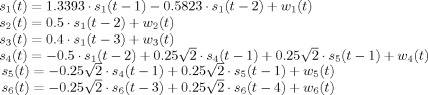 |
(21) |
where w n(τ) are zero‐mean uncorrelated white Gaussian noise processes with identical variance.
The causality structure between the simulated source signals is shown in Figure 3. The activation signals were designed with a nominal frequency of 8 Hz (Alpha band) (Fig. 4) and the time courses were sampled with a sampling frequency of 100 Hz. Time‐series were organized in 20 trials with each trial having a duration of 1 s. Source 6 has no causal interaction with the other sources and has the highest power. This source was chosen in order to investigate if a strong source unconnected to the rest of the network will significantly affect the estimation of causality.
Figure 3.
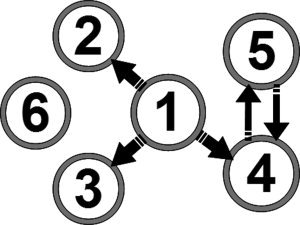
Predefined causality of simulated activation signals. Sources 1 to 5 are part of a causal network while there is no causal interaction between source 6 and the rest of the sources.
Figure 4.
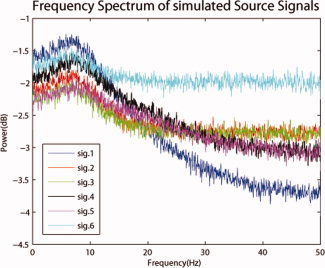
Spectrum of simulated activation signals. All the signals have peak power around 8 Hz. Sources 1 to 5 which are part of the same causal network have similar spectra. Source 6 which is not part of the causal network has in general more power in all frequencies above 10 Hz. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The locations of the six dipoles were defined with respect to the head coordinate system (x‐axis pointing to nasion, y‐axis pointing to left preauricular point, and z‐axis point up) and can be seen in Figure 5. The orientations of the dipoles were chosen to be random unit vectors, orthogonal to the line connecting the center of mass of the brain volume to each activation point. For reference, the locations and orientations of the six dipoles are given in Table I.
Figure 5.
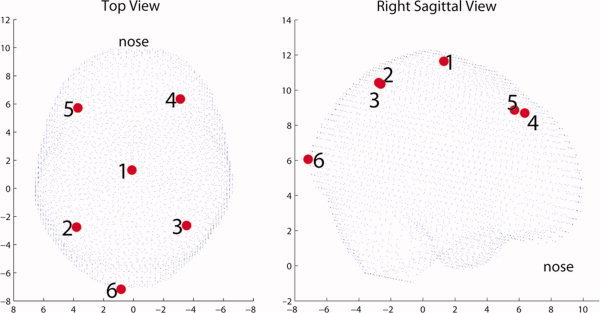
Simulated sources topology. On the left the sources are viewed from a top view while on the right from a right sagittal view. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Table I.
Location inside brain volume and orientation of the 6 simulated activation sources
| Source no | Location (cm) | Orientation (normalized so norm = 1) | ||||
|---|---|---|---|---|---|---|
| x | y | z | x | y | z | |
| 1 | 1.3027 | 0.1034 | 11.6434 | 0.8461 | −0.5329 | 0.0119 |
| 2 | −2.7540 | 3.8043 | 10.4273 | 0.1262 | 0.8494 | −0.5124 |
| 3 | −2.6379 | −3.5436 | 10.3444 | 0.9562 | −0.1412 | 0.2565 |
| 4 | 6.3504 | −3.1215 | 8.6962 | −0.7524 | 0.1079 | 0.6498 |
| 5 | 5.7133 | 3.7076 | 8.8705 | −0.6381 | −0.3040 | 0.7074 |
| 6 | −7.1710 | 0.8328 | 6.0670 | 0.0169 | 0.8770 | −0.4802 |
The overall magnetic field generated by these brain sources was simulated by multiplying the simulated activation signals with their corresponding leadfields, and white Gaussian noise representing noisy environment processes was added. The simulation of the activation sources and the computation of the corresponding magnetic field were performed in MATLAB with the use of the Fieldtrip toolbox [Oostenveld et al.,2010].
PDC deviation from ‘Ideal’ with respect to environment noise, N of samples and model order
We investigated the deviation of PDC from the ‘ideal’ for different values of environment noise level, number of samples per trial and MAR model order.
For the environment noise investigation spatially and temporally uncorrelated white Gaussian noise was added to simulated MEG data. The amplitude of the noise was defined relative to the rms value of the MEG sensor pseudo‐measurements, resulting only from the simulated activation signals. The investigated range was 1–20. The above type of noise was chosen because it is the simplest type and usually represents sensor noise. It suitable to investigate the effect of noise that in spatially and temporally uncorrelated. In section “PDC deviation from ‘Ideal’ for spatiotemporally correlated noise” the same investigation is repeated for spatiotemporally correlated noise.
For the investigation of number of samples per trial, 20 trials were used. The investigated range was 500–4,000 samples/trial. For the investigation of the MAR model order, the number of time lags used in the model was varied. The investigated range was 5–100 lags.
The objective here was to investigate average PDC deviation from ‘Ideal’ PDC for the above parameters, around the frequency of the activation signals (8 Hz) and separately for causal and noncausal pairs. For this purpose two metrics were used. The first metric is defined as the mean (PDC of Projected MAR Model‐'Ideal' PDC) for frequency range 7–12 Hz, averaged for the causal pairs 2‐1, 3‐1, 4‐1, 5‐4, and 4‐5. The second metric is the same as the first one but for noncausal pairs.
PDC deviation from ‘Ideal’ for spatiotemporally correlated noise
In the previous investigation, the added noise at the sensor‐level was white Gaussian. Although, this investigation is valuable in examining the effect of different levels of uncorrelated noise into the recovery of causal information, in reality it mostly represents MEG sensor noise. If one wants to represent realistic brain background noise, emanating from within the brain, it is more realistic to model the noise as spatially and temporally correlated [Bijma et al.,2003; de Munck et al.,2002; Jun et al.,2002; Lütkenhöner,1994]. We have evaluated the performance of PDC for different rms levels of spatiotemporally correlated noise when the activity locations are known.
For creating the spatial noise correlation, an approach similar to Lutkenhoner [1994] and Jun et al. [2002] has been followed. Dipoles (2,184) locations were selected, uniformly distributed within the brain volume. In each of these locations a dipole with random orientation was actuated.
For creating the temporal noise correlation, an approach similar to Bijma et al. [2003] and Nolte et al. [2008] was followed. Each of the 2,184 noise dipoles was activated by a pink noise signal. This is due to the fact that it is well known that the background noise is not white but has a 1/f characteristic [de Munck et al.,2002]. Each pink noise signal was derived by passing a white Gaussian signal through a third‐order low‐pass filter designed with an 1/f frequency spectrum characteristic and with cut‐off frequency of 15 Hz providing most spectrum power in the alpha‐range Bijma et al. [2003].
The designed filter has the following autoregressive representation:
| (22) |
where here y(t) is the output of the filter (temporally correlated pink noise) and x(t) is the input (white Gaussian noise). As it can be seen for this equation, the autocorrelation of the resulting pink noise is temporally extended to three time lags. Through this process the output noise signal had both an 1/f spectrum characteristic and a temporal auto‐regression extended to three time lags in the past. The 2,184 noise dipole time‐series where not temporally cross‐correlated similarly to Nolte et al. [2008]. As the pink noise signal is derived from random white Gaussian Noise, the phase of the temporal correlation is also randomized [Bijma et al.,2003].
