Abstract
Molecular imaging is an emerging discipline which plays critical roles in diagnosis and therapeutics. It visualizes and quantifies markers that are aberrantly expressed during the disease origin and development. Protein molecules remain to be one major class of imaging probes, and the option has been widely diversified due to the recent advances in protein engineering techniques. Antibodies are part of the immunosystem which interact with target antigens with high specificity and affinity. They have long been investigated as imaging probes and were coupled with imaging motifs such as radioisotopes for that purpose. However, the relatively large size of antibodies leads to a half-life that is too long for common imaging purposes. Besides, it may also cause a poor tissue penetration rate and thus compromise some medical applications. It is under this context that various engineered protein probes, essentially antibody fragments, protein scaffolds, and natural ligands have been developed. Compared to intact antibodies, they possess more compact size, shorter clearance time, and better tumor penetration. One major challenge of using protein probes in molecular imaging is the affected biological activity resulted from random labeling. Site-specific modification, however, allows conjugation happening in a stoichiometric fashion with little perturbation of protein activity. The present review will discuss protein-based probes with focus on their application and related site-specific conjugation strategies in tumor imaging.
Keywords: Tumor targeting, Antibody, Human epidermal growth factor receptor (HER), Vascular endothelial growth factor (VEGF)
Introduction
Molecular imaging is an emerging discipline which plays more and more important roles in diagnosis and therapeutics. It emphasizes the visualization, characterization, and measurement of biological processes at the molecular and cellular levels in humans and other living systems (Mankoff 2007). Stemming from the conventional imaging methodologies, molecular imaging techniques include magnetic resonance imaging (MRI), magnetic resonance spectroscopy (MRS), ultrasound, single-photon emission computed tomography (SPECT), positron emission tomography (PET), and in the pre-clinical small animal setting, optical bioluminescence and fluorescence imaging. Compared with traditional methods, it outweighs by providing higher spatial, temporal and even quantitative resolution. Molecular imaging turns early stage diagnosis and treatment into possibility, and is indispensible in the coming era of individualized therapy and therapeutic monitoring.
Molecular imaging typically targets markers that are aberrantly expressed during the disease origination and development. For example, in many cases of tumor imaging, biomarkers are antigens overexpressed on either the tumor cells or tumor vasculatures. Such tumor markers can be used to describe the morphology and stage of tumor. When appropriate probes of imaging activity and antigen targeting specificity are applied, tumor area will be located and lighten up. A large number of tumor markers that could be used for monitoring malignant disease have recently been reviewed (Voorzanger-Rousselot and Garnero 2007).
Protein molecules remain to be one major class of imaging probes, and the option has been widely diversified because of the recent progress in protein engineering techniques. These techniques now allow biologists to engineer proteins for altered, improved, or even novel features. In parallel, techniques of conjugating proteins to imaging motifs have been developed. This is a practically critical topic, which addresses the issue of minimally impacting the protein activity and pharmacokinetics. In the field of molecular imaging, several criteria have to be considered when screening the probes. First, they must have high tumor target specificities but minimal normal tissue affinities. Furthermore, rapid clearance of probes is preferential. Large proteins such as antibodies have a relatively long circulation time and a suboptimal tissue penetration rate therefore are less practical for imaging purpose. It is under this context that certain antibody fragments, protein scaffolds, or even peptides are generated and more favorably utilized, which have sizes below the threshold for kidney filtration (approx. 60 kDa) (Behr et al. 1998; Holliger and Hudson 2005).
Current methods for protein site-specific modification generally fall into three categories. The first category takes advantage of reactions between small molecule probes and specific tags of target proteins. The second category of protein site-specific labeling reactions is based on enzyme-catalyzed protein post-translational modifications. The third one is the intein-based labeling strategy. Site-specific conjugations of radioisotope, fluorophores or other detectable labels onto a protein ligand have been a valuable tool for such molecular imaging because of their stoichiometric and bioactive manners.
In this review, we will first discuss several valuable protein probes which have shown wide applications in molecular imaging, and then we will focus on some useful site-specific modification strategies to conjugate protein ligands with labeling chemicals.
Common tumor markers
Despite the growing population of imaging probes, the number of probes used to target tumor antigens is surprisingly small (Leader et al. 2008). Generally speaking, tumor targeting protein probes can be divided into the following groups based on their targets (Friedman and Stahl 2009), and a brief discussion on the representative tumor markers will be given in this section:
in solid tumors [e.g., epidermal growth factor receptor (EGFR) family, epithelial cell adhesion molecule (EpCAM), carcinoembryonic antigen (CEA) and insulin-like growth factor receptor 1 (IGF-1R)];
in lymphomas (e.g., CD20, CD52, CD33, CD4);
in tumor stroma [e.g., fibroblast activation protein (FAP), tenascin];
in tumor vasculature [e.g., fibronectin ED-B, prostate-specific membrane antigen (PSMA) and vascular endothelial growth factor receptor 2 (VEGFR2)] or
a ligand [e.g., vascular endothelial growth factor (VEGF)].
Human epidermal growth factor receptor (HER) family
The epidermal growth factor (EGF) and EGFR were among the first growth factor ligand–receptor pairs discovered (Lin et al. 1984; Downward et al. 1984). Later on, EGFR was found to be a receptor tyrosine kinase (TK) in the HER family. There are four closely related members in this family, i.e., EGFR (HER1 or ErbB1), HER2 (ErbB2), HER3 (ErbB3), and HER4 (ErbB4) (Casalini et al. 2004; Mass 2004). Together, the HER family controls a complex network of ligand–receptor interactions and cellular responses known as the HER-kinase axis (Gross et al. 2004). Ligands that bind to the HER-kinases include over ten independent proteins such as EGF, transforming growth factor (TGF)-α,β-cellulin, and neuregulins/heregulins, many of which exhibit relatively similar receptor-binding characteristics (Casalini et al. 2004; Mass 2004). Alterations and disruptions in the function of the HER-kinase axis can lead to malignancy. Among the receptors in HER family, EGFR and HER2 are the most popular tumor markers in many types of cancers, such as prostate, lung, gastric, and oral cancers (Hofer et al. 1996; Slamon et al. 1989; Schneider et al. 1989), and their quantitative imaging may aid in lesion detection, new drug development, and treatment monitoring in a noninvasive manner.
EGFR is a 170-kDa cell surface protein overexpressed in many epithelial cancers (Arteaga 2003; Sebastian et al. 2006). Dysregulation of EGFR is associated with several key features of cancer, such as autonomous cell growth, inhibition of apoptosis, angiogenic potential, invasion and metastases (Schlessinger 2000; Normanno et al. 2006). EGFR is activated by EGF and TGF-α on its external domain, and its intracellular TK domain is afterward phosphorylated to initiate downstream cell proliferation, transformation and division. In many cases, the aberrant EGFR activation is an important factor in tumorigenesis and a leading cause for the aggressive growth of cancer cells.
HER2 oncogene encodes a transmembrane TK of 1,255 amino acids which is the receptor for a family of polypeptide growth factors. HER2 is a glycoprotein, with a molecular weight of 185 kDa, normally expressed in the epithelia of different organs such as the lung, bladder, pancreas and prostate. The ectodomain of the HER2 protein can be proteolytically cleaved from the intact receptor and released as a soluble molecule (Wikman et al. 2006). HER2 tissue overexpression and elevated serum HER2 levels have been observed in breast, prostate, ovarian and lung carcinoma (Sandstrom et al. 2003; Kronqvist et al. 2008; Friedman et al. 2008; Gunneriusson et al. 1999).