The rms level of actuation was selected to be the same for all sources. The pseudo MEG sensor measurements were derived by projecting all the 2,184 activation dipole time‐series through the leadfield matrices. The instant spatial correlation and the lagged temporal correlation for each sensor is higher with the neighboring sensors and diminishes with distance from the sensor.
The resulting noise at the sensor level was adjusted in magnitude in order to investigate noise levels 1–20 times the rms value of the sensor time‐series from the six actual activation sources, similarly to the evaluation for the white Gaussian noise in section “PDC deviation from ‘Ideal’ with respect to environment noise, N of samples and model order”. The evaluation was also performed with the same metrics, which is the mean deviation of PDC from ‘ideal’ PDC for the causal and the noncausal pairs separately.
Statistical inference of PDC
As PDC depends on the spectrum of the estimated MAR model coefficients for each pair, which are random for signals containing no causality, it is instructive to be accompanied by statistical inference of significance. The statistical significance of the PDC causality results was accessed through a jackknife method, the trial based leave‐one‐out method (LOOM) [Schlögl and Supp,2006].
One trial is excluded from the sensor data set. The data from all trials is then concatenated and the MAR model is built. Then the MAR coefficients are projected through the inverse solution into the six known simulated source locations inside the brain volume and PDC is computed for the frequency range 1–50 Hz. Then the next trial is excluded from the sensor data set and the procedure is repeated. This procedure is repeated until each of the trials has been left out once.
The LOOM approach provides two main advantages [Schlögl and Supp,2006]. Firstly, LOOM obtains the least‐biased estimates over all other resampling methods. Secondly, no a priori assumption regarding the type of distribution is needed.
Through the LOOM procedure, a sampling distribution
 is obtained for the PDC for each of the frequency bins considered with mean μu and standard deviation σu. However, the standard deviation is not derived from N independent trials. Only the (N−1)th part of each concatenated data vector was independent (one out of N−1 trials). Consequently, the true standard error is
is obtained for the PDC for each of the frequency bins considered with mean μu and standard deviation σu. However, the standard deviation is not derived from N independent trials. Only the (N−1)th part of each concatenated data vector was independent (one out of N−1 trials). Consequently, the true standard error is
 [Schlögl and Supp,2006].
[Schlögl and Supp,2006].
The mean and the standard error are used in a simple t‐test for testing whether the PDC at a specific frequency bin is significantly different from zero or not. From this t‐test, the 95% confidence limits of the mean can be computed according to:
| (23) |
where p is the significance level, N: is the number of trials, t(p/2, N−1) is the upper critical value of the t‐distribution with N−1 degrees of freedom, and significance level p. In this investigation for each frequency bin, N = 20 and p = 0.05 (95% confidence), which yields to confidence limits computed by
| (24) |
These confidence limits have been calculated for each of the 36 investigated voxel pairs and for each integer frequency in the range 1–50 Hz.
Investigation 1:results
'Ideal' reference PDC was calculated directly from the 6 simulated activation signals. It is shown in Figure 6a for all possible pair combinations between signals. The pairs that show distinct PDC are in agreement with the expected causal pairs from the configuration of the simulated activation signals. These pairs are: 2‐1, 3‐1, 4‐1, 5‐4, and 4‐5. Then the PDC was calculated from MEG sensor data.
Figure 6.
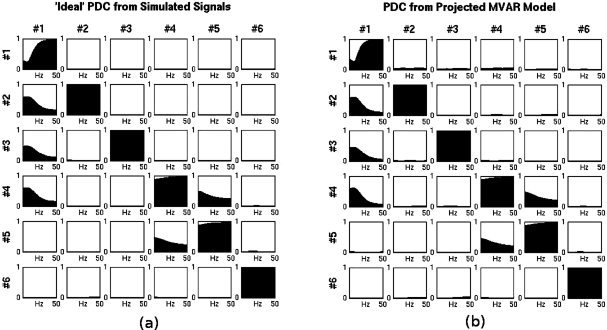
(a) ‘Ideal’ PDC directly from activation signals (b) PDC from MAR Model projected through the spatial filters of LCMV beamformer. In (b) the coefficients have been projected to the precise known locations of the simulated activation signals.
PCA was applied to the sensor data. Twenty‐one of the components explained 99% of the variance so the remaining principal components were discarded. The MAR model was built on the principal components using the Yule‐Walker method [Schlögl and Supp,2006]. Through Akaike's criterion [McQuarrie and Tsai,1998] the model order was selected as six.
After the MAR model was built on the principal components of MEG sensor time‐series, it was projected to the 6 dipole locations with the spatial filters and orientations estimated by a LCMV beamformer for the precise known locations of the activated dipoles. The spatial filters and the dipole orientations were computed in MATLAB with the use of the Fieldtrip toolbox [Oostenveld et al.,2010]. The derived PDC is shown in Figure 6b. PDC calculated from the projected MAR model in approximating the ‘ideal’ reference PDC from the activation signals. This means that a MAR model built from MEG data in the sensor space and projected in the source space preserves causality information for the underlying generating activation processes. Examination of the levels of PDC shows that it is distinguishably high for the causal pairs when compared to all the rest noncausal pairs. Maxima occur around 8 Hz, the nominal frequency of the activation signals.
Subsequently, we examined the average deviation of the reconstructed PDC from ‘ideal’ with respect to environment noise, N of samples, and model order, averaged across the causal and noncausal source pairs. In Figure 7 the 2 metrics described in section “PDC deviation from ‘Ideal’ with respect to environment noise, N of samples and model order” are shown in a comprehensive way describing the variation of all three investigated parameters. Each subplot presents the two metrics for a particular combination of N Samples/Trial, environment noise level, and MAR model order. Up to noise levels four times the strength of the dipoles the deviation of the PDC for the causal pairs remains low. In the presence of higher noise, PDC is deviating significantly from the ideal PDC. When a small number of samples is used, combined with high model order, PDC for both causal and noncausal pairs is inconsistent relative to the ideal. In such cases, one cannot distinguish between causal and noncausal pairs. The number of coefficients in the MAR model is
 where p is the model order and N is the number of variables in the model. When a small number of samples per trial is combined with a high model order, then this means that the number of data points per trial is only a modest multiple of the number of estimated parameters. As seen in Figure 7 in such cases the performance of PDC becomes detrimental.
where p is the model order and N is the number of variables in the model. When a small number of samples per trial is combined with a high model order, then this means that the number of data points per trial is only a modest multiple of the number of estimated parameters. As seen in Figure 7 in such cases the performance of PDC becomes detrimental.
Figure 7.
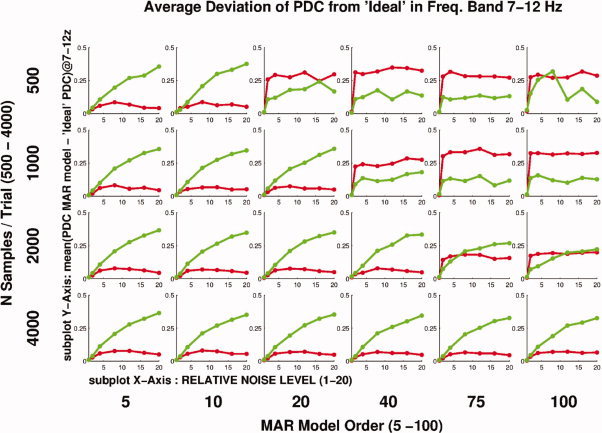
Average deviation of PDC from ‘Ideal’ for causal and noncausal pairs. Causal pairs are represented by green color, noncausal pairs by red. In each subplot PDC is plotted versus N Samples/Trial. Each subplot corresponds to a different combination of environment noise level and MAR model order. For low N samples/Trial and high MAR model order (top right), PDC fails to capture causality correctly and non‐causal pairs appear to have significant causality. For higher N samples/Trial and lower model order (bottom left) PDC recovers information much more consistently and non causal pairs do not appear to have significant causality. In such cases it can be seen that for noise levels below five times the rms value of the brain signals, the average deviation of both causal and noncausal pairs from the ‘Ideal’ PDC remains low. For noise levels above five the average deviation of the causal pairs seems to systematically increase. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The next investigation was the average deviation of the reconstructed PDC from ‘ideal’ for different levels of spatiotemporally correlated noise as described in section “PDC deviation from ‘Ideal’ for spatiotemporally correlated noise”. According to the previous investigation with white Gaussian noise, a robust combination of number of samples per trial and model order was chosen, specifically 2,000 samples per trial and model order 5. The results are shown in Figure 8 where also the same evaluation for the white Gaussian noise case is shown for comparison. In the case of the spatiotemporal noise, the causal pairs seem to be more robust to higher noise levels, as the average deviation from the ideal is lower as compared to the white Gaussian Noise. For the noncausal pairs the average deviation from ideal increases consistently with noise level. This is different from the white Gaussian noise where the average deviation after a noise level of 8, seemed to stabilize around a certain level. The above observations show that because of the spatial and temporal correlation of the noise, PDC for the causal pairs has a tendency for higher rate of correct detection and for the noncausal pairs a higher rate for false detections. These results are in agreement with Nolte et al.2008, where Granger causality appeared to behave in the same way for high noise levels. Another observation from these results is that up to a noise level four times the rms value of the actual brain signal, the mean deviation of PDC from the ‘ideal’ remains in low‐levels and is similar for white Gaussian and spatiotemporally correlated noise.
Figure 8.
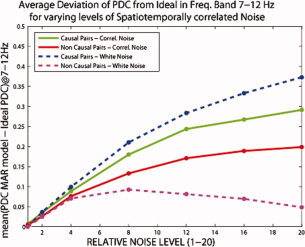
Average deviation of PDC from ‘Ideal’ for causal and noncausal pairs for different levels of spatiotemporally correlated noise added at the sensor‐level. The same deviation is shown for the same levels of white Gaussian noise for comparison. As spatiotemporally correlated noise level increases the noncausal pairs appear in fault to have significant causality. Up to noise level four times the rms of the actual brain signal, deviations from ideal remain low. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Finally, the confidence intervals for PDC were evaluated by the LOOM method as described in section “Statistical inference of PDC”. As it can be seen in Figure 9, the 95% PDC confidence limits for the pairs that have an actual causal relationship are significantly different from 0, especially for the lower frequency range where the most spectral power of the simulated signal is contained. For the pairs that have no actual causal relationship the confidence intervals systematically encompass 0. These results show that the PDC computed from the projected MAR coefficients into the six known source locations is robust and consistent with the actual simulated causality configuration. If the locations of the actual activated sources are known then the PDC can provide consistent representation of the causal interactions within the network of these sources.
Figure 9.
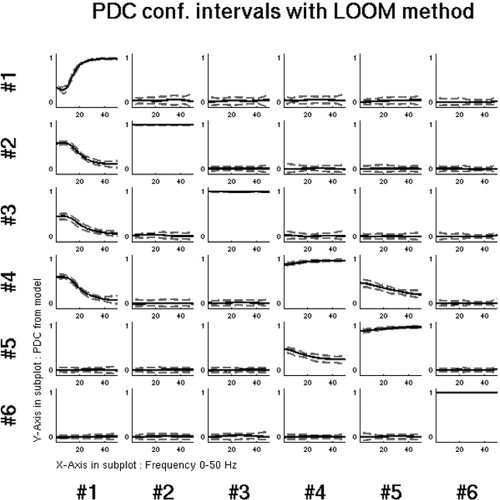
Ninety‐five percent confidence limits of PDC derived by the LOOM method. PDC has been computed by projecting the sensor‐level MAR model into the six known locations of the simulated brain sources. The causal and noncausal pairs can be confidently distinguished.
Investigation 2: Consideration of All Voxels in Brain Volume as Potential Activation Sources
Investigation 1 showed that if the actual activation locations inside the brain are known then the methodology of building the MAR model in the principal component space of sensor data and projecting it into the source space correctly reconstructs the causality structure by PDC. However, though this scenario serves as a good demonstration of the theoretical feasibility of the methodology, it is unrealistic as in real experiments the number and location of activated brain sources are not known and have to be identified. To evaluate the methodology in such a realistic scenario a similar approach to investigation 1 was followed as described in section “Investigation 1: Small Number of Voxels with Known Causality Structure.” The noise used in this investigation was spatiotemporally correlated noise, as described in section “PDC deviation from ‘Ideal’ for spatiotemporally correlated noise”, scaled to two times the rms value of the pseudo‐MEG measurements from the simulated dipoles. The main difference in this investigation is that the MAR model coefficients were projected into all voxels inside the brain volume through the beamformer spatial filters (8,942 locations). Then PDC was computed from the coefficients of the projected MAR model.
In Investigation 1, as only six voxels were considered it was easy to visualize PDC results for all pairs and frequencies simultaneously. Here, as there are 8,942 voxels, it is impossible to visualize all combinations for all frequencies simultaneously. As it is known that the ‘Ideal’ PDC has a peak around 8 Hz, PDC was visualized only for this frequency. The method of visualization chosen was a sliced topological plot of PDC Maps.
Four different types of PDC maps were constructed. First the map of PDC to all voxels from each of the known activity sources (Receive direction). Second the map of the mean of PDC received by each voxel from all voxels (Receive direction). Third the map of PDC from all voxels to each of the known activity sources (Transmit direction). Fourth the map of the mean of PDC transmitted by each voxel to all voxels.
The first case was investigated in order to see if causality information for each of the known simulated sources is preserved when the MAR model is projected in the entire brain volume. Because in this case we still know the locations of actual activity, the second case was investigated as a potential way of identifying causal areas inside the brain without any a priori knowledge about potential source locations. This comes naturally from the fact that PDC is normalized relative to the causal voxel. In the contrary averaging the PDC in the causal direction is not a feasible method due to this semiarbitrary normalization. To demonstrate this fact, cases 3 and 4 have been used.
All four cases represent maps of caused and causal activity in the entire brain either from or to a seed or as an average. These maps can provide a very useful initial view of the causality patterns within the entire brain. Such maps of causality have already been used with fMRI data [Roebroeck et al.,2005]. Deriving such maps also from MEG data with a high spatial resolution scan grid offers the advantage that causality maps for the same phenomenon can be derived and compared between these two modalities for the entire brain. The causality recovered by fMRI is typically in a timescale of seconds while the causality recovered by MEG is in a timescale of milliseconds. Combing causality in these two different timescales can prove very useful in understanding how low‐frequency causal networks modulate high frequency causal networks and vice versa.