VEGFR
Angiogenesis is the formation of new blood vessels, a process highly implicated in tumor progression, wound healing, cardiovascular, inflammatory, ischemic, and infectious diseases (Folkman 1995; Bergers and Benjamin 2003; Carmeliet 2005). One of the most extensively studied angiogenesis-related signaling pathways is VEGF/VEGFR interactions (Ferrara 2002, 2004), which has been intensively investigated for both therapeutic and imaging purposes. The angiogenic actions of VEGF are mainly mediated via two endothelium-specific receptor TKs, Flt-1 (VEGFR-1) and Flk-1/KDR (VEGFR-2) (Hicklin and Ellis 2005). Both VEGF and VEGFR have been evaluated as targets for imaging probe development. This is important for providing an approach to assess anti-angiogenic therapeutics and to better understand the role and expression profile of VEGF/VEGFR in many angiogenesis-related diseases.
CEA
CEA is a well-characterized tumor-associated glycoprotein that is expressed by a wide variety of epithelial malignancies, including colorectal, non-small cell lung cancer (NSCLC) and breast carcinomas. Besides tumor cell surface, CEA is also shed into circulation. CEA has been investigated clinically as the target for radioimmuno-therapy (RIT) and antibody-directed enzyme prodrug therapy (ADEPT), and in radioimmuno-guided surgery (RIGS) (Wong et al. 2006; Francis et al. 2002; Mayer et al. 2000).
Integrins
Integrins are a family of cell adhesion molecules consisting of two transmembrane subunits, α and β. Both subunits are type I membrane proteins which pair to form heterodimers with distinct adhesive capabilities (Hynes 2002). So far, 18 α-subunits and 8 β-subunits have been indentified in mammals, which assemble into at least 24 different receptors. Interests on integrin signaling in the imaging setting mainly arise from its implication in tumor angiogenesis and metastasis (Hood and Cheresh 2002). Integrins expressed on endothelial cells modulate cell migration and survival during tumor angiogenesis, whereas integrins expressed on carcinoma cells potentiate metastasis by facilitating invasion and movement across blood vessels. Especially, integrin ανβ3, which binds to arginine-glycineaspartic acid (RGD)-containing components with high affinity, is proved to be an efficient tumor marker with high expression on tumor vasculature but not on quiescent endothelium (Hood and Cheresh 2002; Xiong et al. 2002). Probes in the forms of monoclonal antibodies (mAbs), cyclic RGD peptide antagonists, and peptidomimetic agents against integrin ανβ3 have been intensively investigated in both imaging and therapy settings with success (Cai and Chen 2006).
Antibodies
Antibodies, or immunoglobulins, are part of the immunosystem which interact with target antigens with high specificity and affinity. They have long been investigated as imaging probes and have been coupled with imaging motifs such as radioisotopes for that purpose.
Characteristics of antibody
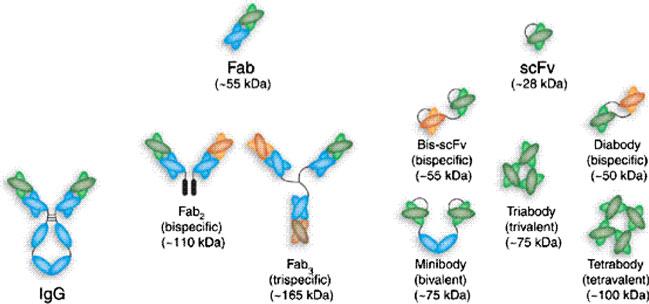
The Y-shaped mAb molecule has two identical antigen-binding fragments (Fab). Each Fab is comprised of one light chain (L) and one heavy chain (H), which are held together by disulfide bond (Fig. 1) (Holliger and Hudson 2005). Both the light and the heavy chains consist of a variable (V) and a constant part (C). The variable regions of H and L chains combine to form two identical antigen-binding sites containing six hypervariable loops, referred to as CDRs. The stem of the Y-shaped mAb, Fc, is responsible for recruiting different effector functions and can provide longer half-lives through its interaction with Fc receptors (Ward and Ghetie 1995; Roopenian and Akilesh 2007).
Fig. 1.
Schematic representation of different antibody formats. Reproduced with permission from Holliger and Hudson (2005)
Previously utilized mAbs were of murine origin, whose clinical applications encountered the problems of immunogenicity, low ability to recruit immune effector cells and short serum half-lives. These issues have been solved with the emerging chimerization (Boulianne et al. 1984) and humanization (Jones et al. 1986) techniques, which, while retaining the targeting specificity and affinity, incorporate human Fc in the protein structure to lower the risk of immunogenicity. The hybridoma technology, together with humanization of antibodies of animal origin or by the use of transgenic mice (Fishwild et al. 1996), has been very successful technologies in generating therapeutic mAbs.
Antibody imaging
SPECT
The use of mAb-based tracers has gained acceptance in its own right for imaging-specific molecular targets in disease. Before PET technology became broadly available, extensive experience was gained with SPECT camera imaging of mAbs. For this purpose, a γ-emitting radionuclide, such as 99mTc, 111In, 131I, or 186Re, was coupled to the mAb (Colnot et al. 2000; Takahashi et al. 1995; De Bree et al. 1994). The first HER2-specific imaging study was performed by Saga et al. (1991) with a class-switched murine mAb SV2-61r. Tumor uptake and localization index of 125I-SV2-61r were lower than those of 111In-labeled SV2-61r, probably due to the internalization and dehalogenation of the formed antibody–antigen complexes. ICR12 mAb is a rat IgG2a protein directed against an epitope of the external domain of HER2 (Styles et al. 1990; Dean et al. 1993). 99mTc-labeled ICR12 was evaluated in a mouse model system and averaged 20% localization of the total injected dose per gram of tumor at 24-h time point. In patients selected by IHC staining, SPECT imaging with 99mTc-ICR12 showed good tumor localization in both the primary lesion and regional node metastases (Allan et al. 1994).
Many anti-EGFR mAbs have been labeled with γ-emitters for SPECT imaging. The mAb ior egf/r3 is an IgG2a protein that recognizes EGFR (Fernandez et al. 1992). The diagnostic efficacy of 99mTc-labeled ior egf/r3 for the detection of epithelial-derived tumors, metastases, and recurrences was evaluated in 148 patients (Ramos-Suzarte et al. 1999). Overall sensitivity, specificity, accuracy, and positive and negative predictive values of the immunoscintigraphic imaging were 84.2, 100.0, 86.5, 100 and 52.4%, respectively. New metastases not identified previously by other diagnostic methods were detected in 50% of the patients. Therefore, this tracer was useful for the diagnosis and follow-up of cancer patients. The type III mutant EGFR is expressed on the cell surface of a subset of glioma instead of normal tissues, and mAb 3C10 specifically recognizes EGFRvIII (Takasu et al. 2003). 99mTc-3C10 was significantly accumulated to tumor xenografts transplanted subcutaneously or intracranially in nude mice, showing high tumor-to-blood ratios of 10.30 and 4.01, respectively. In scintigrams, intracranially transplanted tumor xenografts were detectable at 3 h after injection of 99mTc-labeled 3C10 mAb. These results suggest that 3C10 mAb is a potential diagnostic and therapeutic agent for patients with gliomas expressing EGFRvIII (Takasu et al. 2003).
Several other anti-EGFR antibodies have been labeled with 99mTc, using various chelators and bifunctional agents, and tested in animal models (Scopinaro et al. 1997; Schechter et al. 2003). The mouse anti-EGFR mAb C225 has been labeled with 111In through diethylenetriamine penta-acetic acid (DTPA) chelator and investigated for its capacity to localize in human tumor xenografts (Goldenberg et al. 1989).