Investigation 2: Results
For a clearer interpretation of Figures 10 and 11 one should first get familiar with the topology of the simulated sources inside the brain shown in Figure 5 and with the causality configuration of those signals shown in Figure 3.
Figure 10.
PDC sliced maps for f = 8 Hz. PDC to all voxels from source (a) 1, (b) 2, (c) 3, (d) 4, (e) 5, (f) 6, (g) map of mean PDC caused to each voxel, (h) map indicating the location of the simulated sources with spheres of 1.5 cm radius, and (i) original causal configuration between the six sources. The position of each source represents coarsely its expected location in the topographic maps. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 11.
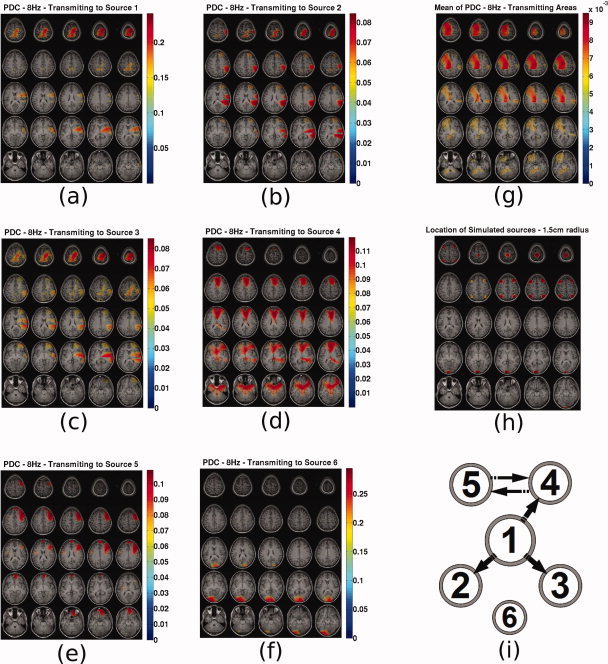
PDC sliced maps for f =8 Hz. PDC from all voxels to source (a) 1, (b) 2, (c) 3, (d) 4, (e) 5, (f) 6, (g) map of mean PDC caused by each voxel, (h) map indicating the location of the simulated sources with spheres of 1.5 cm radius and (i) original causal configuration between the six sources. The position of each source represents coarsely its expected location in the topographic maps. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
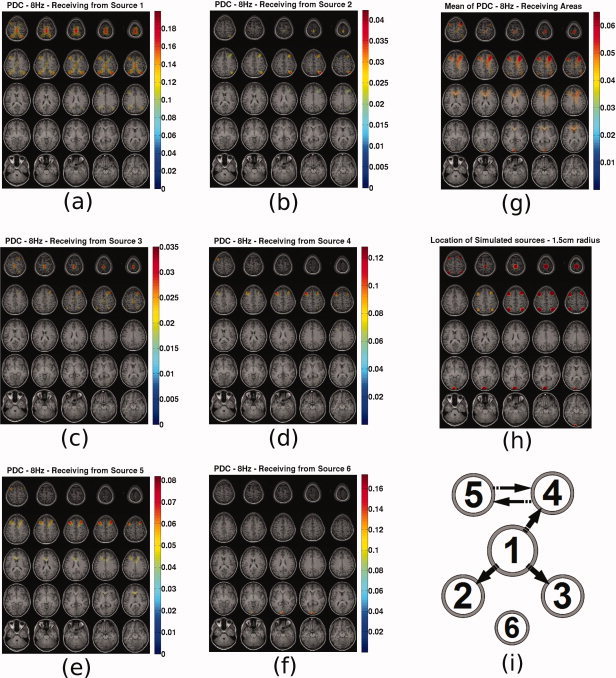
In Figure 10 the following are presented. Subfigures (a–f) present the PDC maps from the known sources 1–6 to all voxels. Subfigure (g) presents the mean PDC caused to each voxel. Subfigure (h) presents the actual locations of the simulated sources with spheres of 1.5 cm radius for ease of reference. Subfigure (i) present the original causal configuration between the six sources. The position of each source represents coarsely its expected location in the topographic maps. Depth information is encoded in the radius of each source with bigger radius corresponding to sources closer to the top of the head. This diagram has been included in order to assist the reader to infer the results in the PDC maps.
From these plots the following can be observed. Source 1 is in reality causing sources 2, 3, and 4 and itself through autoregression. This is well represented in Figure 10a, where also source 5 appears to be caused by 1. Sources 2 and 3 in reality do not cause any activity in any voxel. However, in Figure 10b,c PDC appears to be prominent to voxels 1, 2, 3, and 4 for both cases. It seems that PDC from source 2 and 3 is a ghost of the PDC from source 1 which causes these 2 sources. The PDC map values for these two sources remain significantly lower from all other voxels. In Figure 10d, source 4 appears to be causing source 5 and itself which is in accordance with the real causality. In Figure 10e, source 5 appears to be causing source 4 and itself, which is in accordance with the real causality. In Figure 10f, source 6 appears to be caused only by itself, which is also in accordance with reality. An important observation is that in Figure 10a–e for sources 1–5, source 6 does not appear to interfere in the recovered causality maps. To summarize, firstly, PDC seems to indicate the correct areas where real causal connections exist. Secondly, PDC does not seem to always correctly reconstruct the individual causal pathways, and ghosts of real connections appear in other voxels of the causal network. Ghost connections do not appear in locations without activity.
Averaging all the maps of PDC from each voxel to all other voxels, provides a map on which are highlighted all areas that are on average caused. This map is presented in Figure 10g. The sources that are actually caused by other sources are 2, 3, 4, and 5. Sources 1 and 6 are auto‐correlated. All these sources appear on this map which could serve as an initial indicator of the areas that are involved in a causal functional network. Local maxima or cluster centers could then be selected, at which the MAR model would be projected and PDC (or any other causality metric) would be recalculated only for these points.
In Figure 11 the following are presented. Subfigures a–f present the PDC maps from all voxels to the known sources 1 to 6 Subfigure (g) presents the mean PDC caused by each voxel. Subfigure (h) presents the actual locations of the simulated sources with spheres of 1.5 cm radius for ease of reference. Subfigure (i) present the original causal configuration between the six sources. The position of each source represents coarsely its expected location in the topographic maps.
Following the analysis in the same fashion as before, it can be seen that in this case, as expected, that PDC from all voxels to a single voxel cannot serve as a useful causality map because PDC is normalized with reference to the causal voxel. So each PDC value from every voxel to a single voxel has been differently normalized. This is evident in the plots, where causality maps fail to resemble the real causality connections. This is a very significant drawback of using PDC in cases when the entire brain volume is considered as potential activity sources. Using the map of mean PDC received by each voxel, one can create a map representing the areas that are in generally caused and in which causal areas might also appear due to mixing. However, a similar map of causal areas cannot be constructed based on PDC.
Investigation 3: Using the MAR Model Coefficients Directly for Causality Identification
In Investigation 2, it was seen that when a very large number of voxels is considered as potential source locations, PDC can provide a map of causal areas in which also caused areas might appear because of ghost causality connections identified by PDC. Also it was seen that a map of caused locations cannot be constructed on account of the way PDC is normalized. The above limitations have instigated the interest to investigate if causal connections can be recovered directly from the MAR model coefficients without the use of PDC.
The simplest metric that could be used for such a purpose is the norm of the MAR model coefficients across time lags of the model. This can be represented as:
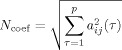 |
(25) |
The main advantage of using this metric is that all MAR model coefficients are derived simultaneously for all voxel pairs, and thus relative comparison between different voxel pairs within the same model is feasible. Additionally, the norm of the coefficients is not normalized relative to causal or caused voxel and thus both causal and caused maps can be constructed from this metric. A similar metric has already been used to infer causality in MEG data [Ramirez and Baillet,2010].
The main disadvantage of this metric is that it is not frequency‐specific and if frequency‐specific information is needed then the most feasible solution would be to build a model on a narrowband filtered version of the data. An additional disadvantage is that because the norm of the coefficients is not normalized, comparison between different conditions (MAR models for different data sets) on the relative causality strength cannot be confidently performed only by this metric.
The evaluation of this methodology followed a similar approach to investigation 2 as described in section “Investigation 2: Consideration of All Voxels in Brain Volume as Potential Activation Sources”. The noise used in this investigation was spatiotemporally correlated noise, as described in section “PDC deviation from ‘Ideal’ for spatiotemporally correlated noise,” scaled to two times the rms value of the pseudo‐MEG measurements from the simulated dipoles. The main difference in this investigation is that the MAR model coefficients were projected into all voxels inside the brain volume through the beamformer spatial filters (8,942 locations). Then, N coef was computed from the coefficients of the projected MAR model for all voxel pairs.
Four different types of N coef were constructed. First the map of N coef to all voxels from each of the known activity sources (Receive direction). Second the map of the mean of N coef received by each voxel from all voxels (Receive direction). Third the map of N coef from all voxels to each of the known activity sources (Transmit direction). Fourth the map of the mean of N coef transmitted by each voxel to all voxels (Transmit direction).
Investigation 3: results
In Figure 12 the following are presented. Subfigures (a–f) present the N coef maps from the known sources 1–6 to all voxels. Subfigure (g) presents the mean N coef caused to each voxel. Subfigure (h) presents the actual locations of the simulated sources with spheres of 1.5 cm radius for ease of reference. Subfigure (i) present the original causal configuration between the six sources. The position of each source represents coarsely its expected location in the topographic maps. Depth information is encoded in the radius of each source with bigger radius corresponding to sources closer to the top of the head. This diagram has been included in order to assist the reader to infer the results in the PDC maps.
Figure 12.
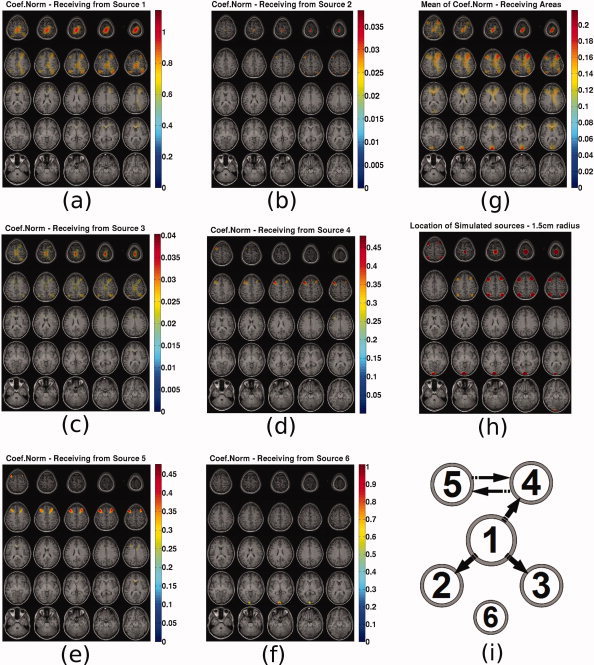
N coef sliced maps: N coef to all voxels from source (a) 1, (b) 2, (c) 3, (d) 4, (e) 5, (f) 6, (g) map of mean N coef caused to each voxel, (h) map indicating the location of the simulated sources with spheres of 1.5 cm radius and (i) original causal configuration between the six sources. The position of each source represents coarsely its expected location in the topographic maps. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
In this case when the metric represents the caused voxels, similar observations as for the PDC can be made. For sources 4, 5, and 6 causality is correctly reconstructed while for the other three known sources, ghost causal connections appear in the maps in addition to the correct connections. Again source 6 does not seem to interfere with the causality maps of the other sources. By examining the map of mean N coef received by each voxel one can see that all areas that have auto‐ or cross‐correlated activity are highlighted similarly to PDC.
In Figure 13 the following are presented. Subfigures (a–f) present the N coef maps from all voxels to the known sources 1–6. Subfigure (g) presents the mean N coef caused by each voxel. Subfigure (h) presents the actual locations of the simulated sources with spheres of 1.5 cm radius for ease of reference. Subfigure (i) present the original causal configuration between the six sources. The position of each source represents coarsely its expected location in the topographic maps.
Figure 13.
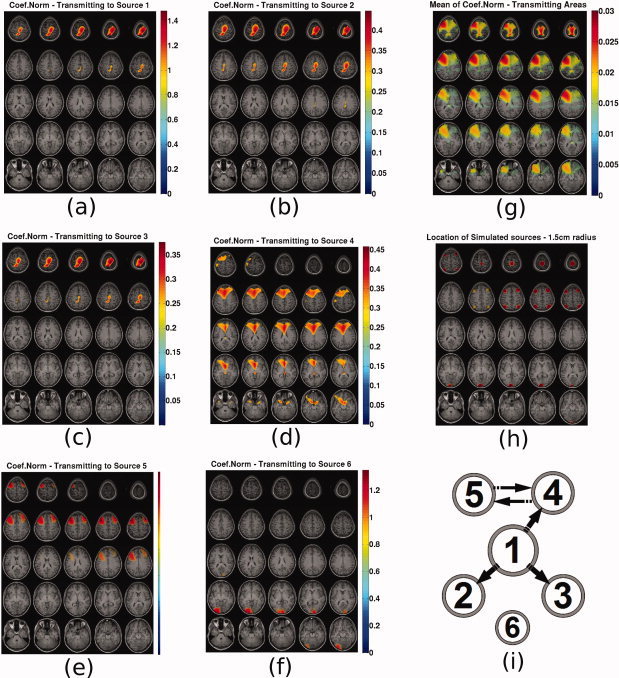
N coef sliced maps: N coef from all voxels to source (a) 1, (b) 2, (c) 3, (d) 4, (e) 5, (f) 6, (g) Map of mean N coef caused by each voxel, (h) map indicating the location of the simulated sources with spheres of 1.5 cm radius and (i) original causal configuration between the six sources. The position of each source represents coarsely its expected location in the topographic maps. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
These maps represent the areas from which activity is caused. Source 1 seems to be caused only by itself which is in accordance with reality. Sources 2 and 3 appear correctly to be caused by source 1. Source 4 seems correctly to be caused by itself and source 5 but not from source 1. Source 5 seems also correctly to be caused by sources 4 and 5. Source 6 is also correctly appearing to be causing itself and not interfering with the causal maps of the other voxels. In summary, the causal activity maps highlighted the areas where caused activity was actually present. Most of the recovered individual causal connections were correctly recovered.