PET
Positron emitters
Positron emitters have been coupled to mAb through various mechanisms to arrive at antibody-based PET imaging probes. Appropriate positron-emitting radionuclides for immuno-PET should comply with several criteria, such as proper decay characteristics, low cost, and facile conjugation techniques with minimal impact to the properties of antibodies. A list of the commonly used positron emitters includes 68Ga, 18F, 64Cu, 86Y, 76Br, 89Zr, and 124I. Among them, 68Ga and 18F with relatively short half-lives (68 and 109.7 min) are usually coupled to mAb fragments; and 89Zr and 124I with long half-lives (3.3 and 4.2 days) are used to couple intact antibodies for long-term imaging. It is noteworthy that only 76Br and 124I can be coupled directly to mAbs. The remaining isotopes require the use of either bifunctional chelates or prosthetic groups.
Another concern that might affect the isotope selection is the stability of the label during circulation and within the target cells. It is possible that the mAbs may be internalized into the cells after binding to the target antigen, which is usually followed by protein degradation. In the case of 76Br and 124I labeling, this could result in the loss of cellular radionuclides, which are effluxed and thereafter not reflective of the mAb distribution. On the other hand, the metal isotopes, i.e., 68Ga, 64Cu, 86Y, and 89Zr, have higher preference of lysosome trapping. Thus, imaging of trastuzumab, cetuximab, and bevacizumab is best performed using a residualizing positron emitters.
Antibodies for PET imaging
Cetuximab, a chimeric mAb targeting EGFR, was approved by the FDA to treat patients with metastatic colorectal cancer (Cai et al. 2007a). Clinical and pre-clinical trials utilizing cetuximab for the treatment of other solid tumors are also under intensive investigations. Coupling PET isotopes to antibody and use the conjugates for tumor imaging has been reported by several groups. For instance, Cai et al. reported PET imaging of EGFR expression in xenograft-bearing mice using 64Cu-DOTA-labeled cetuximab. Highly specific EGFR targeting was observed as 64Cu-DOTA-cetuximab showed increased tumor activity accumulation over time in EGFR-positive tumors but relatively low uptake in EGFR-negative tumors (Fig. 2). The specificity was validated by western blotting analysis, which found good correlation between the tracer uptake and the EGFR expression level. It is expected that such imaging can be translated into clinics to favor the selection of right population of patients for EGFR-targeted therapy and to monitor the therapeutic efficacy of anti-EGFR treatment (Cai et al. 2007a).
Fig. 2.
Serial microPET images of different xenograft tumor models after intravenous injection of 64Cu-DOTA-cetuximab. Decay-corrected whole-body coronal images that contain the tumor were shown and the tumors are indicated by white arrows. Reproduced with permission from Cai et al. (2007a)
Similar 64Cu-DOTA technique has been utilized to label trastuzumab for HER2-related imaging. For example, Niu et al. utilized such conjugates to monitor the therapeutic response of 17-dimethylaminoethylamino-17-demethoxygeldanamycin (17-DMAG) on xenograft mouse models. 17-DMAG is a heat-shock protein 90 (Hsp 90) inhibitor that has been intensively investigated for cancer therapy and is undergoing clinical trials. It is expected that HER2, as one client protein of Hsp 90, may be down-regulated upon 17-DMAG treatment therefore can be used as a marker to monitor 17-DMAG therapy. In this study, therapeutic and imaging studies were performed on both SKOV-3 xenograft models and controls. Quantitative small animal PET imaging showed that 64Cu-DOTA-trastuzumab had prominent tumor accumulation in untreated SKOV-3 tumors, which was significantly reduced in 17-DMAG-treated tumors. Such reduction was attributed to the 17-DMAG treatment as immunofluorescence staining confirmed the significant reduction in tumor HER2 level. It was noteworthy that such uptake difference was not observed in the parallel FDG-PET imaging, indicating a unique role 64Cu-DOTA-trastuzumab may play in monitoring 17-DMAG treatment response (Niu et al. 2009). Also, in clinical trials, 89Zr-trastuzumab has been administrated into patients with HER2-overexpressing breast cancer to assess and quantify HER2 expression of all lesions, including non-accessible metastases (Dijkers et al. 2009). The comparative biodistribution study showed a higher level of 89Zr-trastuzumab in HER2-positive tumors than in HER2-negative tumors, especially at day 6 [33.4 ± 7.6 (mean ± SEM) vs. 7.1 ± 0.7 percentage injected dose per gram of tissue]. Of the tested protein doses, it was found that a minimal dose of trastuzumab (100 μg) was sufficient to provide good imaging quality.
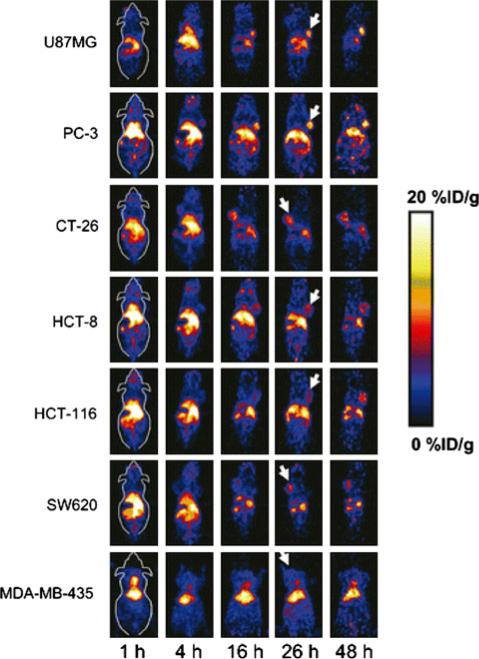
Abegrin (MEDI-522 or Vitaxin), a humanized mAb against human integrin ανβ3, is in clinical trials for cancer therapy (Cai et al. 2006a). Cai et al. conjugated Abegrin with 64Cu-DOTA at five DOTA/Abegrin ratios and tested conjugates in three human (U87MG, MDA-MB-435, and PC-3) and one mouse (GL-26) tumor models. Such 64Cu-DOTA-Abegrin has the clinical translation potential to help characterize the pharmacokinetics, tumor targeting efficacy, dose optimization, and dose interval of Abegrin and/or Abegrin conjugates.
A number of radiolabeled anti-VEGF antibodies have been reported. For example, VG76e, an IgG1 mAb that binds to human VEGF, was labeled with 124I for PET imaging of solid tumor xenografts in immunodeficient mice (Collingridge et al. 2002). Although VEGF specificity in vivo was demonstrated, a poor immunoreactivity (<35%) of the radiolabeled antibody was observed and therefore limited the perspective of such tracer. HuMV833, a humanized anti-VEGF mAb, has also been labeled with 124I and investigated in a phase I clinical trial (Jayson et al. 2002). Patients with progressive solid tumors were treated with various doses of HuMV833, and PET imaging with 124I-HuMV833 was performed to measure the antibody distribution and clearance from tissues. The antibody distribution and clearance were found to be quite heterogeneous, not only among patients but also within individual tumors. These results suggest that intrapatient dose escalation approaches or more precisely defined patient cohorts will be needed in the design of future phase I studies with anti-angiogenic antibodies such as HuMV833.
In a recent study, bevacizumab was labeled with 111In and 89Zr for SPECT and PET, respectively (Nagengast et al. 2007). Nude mice xenografted with SKOV-3 tumors were injected with 89Zr-bevacizumab, 111In-bevacizumab, or 89Zr-IgG. PET revealed tracer uptake in well-perfused organs up to 24 h after injection and clear tumor localization at 72 h after injection and beyond. However, the tumor uptake of 89Zr-bevacizumab, although higher than that of 89Zr-IgG, was much lower than that of other radiolabeled antibodies reported in the literature (Cai et al. 2006a, 2007a, b). Also, there was no clear evidence that VEGF expression was upregulated in the tumors. The higher uptake of 89Zr-bevacizumab than 89Zr-IgG may have been attributed to the different levels of passive targeting of individual antibodies, even though they were isotype matched IgGs.