When plotting the map of the mean N coef caused by each voxel, the correct locations are highlighted. The fact that a consistent causal connectivity map is available in addition to the caused connectivity map, is a significant advantage of using the MAR coefficient norm metric.
By using these two maps from Figures 12g and 13g one could infer that areas around sources 1, 4, and 5 are the main causal hubs and areas around sources 1, 2, 3, 4, 5, and 6 are caused hubs, which corresponds to reality. This is a very significant advantage as compared to the inference about causality one could make by only looking at the PDC map in Figure 10g.
By selecting voxels at the centers of these hubs and examining the individual N coef maps could lead to ghost connectivity being identified as real. Probably a more consistent approach would be to project the MAR model only into these selected hub centers and recompute N coef and PDC.
Local Maxima
Causality information as recovered by the MAR model coefficients is represented in maps as hubs of activity. The most obvious choice for quantifying how accurately the locations of actual activity are recovered by these hubs, is to estimate the local maxima.
The local maxima of mean N coef caused by and to each voxel were computed using multidirectional derivation with the MinimaMaxima3D toolbox in MATLAB [Pichard,2007]. The distance between the actual sources of activity and the corresponding hub maxima were computed and presented in Table II. The accuracy is in most cases better than 1 cm and in one case it exceeds 3 cm.
Table II.
Distance of local maxima from actual activation points in centimeters
| Mode | Source 1 | Source 2 | Source 3 | Source 4 | Source 5 | Source 6 |
|---|---|---|---|---|---|---|
| Caused | 0.7952 | 0.9351 | 1.2368 | 0 | 0.5351 | 0.1612 |
| Causal | 0.5733 | – | – | 0 | 3.2342 | – |
Real Data
To do a preliminary indicative evaluation with real data, datasets for 2 subjects from the following experiment were used. A cue indicating left or right was presented to the subject, and after 2 s a ‘go’ cue indicated to the subject to press a button with the cued hand. The actual data sets used here are from the interval 0–500 ms, from the onset of the left–right visual cue. Within this interval, areas involved in motor planning should be activated.
The sampling frequency for the measurements was 500 Hz. For subject 1, 146 trials were selected and for subject 2, 162 trials were selected after removal of artifacts. The subjects' brain volumes were segmented in 6 mm grids. By applying PCA it was found that for subject 1, 24 principal components and for subject 2, 27 principal components explained 99% of the variance.
To introduce statistical inference in the estimation of causality maps, a LOOM approach similar to the one described in section “Statistical inference of PDC” was used. One trial was removed from the sensor data. The spatial filters and dipole orientations were estimated through an LCMV beamformer. The MAR model was built on the selected principal components of the sensor data by the Yule‐Walker method and the coefficients were projected in source space. Akaike's criterion instructed a model order of 14 time lags. The projected N coef for each voxel pair was computed and the caused and causal maps of its mean were computed. Then the next trial was excluded from the sensor data set and the procedure was repeated. This procedure was repeated until each of the trials has been left out once.
Because of the fact that the coefficient norm is always greater than zero a similar LOOM approach was used to derive the level of random causality against which the observed causality was compared. A randomized data set was derived by randomly shuffling data in each channel across all data points and then reslicing it in trials. Then the procedure was the same as above, excluding one trial and repeating until each trial has been excluded once.
Welch's t‐test for samples with different variances and with P‐value 0.05 was used in each voxel for both the caused and causal maps in order to estimate if the mean N coefis above the randomized case. The voxels for which the null hypothesis (estimated causality same as randomized causality) was not rejected were assigned a causality metric of zero.
The resulting maps of mean N coef caused by and to each voxel were plotted on a 3D mesh of each subject's cortex. The causal maps have a distinct maximum on the visual cortex for both subjects. This is shown in Figure 14a,c where the posterior view for both subjects is shown. The caused map had two local maxima both on the sensory motor areas. These are shown in Figure 14b,d where the dorsal view for both subjects is shown.
Figure 14.
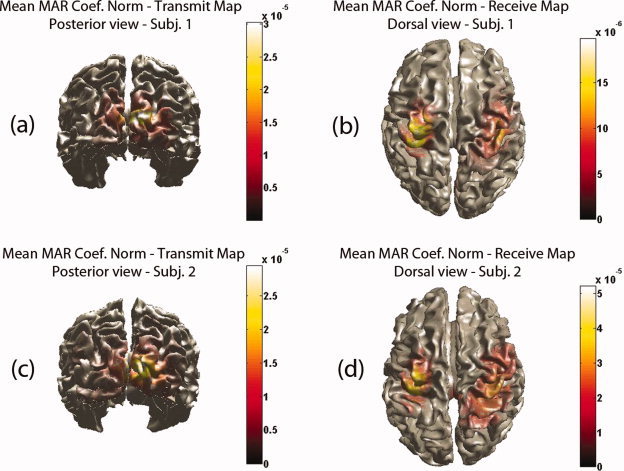
Real data: mean N coef (a) from each voxel to all voxels for subject 1—Posterior view (b) from all voxels to each voxel for subject 1—Dorsal view (c) from each voxel to all voxels for subject 2—Posterior view (b) from all voxels to each voxel for subject 2—Dorsal view. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
For this relatively simple motor planning task, one should expect to identify activated areas involved mainly in the dorsal visuomotor stream as the specific action is underlain by a ‘goal‐directed’ rather than a ‘matching’ representation [Milner and Dijkerman,2002; Prinz and Hommel,2002]. The dorsal stream is considered to emanate from the primary visual cortex and terminates on the motor cortex by projections through the premotor areas [Hoshi and Tanji,2007; Prinz and Hommel,2002]. Indeed by using the maps of N coef mean, the visual cortex has been identified as the causal area and the motor cortex as the caused area. The results of this preliminary investigation show that the use of the MAR coefficients directly provides a meaningful functional network of causality.
CONCLUSIONS
In this work it was investigated how feasible it is to build a MAR model on the principal components of the MEG sensor data, then project the model coefficients through an inverse solution to brain locations and derive from it meaningful causality information. Theoretically, the projection is feasible and the main uncertainty factor is the mixing resulting from the nonuniqueness of the various inverse solutions. The feasibility of this approach was investigated through three different investigations with simulated MEG data from known activity locations inside the brain. From these three investigations the following conclusions were drawn:
Investigation 1: Six dipoles with predefined causality configuration were simulated as a network of dumped oscillators with nominal frequency of 8 Hz. Causal connections existed between sources 1–5. Source 6 had the highest power but no causal connections to any of the other sources. It was used to infer if the presence of a strong unconnected source can affect the recovery of causality between the other sources.