Optical imaging
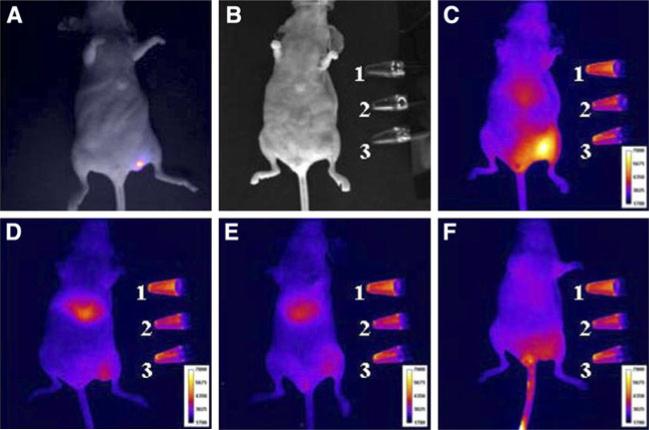
In pre-clinical studies, fluorescent dyes have been conjugated with antibodies for small animal tumor imaging. For example, a cocktail of three fluorescently labeled mAbs, namely cetuximab-Cy5 (targeting HER-1), trastuzumab-Cy7 (targeting HER2), and daclizumab-AlexaFluor-700 (targeting IL-2Ra), were injected into mice with three xenografts that overexpressing different subtypes of EGFR receptor, which were HER-1 (A431), HER2 (NIH3T3/HER2(+)), and interleukin-2 receptor alpha-subunit receptor (IL-2Ralpha; SP2/Tac). It was revealed that, following multifilter spectrally resolved imaging, different tumor types can be simultaneously distinguished and diagnosed in vivo (Koyama et al. 2007). Also, paired fluorophores with mutual-quenching effect can be coupled to antibodies to generate activatable probes. With this system, fluorescence is quenched by the fluorophore–quencher interaction outside cancer cells, but is activated within the target cells by dissociation of the fluorophore–quencher pair. In one example, TAMRA (fluorophore) and QSY7 (quencher) were coupled to trastuzumab and the resulting conjugates were found to be able to detect tumors (Ogawa et al. 2009). Furthermore, more than one imaging motifs can be coupled simultaneously onto antibodies to achieve imaging probes with multiple imaging capability. For example, IRDye800 was coupled to trastuzumab along with 111In-DTPA, and the resulting conjugates were injected on SKBr3 tumor model for NIRF/PET dual modality imaging (Fig. 3) (Sampath et al. 2007).
Fig. 3.
SKBR3-luc cancer cells were inoculated into left flank of athymic nu/nu mice. a Bioluminescence image overlaid on white-light image. White-light image of b mouse taken before fluorescence imaging of (111In-DTPA)n-trastuzumab-(IRDye800)m (c), 200-fold molar excess of trastuzumab followed by (111In-DTPA)n-trastuzumab-(IRDye800)m (d), (111In-DTPA)p-IgG-(IRDye800)q (e), or equivalent dose of IRDye800CW (f) 48 h after administration. Standards placed on side include: (1) (111In-DTPA)n-trastuzumab-(IRDye800)m, (2) (111In-DTPA)p-IgG-(IRDye800)q, and (3) IRDye800CW. Reproduced with permission from Sampath et al. (2007)
Antibodies have also been coupled with nanoparticles with various kinds of imaging activities. For instance, antibodies were coupled to Au nanorods and the resulting conjugates were used for photoacoustic (Li et al. 2008) and SERS imaging (Park et al. 2009). HER2 antibodies were coupled to single-walled carbon nanotubes for the detection and selective destruction of breast cancer cells (Xiao et al. 2009). However, most of these studies are still at proof-of-concept level and their clinical relevance or translation perspectives are unclear at the current stage.
Summary
Antibody-based therapy is emerging as an important discipline, and under this topic, antibody-based imaging has been extensively studied. Although antibodies have superior targeting selectivity and affinity, they are not necessarily the ideal candidate probes for molecular imaging. First of all, the relatively large size of antibodies leads to a half-life that is too long for common imaging. Besides, it also cause a poor tissue penetration rate that may compromise some medical applications (Holliger and Hudson 2005; Beckman et al. 2007). Second, the Fc-mediated immunological effector functions are only desirable for certain applications, and an inappropriate activation of Fc-receptor-expressing cells, such as neutrophils, NK (natural killer) cells and macrophages, can lead to undesired side effects. Moreover, it is economically impractical to use antibody-based probes for imaging purposes in daily practice.
Antibody derivatives
As mentioned above, despite the excellent target specificity and affinity, the long circulation half life and the associated effector functions of the antibodies have limited their usages in clinical imaging. To address these issues, engineered protein probes, essentially antibody fragments, have been developed, which possess more compact size in relation to the parent antibodies. Along this line, these fragments have been rebuilt into multivalent reagents to achieve higher avidity.
Fab and scFv
In the imaging setting, the inclusion of Fc is frequently regarded as an undesirable feature of antibodies. Since the targeting binding avidity mainly comes from the Fab region, it naturally raises the interest of dissecting the antibody to obtain Fab fragment, or even scFv, which is half of the Fab fragment. This was achieved initially through proteolysis with papain and pepsin, and later by genetic engineering. The resulting Fab and scFv fragments usually retain the specific, monovalent, antigen-binding affinity of the parent mAbs, only with improved pharmacokinetics for tissue penetration. Several recombinant Fabs and scFvs are currently approved by the FDA or are in late-stage clinical development (Holliger and Hudson 2005; Leader et al. 2008).
For example, trastuzumab Fab was prepared by digestion of intact IgG with immobilized papain. The resulting trastuzumab Fab exhibited preserved immunoreactivity in binding to HER2-positive SKBR-3 human breast cancer cells. Such Fab fragment of trastuzumab has been labeled with 99mTc and 111In through hydrazinenicotinamide (HYNIC) and DTPA chelators, respectively (Tang et al. 2005; Tolmachev et al. 2007). Both the radiolabeled probes were tested on a HER2-positive breast cancer xenograft model and demonstrated good targeting specificity (Tang et al. 2005; Tolmachev et al. 2007). However, it also showed that the modification with HYNIC or DTPA had reduced its receptor-binding affinity by several-fold.
Single chain Fv antibody fragment (scFv) is an even smaller format of antibody derivative which comprises one VH and one VL that are linked by polypeptide of at least 12 residues. Its advantages in practice rise from its compact size (~27 kDa) and its ready production in E. coli. Especially, while most other antibody-based probes are directing extracellular targets, scFvs are capable of reaching intracellular targets and affecting their functions. Studies on using such strategy to inhibit disease progression have been reported (Wolfgang et al. 2005; Lobato and Rabbitts 2004).
The relatively compact size of scFv suggests its more susceptibility to bioconjugation. Regarding this, several site-specific conjugation techniques have been developed. Ramakrishnan et al. (2009) have constructed a human anti-HER2 scFv, with a C-terminal fusion polypeptide containing 1, 3, or 17 threonine residues. These fusion scFv proteins with the modified galactose were then conjugated with a fluorescence probe, Alexa488, which carries an orthogonal reactive group. The fluorescently labeled scFv proteins bind specifically to a human breast cancer cell line (SKBR-3) that overexpresses HER2, indicating that the in vitro folded scFv fusion protein is biologically active and the presence of conjugated multiple Alexa488 probes at the C-terminal end does not interfere with its binding to the antigen.
Multivalent antibody fragments
Protein engineering work can be made on Fab and scFv to yield multivalent antibody fragments. Such multimerization can lead to dramatically improved avidity and dissociation rates and the resulting agents have found wide uses in immuno-diagnostic and immuno-therapeutic applications (Chames and Baty 2000; Funaro et al. 2000; Hudson 2000). If the multimerization occurs between fragments from different parent molecules, the newly constructed proteins may be gifted with different functionalities or secondary activity. The multimerization can be achieved by chemical cross-linking or more commonly, by the use of a variety of recombinant fusions using adhesive protein domains or peptides (Hudson 1999, 2000; Hudson and Kortt 1999; Pluckthun and Pack 1997; Hu et al. 1996; Adams et al. 1993).
Multivalent Fab
F(ab′)2 is formed by the non-specific digestion of whole IgG to remove the Fc portion. This fragment is comprised of a pair of Fab units connected by two disulfide bonds. Fab fragments can also be connected by more stable chemical cross-link to form dimer (divalent-Fab′) or trimer (Fab3), and the resulting probes have improved retention time and internalization properties over the Fab. These multimers show significant increases in avidity and much slower dissociation rates for cell surface receptors or antigens.
As mentioned above, different antibody fragments can be assembled to achieve reagents with multiple functions. For instance, Fab from different sources can be linked together to yield derivatives of bi-specific targeting specificity. In one study, the F(ab′)2 fragments of the anti-MUC1 MAb 12H12, which reacts with the vast majority of breast tumors, and the F(ab′) fragments of an anti-gallium (Ga) chelate mAb were linked via a mixed functional chemical linker (Schuhmacher et al. 2001). The resulting bi-specific antitumor/antimetal chelate antibody (BS-MAb) was used as a pretargeting probe in a two-step breast cancer imaging paradigm. In brief, BS-MAb was first administrated, which binds to the targets, in this case breast cancer tumor cells. This was followed by the injection of 68Ga chelate, which homes to the tumor site via the interaction with the BS-MAbs for PET imaging. Such idea was successfully demonstrated in small animal studies and has advanced into clinical trials with success (Schuhmacher et al. 2001).
A humanized divalent-Fab′ cross-linked with a bismaleimide linker referred to as humanized divalent-Fab′ maleimide was produced (Casey et al. 2002; Weir et al. 2002). It is a humanized divalent antibody with no Fc, which can be produced in bacteria and has enhanced stability compared with F(ab′)2. A clinical study was performed in patients with colorectal cancer using this divalent-Fab′ generated from the 131I-labeled anti-CEA antibody A5B7. Positive tumor images were obtained by gamma camera imaging in 80% patients (8/10) with known lesions, and one previously undetected lesion was identified. Area under the curve analysis of serial blood gamma counting and gamma camera images showed a higher tumor-to-blood ratio compared to A5B7 mF(ab′)2 used previously in the clinic, implying that this new molecule may be superior to RIT (Casey et al. 2002; Weir et al. 2002).
Multivalent scFv
As mentioned previously, a linker between VH and VL of at least 12 resides is necessary to form scFv with a functional Fv domain. Reducing the length of linker will make it impossible to form functional domain by single scFv; rather, one fragment will be associated with a second scFv to form a bivalent dimer, called diabody. Further reducing the linker length to three residues or less may result in the formation of trimers (triabodies) or tetramers (tetrabodies), with the specific structures dependent on linker length, composition and V-domain orientation. Another promising antibody derivative is minibody, a dimer formed by scFv-CH3. It is a bivalent format with a molecular weight comparable to dia/triabodies.
scFv multimers possess a size of 60 kDa (diabodies) to 90 kDa (triabodies), which are significantly larger than the scFv monomers (~30 kDa) yet much smaller than intact mAb (~150 kDa). Such intermediate size makes scFv multimers appealing probes in imaging for providing an appropriate circulation half life and tumor penetration rate (Colcher et al. 1999). In addition, the multivalency feature of dia/triabodies confers the antibody complexes high-avidity and high target retention. Indeed, accumulating data have indicated the advantages of radiolabeled diabodies over their monomeric peers and intact mAb in tumor imaging (Wu et al. 1996; Adams et al. 1998; Adams and Schier 1999; Viti et al. 1999; Antoniw et al. 1996; Casey et al. 1996).
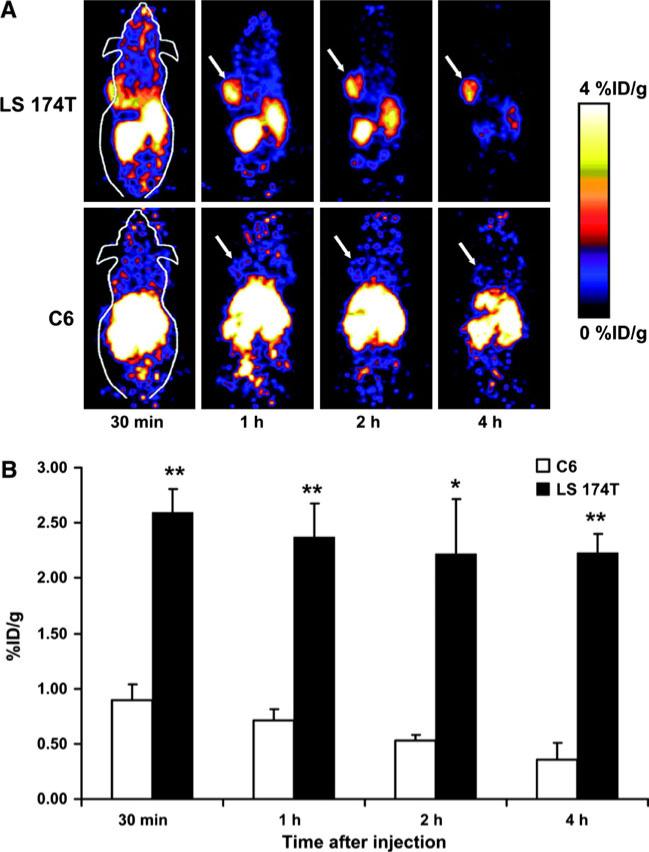
The actual benefit of tumor targeting can be estimated by an ‘Imaging Figure of Merit’ (IFOM) score (Todorovska et al. 2001). For short-lived radioisotopes (123I, 18F), 60 kDa diabodies are ideal for in vivo imaging due to the faster clearance rates and greater IFOM score over 90 kDa triabodies and minibodies (scFv-CH3 dimers) (Sundaresan et al. 2003). In one such study, Cai et al. (2007c) investigated 18F-labeled CEA T84.66 diabody for small animal PET imaging of CEA expression in xeno-graft-bearing mice (Fig. 4). 18F labeling of the anti-CEA T84.66 diabody (molecular mass 55 kDa) was achieved with N-succinimidyl-4-18F-fluorobenzoate (18F-SFB). The biodistribution of 18F-fluorobenzyl-T84.66 diabody (18F-FB-T84.66 diabody) was evaluated in nude mice bearing subcutaneous LS174T human colon carcinoma and C6 rat glioma tumors for PET imaging. The 18F-FB-T84.66 diabody showed rapid and high tumor uptake and fast clearance from the circulation in the LS174T xenograft model, as evidenced by both small animal PET imaging and biodistribution studies. High-contrast small animal PET images were obtained as early as 1 h after injection of the 18F-FB-T84.66 diabody, and only a background level of activity accumulation was found in CEA-negative C6 tumors. The tracer exhibited predominantly renal clearance, with some activity in the liver and spleen at early time points.