When such a small number of known activity locations are considered, projecting the MAR model from sensor principal component space to these locations and using PDC as causality metric, correctly recovers the causality information between different pairs. In this case, it was shown that this methodology is quite tolerant of sensor noise (white Gaussian) and background brain noise (spatiotemporally correlated) up to levels of four times the rms value of the brain signal, but can suffer when low‐sampling rate is combined with high model order, in other words when the total number of sampling points is low with respect to the number of coefficients that have to be estimated. It was also shown that significance levels estimated by a jackknife method (LOOM) can be used to distinguish significant PDC values. Investigation 2: When the method is used with a very large number of voxels considered as potential activity locations, PDC seems to indicate the correct areas where real causal connections exist but does not seem to correctly reconstruct the individual causal pathways and ghosts of real connections appear between other voxel pairs. Maps of the mean of PDC caused to each voxel seem to provide an acceptable initial indication of the causal areas in which also caused locations might show up because of ghost connections. Maps of the mean of PDC caused by each voxel are not feasible as PDC is normalized with reference to the causal voxel.Investigation 3: When the method is used with a very large number of voxels, a new causality metric was investigated. This is the norm of MAR model coefficients across time‐lags, termed here as N coef. The norm metric can be used to construct maps of both causal and caused connections due to the fact that the MAR model coefficients between different pairs are comparable within the same model. The causal and caused maps of the norm metric for individual voxels seem to indicate the correct areas where real causal connections exist but again ghost connections might appear. However, maps of the mean of norm metric from each voxel to all voxels, and from all voxels to each voxel, showed that ghost connections are weak relative to the real connections and both the causal and the caused maps seem to correctly resemble reality.
In an attempt to quantify the accuracy of the N coef maps with respect to the known locations of the six simulated dipoles, the local maxima were computed and their distance from the actual locations was calculated. The local maxima accuracy was in most cases in the order of 1 cm and in one case 3 cm.
From the above three investigations it was concluded that the MAR model coefficient Norm metric is more appropriate to be used when a very large number of voxels is used as potential activity sources and when no a priori assumptions are made about activity locations. This is due to the fact that PDC employs a semiarbitrary normalization based on the causal voxel. This means that pairs with different causal pairs cannot be compared in terms of PDC. The MAR coefficients contain the linear weights between variables at different time lags. The higher the coefficient values are, the strongest the linear interaction. The main issue regarding the coefficients is that they are not normalized and thus they are not bounded. But within the same model the coefficients are scaled according to the strength of linear interactions. The MAR coefficients although unbounded, do not have the normalization problem of PDC and that is why their maps are much more consistent.
Maps of N coef from each voxel to all voxels, and from all voxels to each voxel, can provide a good initial indication of the causal and caused hubs inside the brain. Such maps based on PDC appear to be less reliable. On the basis of these maps the hub or cluster centers can be selected, so that few voxels will represent the functional network topology. Then the MAR model can be projected in only these few locations of activity and the PDC can then be used to infer causality, as in such cases of few activated areas, the use of PDC was shown to be relatively robust. Also in such cases confidence intervals of PDC can be used to estimate significant levels. From all three investigations it was also observed that source 6 did not appear in the caused and causal maps for each of the other five sources. This means that causality recovered by PDC and N coef for each of the five connected sources was not significantly affected by the unconnected source 6, although it had the highest power.
To further demonstrate the feasibility of the N coef maps, real data for two subjects from a simple motor planning MEG experiment was used. Through the N coef maps, the visual cortex was identified as the causal end of the functional network and the sensorimotor cortex as the caused end. This seems to be in agreement with the functional structure of the dorsal stream, which is activated during such goal‐directed visuomotor tasks.
On the basis of the above conclusions, the methodology of building the MAR model in the sensor space and projecting it, even in a very large number of voxels, inside the brain in order to estimate causality is a feasible approach and can be used to provide entire brain maps of causality.
Re‐use of this article is permitted in accordance with the Terms and Conditions set out at http://wileyonlinelibrary.com/onlineopen#OnlineOpen_Terms
REFERENCES
- Astolfi L, Cincotti F, Mattia D, de Vico Fallani F, Lai M, Baccalá LA, Salinari S, Ursino M, Zavaglia M, Babiloni F ( 2005): Comparison of different multivariate methods for the estimation of cortical connectivity: Simulations and applications to EEG data. Conf Proc IEEE Eng Med Biol Soc 5: 4484–4487. [DOI] [PubMed] [Google Scholar]
- Baccalá LA, Sameshima K ( 2001): Partial directed coherence: A new concept in neural structure determination. Biol Cybern 84: 463–474. [DOI] [PubMed] [Google Scholar]
- Baccalá LA, Sameshima K ( 2006): Comments on is partial coherence a viable technique for identifying generators of neural oscillations?: Why the term gersch causality is inappropriate: Common neural structure inference pitfalls. Biol Cybern 95: 135–141. [DOI] [PubMed] [Google Scholar]
- Baillet S, Riera JJ, Marin G, Mangin JF, Aubert J, Garnero L ( 2001): Evaluation of inverse methods and head models for eeg source localization using a human skull phantom. Phys Med Biol 46: 77–96. [DOI] [PubMed] [Google Scholar]
- Bijma F, de Munck JC, Huizenga HM, Heethaar RM ( 2003): A mathematical approach to the temporal stationarity of background noise in MEG/EEG measurements. NeuroImage 20: 233–243. [DOI] [PubMed] [Google Scholar]
- Chen Y, Bressler SL, Ding M ( 2006): Frequency decomposition of conditional Granger causality and application to multivariate neural field potential data. J Neurosci Methods 150: 228–237. [DOI] [PubMed] [Google Scholar]
- Chen Y, Bressler SL, Ding M ( 2009): Dynamics on networks: assessing functional connectivity with Granger causality. Comput Math Organ Theory 15: 329–350. [Google Scholar]
- David O, Kiebel SJ, Harrison LM, Mattout J, Kilner JM, Friston KJ ( 2006): Dynamic causal modeling of evoked responses in EEG and MEG. Neuroimage 30: 1255–1272. [DOI] [PubMed] [Google Scholar]
- de Munck JC, Huizenga HM, Waldorp LJ, Heethar RM ( 2002): Estimating stationary dipoles from MEG/EEG data contaminated with spatially and temporally correlated background noise. IEEE Trans Signal Process 50: 1565–1572. [Google Scholar]
- Faes L, Porta A, Nollo G ( 2010): Testing frequency‐domain causality in multivariate time series. IEEE Trans Biomed Eng 57: 1897–1906. [DOI] [PubMed] [Google Scholar]