Fig. 4.
a Dynamic small animal PET scans obtained for 18F-FB-T84.66 diabody with LS 174T tumor-bearing mice and C6 tumor-bearing mice. Coronal whole-body slices that contained tumors are shown; arrows indicate tumors. b Comparison of LS 174T tumor uptake and C6 tumor uptake. Values were determined from ROI analysis of small animal PET imaging data. Differences were significant at all time points examined. *P < 0.05; **P < 0.01. Reproduced with permission from Cai et al. (2007c)
For radioisotopes with long half lives, however, the advantages of diabodies over minibodies are debatable. The anti-CEA antibody fragments, T84.66 minibody and diabody, were both labeled with 124I and were evaluated in a tumor xenograft model by PET (Sundaresan et al. 2003). Fast blood clearance, combined with metabolism and clearance of radioiodine activity from peripheral tissues, leads to high ratios of tumor to normal activity and excellent visualization using either fragment by 18 h. The T84.66 minibody reaches higher absolute activity levels in tumor, but takes longer for the background activity to clear and is still apparent at 18 h in the murine model. By contrast, radioactivity delivered to the CEA-positive xenografts by diabody is substantially lower; however, this apparent disadvantage is ameliorated by the much lower background activity seen in the mouse.
Affibodies
Affibodies are an emerging class of probes which mimic antibodies in many ways and can be engineered to bind specifically to a large number of target proteins. The original affibody protein scaffold was designed based on the Z domains of staphylococcal surface protein A. The staphylococcal surface protein A binds to the Fc portion of immunoglobulins from most mammalian species, including human (Langone 1982), and has been widely used as an immunological tool in purification for its binding to immunoglobulins (Uhlen et al. 1983). The selected domain is structurally rigid and can withstand amino acid substitutions or inserts. These molecules underwent combinatorial randomization of 13 solvent-accessible residues, including those involved in the Fc binding of domain Z, to generate a synthetic library. Such a pool of molecules was then subjected to a screening step to obtain variants with desired affinity to the targets.
The first isolation of affibody molecules was performed by Nord et al. (1997). Specific binders to three target proteins (Taq DNA polymerase, human insulin and human apolipoprotein A-1 variant) were selected from an affibody library (~4 × 107 variants) presented on phages and the affinities were in the micromolar range. Since then, a larger affibody library consisting of 3 × 109 variants (Gronwall et al. 2007a) has been used in panning to select binding molecules. The targeted molecules include HER2 (Wikman et al. 2004), transferrin (Gronwall et al. 2007b), amyloid β peptide (Gronwall et al. 2007a), EGFR (Friedman et al. 2007), Factor VIII (Nord et al. 2001), CD25 (Gronwall et al. 2008), HIV gp120 (glycoprotein of 120 kDa) (Wikman et al. 2006) and CD28 (Sandstrom et al. 2003), with affinities in the mid- to low-nanomolar range.
The robust structure of the affibody molecules makes them suitable for a wide range of biotechnological applications, such as bioprocess- and laboratory-scale bioseparations (Nord et al. 2000; Graslund et al. 2002), specific molecular detection (Lundberg et al. 2007; Ronnmark et al. 2002; Karlstrom and Nygren 2001; Nord et al. 2005), and for inhibition of receptor interactions (Sandstrom et al. 2003). Affibody molecules have also been reported as targeting agents in molecular recognition-based therapy, e.g., adenoviral targeting for gene therapy or as affinity handles for extracorporeal removal of pathogenic structures in plasma (Wikman et al. 2006; Gronwall et al. 2007a; Henning et al. 2002; Myhre et al. 2007).
Affibodies are characteristic of their much smaller size (~7 kDa), more rigid structure and rapid folding properties, especially when compared with antibodies and their derivatives. These features allow affibody molecules to be fully produced by chemical peptide synthesis methods. Therefore, fluorescent or radio labels, such as dye molecules and chelates, can be introduced into the affibodies site-specifically in a single chemical production process (Engfeldt et al. 2005, 2007).
HER2-binding affibodies are probably the most studied affibody probes in imaging. They have been selected by phage display from a combinatorial protein library based on the 58-amino-acid-residue staphylococcal protein A-derived Z domain (Wikman et al. 2004). One of the selected variants, denoted His6-ZHER2, was demonstrated to bind with nanomolar affinity (~50 nM) to the HER2-ECD molecule, but at a different site than the mAb trastuzumab. Radiolabeled His6-ZHER2 affibody showed specific binding to native HER2, overexpressed on the SKBR-3 tumor cell line (Wikman et al. 2004). To achieve better binding affinity, (Z(HER2:4))2, a bivalent form of the affibody ligand, was created and was compared with its monovalent peer in various assays (Steffen et al. 2005). The new bivalent affibody has a molecular weight of 15.6 kDa and an apparent affinity (Kd) against HER2 of 3 nM. Internalization of 125I was shown after delivery with both the monovalent and the bivalent affibodies, but the cellular retention was found longer in the bivalent case. A cysteine-containing variant of the bivalent affibody, (Z(HER2:4))2-Cys, was labeled with 76Br (Mume et al. 2005). It was found that HPEM can be radiobrominated with an efficiency of ~83% and thereafter coupled to freshly reduced affibody with a yield of ~65%. A “one-pot” labeling enabled the radiochemical purity of the conjugate to exceed 97%. The label was stable against large excess of nonlabeled bromide and high molar strength. In vitro cell tests demonstrated that radiobrominated affibody binds specifically to the HER2-expressing cell line, SKOV-3. Biodistribution studies in nude mice bearing SKOV-3 xenografts have shown tumor accumulation of 4.8 ± 2.2% IA/g and good tumor-to-normal tissue ratios.
By truncating one α-helix of affibody and maturating the protein affinity through synthetic strategies, Ren et al. (2009) have successfully identified several small 2-helix proteins with excellent binding affinities to HER2. In this study, a 2-helix small protein, MUT-DS, was chemically modified with DOTA followed by site-specific radiolabeling with 68Ga. The DOTA-MUT-DS displayed high stability in mouse serum and specificity toward HER2 in cell cultures. Biodistribution and small animal PET studies further showed that 68Ga-DOTA-MUT-DS had rapid and high SKOV-3 tumor accumulation and quick clearance from normal organs.
Kramer-Marek et al. (2009) described the in vivo characterization of the HER2-specific N-2-[(4-18F-fluorobenzamido)ethyl]maleimide (18F-FBEM)-ZHER2:342 affibody molecule and its application to study the effect of 17-DMAG on HER2 expression by PET. The tracer was eliminated quickly from the blood and normal tissues, providing high tumor-to-blood and tumor-to-muscle ratios as early as 20 min after injection. The high-contrast images between normal and tumor tissue were recorded for BT474 and MCF7/clone18 tumors. Kramer-Marek's results suggest that the described 18F-FBEM-ZHER2:342 affibody can be used to assess HER2 expression in vivo by PET and monitor possible changes of receptor expression in response to therapeutic interventions.
Lyakhov et al. (2010) recently created novel photostable imaging probes for the in vitro staining of EGFR and HER2. These new reagents, called affiprobes, consist of a HER2- or EGFR-specific affibody molecule, and a fluorescent moiety, mCherry (red) or EGFP (green). The flow cytometry and confocal microscopy experiments demonstrated high specificity and signal/background ratio of affiprobes. Affiprobes are able to stain both live cells and frozen tumor xenograph sections. This type of optical probe can easily be extended for targeting other cell surface antigens/receptors.
Affibody has also been intensively studied in SPECT imaging of HER2 expression. His6-(Z(HER2:4))2 was site-specifically labeled with 99mTc and the tumor targeting ability was compared with 125I-labeled His6-(Z(HER2:4))2 (Orlova et al. 2006a). Biodistribution study demonstrated moderate tumor uptake for both tracers with reasonable tumor-to-background contrast. Both conjugates provided clear imaging of SKOV-3 xenografts at 6 h after injection, but more favorable tumor-to-non-tumor ratios were found with the radioiodinated affibody.
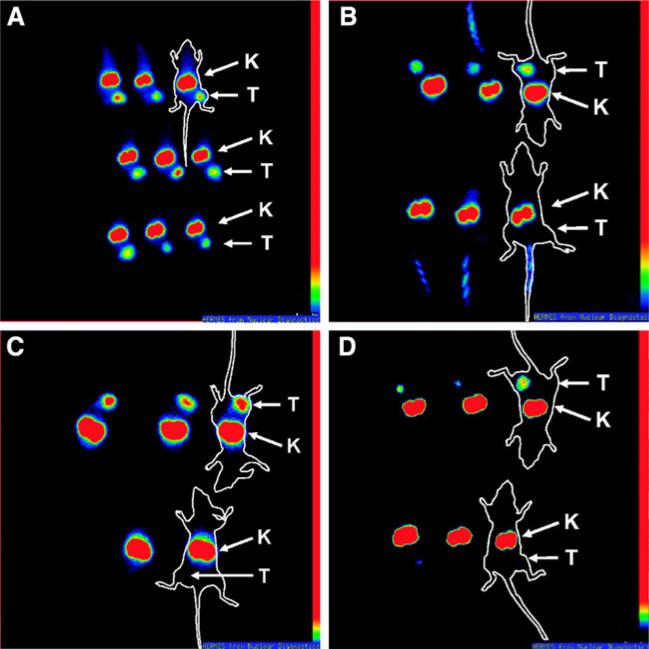
Another type of HER2 affibody, ZHER2:342, which was selected by phage display with subnanomolar affinity with HER2 (Orlova et al. 2006b), was also labeled with 99mTc by site-specific labeling. In brief, a mercaptoacetyltriglycine (MAG3) chelator was incorporated into ZHER2:342 during the peptide synthesis by Fmoc/tBu solid-phase approach (Engfeldt et al. 2007; Fields and Noble 1990). 99mTc-MAG3-ZHER2:342 showed specific tumor targeting with a contrast similar to a radioiodinated analog in mice bearing LS174T xenografts. Gamma camera imaging demonstrated clear and specific visualization of HER2 expression. In a similar way, site-specific DOTA-conjugated affibody was also prepared by peptide synthesis (Fig. 5) (Orlova et al. 2007). The radiolabeling was efficient with >95% incorporation of 111In within 30 min. Tumor uptake of 111In-DOTA-ZHER2:342-pep2 was found specific for HER2-positive xenografts. A high tumor uptake of 23% injected activity per gram tissue, a tumor-to-blood ratio of >7.5, and high-contrast gamma camera images were obtained already 1 h after injection. Pre-treatment with Herceptin did not interfere with tumor targeting, whereas degradation of HER2 using the Hsp 90 inhibitor 17-allylamino-geldanamycin before administration of 111In-DOTA-ZHER2:342-pep2 obliterated the tumor image.
Fig. 5.
Gamma camera images of HER2-expressing SKOV-3 xenograft tumors in BALB/c nu/nu mice. The time to image acquisition (a) and controls to prove imaging specificity for HER2 using competition with unlabeled ZHER2:342 (b), a negative control affibody molecule not targeting HER2 (c), and drug-induced HER2 degradation (d) are shown. All animals were i.v. injected with 3 MBq (3 μg) of 111In-DOTA-ZHER2:342-pep2 or 111In-DOTA-Ztaq4:5. To facilitate interpretation, white contours were superimposed around some animals to indicate the location of the animals on the gamma camera screen. Arrows indicate positions of kidneys (K) or tumors (T). a Imaging of mice at different time points after injection, 1 h (top), 2 h (middle), and 4 h (bottom) after injection of 111In-DOTA-ZHER2:342-pep2. b Imaging of mice pre-blocked (bottom) or not blocked (top) with 0.9 mg unlabeled ZHER2:342 45 min before injection of 111In-DOTA-ZHER2:342-pep2. Imaging was done 1 h after injection. c Imaging of mice injected with 111In-DOTA-ZHER2:342-pep2 (top) or 111In-DOTAZtaq4:5 (bottom) 4 h after injection. d Imaging of mice 4 h after injection, pretreated with 17-AAG (bottom) and controls (top). Reproduced with permission from Orlova et al. (2007)
Other than the traditional labeling, the small size and cysteine-free structure of affibody molecules also allow direct incorporation of radionuclide chelators. For example, the incorporation of the natural peptide sequences cysteine-diglycine (CGG) and cysteine-triglycine (CGGG) sequences would enable directly labeling of affibody molecules with 99mTc (Tran et al. 2007). The chelating sequences were incorporated by peptide synthesis. Conjugates were directly labeled with 99mTc and preserved the capacity to bind specifically to HER2-expressing cells. The tumor uptake of 99mTc-CGG-ZHER2:342 was HER2-specific and a tumor-to-blood ratio of 9.2 was obtained at 6-h post-injection. Gamma camera imaging with 99mTc-CGG-ZHER2:342 clearly visualized tumors. 111In labeling of the same affibody molecule was also reported (Tolmachev et al. 2006).
Natural ligands
Cytokines and chemokines are a group of signaling molecules that are used extensively in cellular communication. They are implicated in the regulation of a broad range of biological processes, including cross-talk between immunocompetent cells, upregulation of adhesion molecules, wound healing and control of the neoplastic process (Belardelli and Ferrantini 2002; Mantovani et al. 2002). Such implications have made radiolabeled cytokines candidate probes for understanding immune processes in several conditions, such as detecting the immune reaction against tumors in cancer (Signore et al. 2003a; Herberman 2002).
Although cytokines may induce biological side effects, for imaging purposes, they are used in such a small amounts (few micrograms) that they are regarded as safe probes. They are human in origin and produced by recombinant DNA technology and are not immunogenic, even if used on repeated occasions for follow-up studies (Signore et al. 2002). Their low molecular weight makes them rapidly clearing probes. And they are easily concentrated into affected tissues infiltrated by inflammatory cells to delineate the pathological areas. Indeed, several cytokines and chemokines, such as interleukin-2 (IL-2), TGF-β, VEGF, EGF etc., have also been radiolabeled and used as radiopharmaceuticals in patients with cancer, mainly for studying in vivo the presence and extent of peritumoral lymphocytic infiltration.
EGF
EGF (6 kDa) is the natural ligand of EGFR and was evaluated as a perfect targeting probe for EGFR imaging (Cohen 1983). DOTA-human EGF (hEGF) was labeled with 68Ga using microwave heating (Velikyan et al. 2005), and the biodistribution and microPET studies were performed in BALB/c nu/nu mice bearing A431 carcinoma xenografts. Radioactivity accumulation in the tumor and EGFR-expressing organs was observed within 5 min after injection. However, despite the stability of 68Ga-DOTA-hEGF in aqueous buffer for up to a few hours, its in vivo stability is unclear and remains to be evaluated. More critically, 68Ga-DOTA-hEGF has prominent liver and kidney uptake, which limits its potential in clinical applications. But in such study, the DOTA coupling was conducted in a random way. The hEGF contains one terminal and two lysine side chain amino groups; therefore, direct chemical labeling may lead to a mixture of molecules and consequently affect the binding affinity and functional activity. Site-specific labeling of a particular amino group or an additional engineered residue may give better imaging results and should be studied in the future.