- Geweke J ( 1982): Measures of conditional linear dependence and feedback between time series. J Am Stat Assoc 77: 304–313. [Google Scholar]
- Geweke J ( 1984): Measures of conditional linear dependence and feedback between time series. J Am Stat Assoc 79: 907–915. [Google Scholar]
- Gómez‐Herrero G, Atienza M, Egiazarian K, Cantero JL ( 2008): Measuring directional coupling between eeg sources. Neuroimage 43: 497–508. [DOI] [PubMed] [Google Scholar]
- Gross J, Ioannides AA ( 1999): Linear transformations of dataspace in MEG. Phys Med Biol 44: 2081–2097. [DOI] [PubMed] [Google Scholar]
- Gross J, Kujala J, Hämäläinen M, Timmermann L, Schnitzler A, Salmelin R ( 2001): Dynamic imaging of coherent sources: Studying neural interactions in the human brain. Proc Natl Acad Sci U S A 98: 694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Timmermann L, Kujala J, Dirks M, Schmitz F, Salmelin R, Schnitzler A ( 2002): The neural basis of intermittent motor control in humans. Proc Natl Acad Sci U S A 99: 2299–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämäläinen MS ( 1992): Magnetoencephalography: A tool for functional brain imaging. Brain Topogr 5: 95–102. [DOI] [PubMed] [Google Scholar]
- Haufe S, Tomioka R, Nolte G, Mller K‐R, Kawanabe M.( 2010): Modeling sparse connectivity between underlying brain sources for EEG/MEG. IEEE Trans Biomed Eng 57: 1954–1963. [DOI] [PubMed] [Google Scholar]
- Hoechstetter K, Borneth H, Weckesser D, Ille N, Berg P, Scherg M ( 2004): Besa source coherence: A new method to study cortical oscillatory coupling. Brain Topogr 16: 233–238. [DOI] [PubMed] [Google Scholar]
- Hoshi E, Tanji J ( 2007): Distinctions between dorsal and ventral premotor areas: Anatomical connectivity and functional properties. Curr Opin Neurobiol 17: 234–242. [DOI] [PubMed] [Google Scholar]
- Jerbi K, Lachaux J‐P, N'Diaye K, Pantazis D, Leahy RM, Garnero L, Baillet S ( 2007): Coherent neural representation of hand speed in humans revealed by meg imaging. Proc Natl Acad Sci U S A 104: 7676–7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe IT ( 2002): Principal Component Analysis. New York: Springer‐Verlag. [Google Scholar]
- Jun SC, Pearlmutter BA, Nolte G ( 2002): Fast accurate MEG source localization using a multilayer perceptron trained with real brain noise. Phys Med Biol 47: 2547–2560. [DOI] [PubMed] [Google Scholar]
- Kaminski M, Ding M, Truccolo WA, Bressler SL ( 2001): Evaluating causal relations in neural systems: Granger causality, directed transfer function and statistical assessment of significance. Biol Cybern 85: 145–157. [DOI] [PubMed] [Google Scholar]
- Kaminski MJ, Blinowska KJ ( 1991): A new method of the description of the information ow in the brain structures. Biol Cybern 65: 203–210. [DOI] [PubMed] [Google Scholar]
- Kiebel SJ, Garrido MI, Moran R, Chen C‐C, Friston KJ ( 2009): Dynamic causal modeling for EEG and MEG. Hum Brain Mapp 30: 1866–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaux JP, Rodriguez E, Martinerie J, Varela FJ ( 1999): Measuring phase synchrony in brain signals. Hum Brain Mapp 8: 194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F‐H, Witzel T, Hämäläinen MS, Dale AM, Belliveau JW, Stufflebeam SM ( 2004): Spectral spatiotemporal imaging of cortical oscillations and interactions in the human brain. Neuroimage 23: 582–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütkenhöner B ( 1994): Magnetic field arising from current dipoles randomly distributed in a homogeneous spherical volume conductor. J Appl. Phys. 75: 7204–7210. [Google Scholar]
- McQuarrie AD, Tsai C‐L ( 1998): Regression and Time Series Model Selection. Singapore: World Scientific Publishing. [Google Scholar]
- Milner D, Dijkerman C ( 2002): Out of Mind: Varieties of Unconscious Processes. Oxford: Oxford University Press; pp 241–265. [Google Scholar]
- Nolte G, Ziehe A, Nikulin VV, Schlögl A, Krämer N, Brismar T, Müller KR ( 2008): Robustly estimating the flow direct ion of information in complex physical systems. Phys Rev Lett 100: 23410. [DOI] [PubMed] [Google Scholar]
- Oostenveld R, Fries P, Maris E, Schoffelen JM ( 2011): FieldTrip: Open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput Intell Neurosci 2011: 156869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmero‐Soler E, Dolan K, Hadamschek V, Tass PA ( 2007): Swloreta: A novel approach to robust source localization and synchronization tomography. Phys Med Biol 52: 1783–1800. [DOI] [PubMed] [Google Scholar]
- Pichard S. Minimamaxima3D matlab toolbox. (2007): URL: http://www.mathworks.com/matlabcentral/fileexchange/17997
- Pollok B, Gross J, Dirks M, Timmermann L, Schnitzler A ( 2004): The cerebral oscillatory network of voluntary tremor. J Physiol 554( Pt 3): 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollok B, Südmeyer M, Gross J, Schnitzler A ( 2005): The oscillatory network of simple repetitive bimanual movements. Brain Res Cogn Brain Res 25: 300–311. [DOI] [PubMed] [Google Scholar]
- Prinz W, Hommel B, editors ( 2002): Common Mechanisms in Perception and Action. Oxford: Oxford University Press; pp 73–118. [Google Scholar]
- Ramirez R, Baillet S ( 2010): Efficient computation of high‐dimensional MEG/EEG source connectivity and causality measures. BIOMAG 2010, poster number: 317. [Google Scholar]
- Roebroeck A, Formisano E, Goebel R ( 2005): Mapping directed influence over the brain using Granger causality and fMRI. Neuroimage 25: 230–242. [DOI] [PubMed] [Google Scholar]
- Schelter B, Winterhalder M, Eichler M, Peifer M, Hellwig B, Guschlbauer B, Lücking CH, Dahlhaus R, Timmer J ( 2006): Testing for directed influences among neural signals using partial directed coherence. J Neurosci Methods 152: 210–219. [DOI] [PubMed] [Google Scholar]
- Schelter B, Timmer J, Eichler M ( 2009) Assessing the strength of directed influences among neural signals using renormalized partial directed coherence. J Neurosci Methods 179: 121–130. [DOI] [PubMed] [Google Scholar]
- Schlögl A, Supp G ( 2006): Analyzing event‐related EEG data with multivariate autoregressive parameters. Prog Brain Res 159: 135–147. [DOI] [PubMed] [Google Scholar]
- Schnitzler A, Gross J ( 2005a): Functional connectivity analysis in magnetoencephalography. Int Rev Neurobiol 68: 173–195. [DOI] [PubMed] [Google Scholar]
- Schnitzler A, Gross J ( 2005b) Normal and pathological oscillatory communication in the brain. Nat Rev Neurosci 6: 285–296. [DOI] [PubMed] [Google Scholar]
- Schnitzler A, Gross J, Timmermann L ( 2000) Synchronised oscillations of the human sensorimotor cortex. Acta Neurobiol Exp (Wars) 60: 271–287. [DOI] [PubMed] [Google Scholar]
- Schoffelen J‐M, Gross J ( 2009): Source connectivity analysis with MEG and EEG. Hum Brain Mapp 30: 1857–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoffelen J‐M, Oostenveld R, Fries P ( 2005): Neuronal coherence as a mechanism of effective corticospinal interaction. Science 308: 111–113. [DOI] [PubMed] [Google Scholar]
- Schoffelen J‐M, Oostenveld R, Fries P ( 2008) Imaging the human motor system's beta‐band synchronization during isometric contraction. Neuroimage 41: 437–447. [DOI] [PubMed] [Google Scholar]
- Timmermann L, Gross J, Dirks M, Volkmann J, Freund H‐J, Schnitzler A ( 2003): The cerebral oscillatory network of parkinsonian resting tremor. Brain 126( Pt 1): 199–212. [DOI] [PubMed] [Google Scholar]
- Valdés‐Sosa PA, Sánchez‐Bornot JM, Lage‐Castellanos A, Vega‐Hernández M, Bosch‐Bayard J, Melie‐García L, Canales‐Rodríguez E.( 2005): Estimating brain functional connectivity with sparse multivariate autoregression. Philos Trans R Soc Lond B Biol Sci 360: 969–981. [DOI] [PMC free article] [PubMed] [Google Scholar]