The very first SPECT imaging of labeled EGF in patients used 123I as the radiolabel (Schatten et al. 1991). It was used to help identify lymph node involvement in patients with squamous-cell carcinoma of the cervix. Abnormal lymph node imaging was seen in 11 of 14 patients with advanced cervical cancer. However, conventional radiology (CT and ultrasound) did not confirm the presence of disease in 7 of the 11 patients. Hence, it was concluded that 123I-EGF did not fulfill the theoretical expectations of excellent accumulation in lymph node metastases and could not be recommended for routine clinical use (Pateisky et al. 1991). On the other hand, 111In-DTPA-EGF was evaluated firstly as imaging probes (Reilly et al. 2000), and was more recently evaluated as anti-proliferative agents (Chen et al. 2002), on human breast cancer. Also, 131I-hEGF has advanced into clinic trials to treat patients with advanced squamous lung cancer (Cuartero-Plaza et al. 1996). In all cases, tumors were visualized on the same day of the infusion and the best tumor-to-background contrast was observed 2–3 days later.
VEGF
As previously described, the VEGF family is composed of seven members: VEGF-A, VEGF-B, VEGF-C, VEGF-D, VEGF-E, VEGF-F, and placenta growth factor (PlGF) (Ferrara 2004). The targets of VEGF are mainly Flt-1/FLT-1 (VEGFR-1) and Flk-1/KDR (VEGFR-2) (Hicklin and Ellis 2005), two endothelium-specific receptor TKs, both are highly implicated in the process of angiogenesis (Ferrara 2004, 2005). VEGFR-targeted molecular imaging probes are useful for their roles in the assessment of anti-angiogenic therapeutics and for better understanding of the roles and expression profiles of VEGFR(s) in angiogenesis-related diseases. Up to now, most VEGFR imaging was achieved using VEGF-A-based tracers, which, along with its isoforms, mostly bind to both VEGFR-1 and VEGFR-2 (Ferrara 2004).
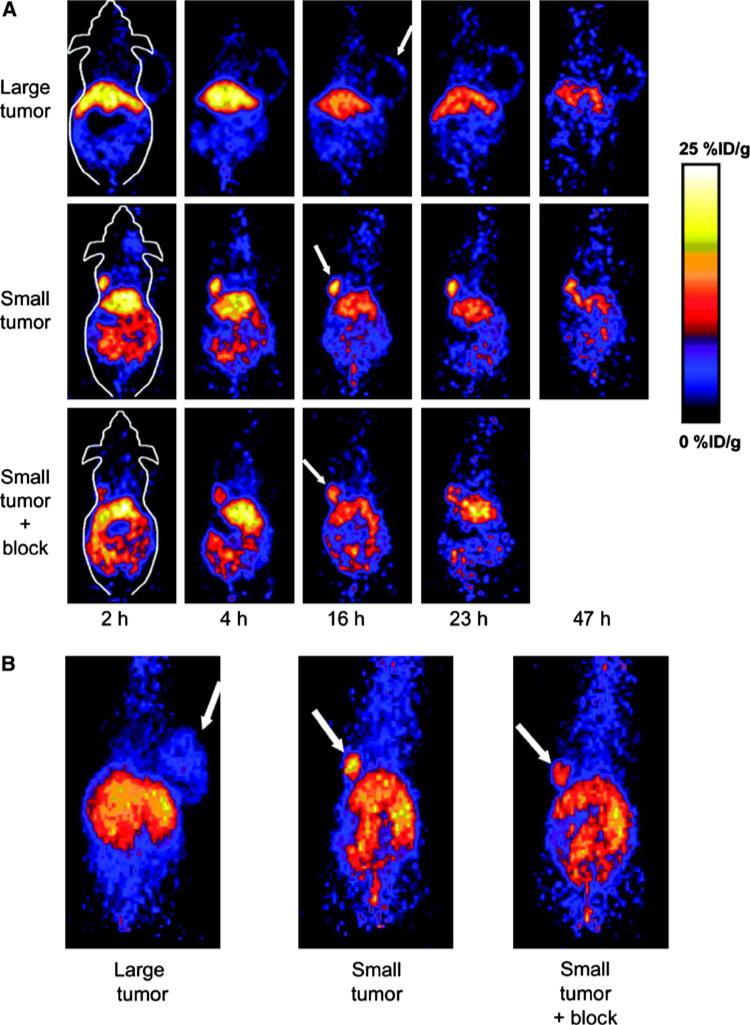
Cai et al. (2006b) labeled VEGF121 with 64Cu for PET of VEGFR expression (Fig. 6). The DOTA-VEGF121 exhibited nanomolar VEGFR-2-binding affinity (comparable to that of VEGF121) in vitro. In a follow-up study, a VEGFR-2-specific fusion protein, VEGF121/rGel (VEGF121 linked to recombinant plant toxin gelonin), was established and was evaluated for its imaging/therapy dual function on an orthotopic glioblastoma mice model (Hsu et al. 2007). Site-specific labeling of VEGF121 with 64Cu was recently reported (Ward and Ghetie 1995). Compared with the 99mTc-labeled analog, for which tumor uptake (~3% ID/g) was lower than that in most of the normal organs and kidney uptake was about 120% ID/g, such 64Cu-labeled VEGF121 showed similar tumor uptake (~2.5% ID/g) and lower kidney uptake, about 65% ID/g.
Fig. 6.
MicroPET of 64Cu-DOTA-VEGF121 in U87MG tumor-bearing mice. a Serial microPET scans of large and small U87MG tumor-bearing mice injected intravenously with 5–10 MBq of 64Cu-DOTA-VEGF121. Mice injected with 64Cu-DOTA-VEGF121 30 min after injection of 100 μg VEGF121 are also shown (denoted as “Small tumor + block”). b Two-dimensional whole-body projection of the three mice shown in a at 16 h after injection of 64Cu-DOTA-VEGF121. Tumors are indicated by arrows. Reproduced with permission from Cai et al. (2006b)
As mentioned above, most VEGF-A isoforms bind to both VEGFR-1 and VEGFR-2. This is problematic in imaging since high level VEGFR-1 expression was found in rodent kidneys, making kidneys the dose-limiting organs (Cai et al. 2006b; Backer et al. 2007; Simon et al. 1998). It is, therefore, ideal to achieve probes of VEGFR-2 targeting specificity, which is generally regarded to be more functionally important than VEGFR-1 in cancer progression (Ferrara 2004; Underiner et al. 2004). Alanine-scanning mutagenesis has revealed that Arg82, Lys84, and His86, located in a hairpin loop of VEGF165, are critical for VEGFR-2 binding and that some negatively charged residues, Asp63, Glu64, and Glu67, are associated with VEGFR-1 binding (Keyt et al. 1996). Based on such knowledge, Wang et al. (2007) developed a VEGFR-2-specific PET tracer based on mutated VEGF121. Such D63AE64AE67A mutant of VEGF121 (VEGFDEE) was generated by recombinant DNA technology, which, while retaining the VEGFR-2-binding affinity, has a VEGFR-1 affinity that is 20-fold lower than VEGF121. Small animal PET imaging revealed that both 64Cu-DOTA-VEGF121 and 64Cu-DOTA-VEGFDEE had rapid, prominent, and comparable levels of activity accumulation in VEGFR-2-expressing tumors. Meanwhile, the renal uptake of 64Cu-DOTA-VEGFDEE was much lower than that of 64Cu-DOTA-VEGF121. The lower kidney uptake confers 64Cu-DOTA-VEGFDEE much lower renal toxicity than other VEGF-A-based tracers, and makes it highly amenable for translation to clinical applications.
Other chemokine-based imaging probes, such as radio-labeled IL-8, IL-2, CCL3 etc., have been investigated for imaging inflammation, infection and cancer, which have been reviewed elsewhere (Signore et al. 2003b).
Annexin V
Apoptosis is closely correlated with tumor sensitivity to radiation and its early detection is highly valuable in predicting tumor response to certain treatments. Annexin V has been proved as an important protein probe for apoptosis by binding phosphatidylserine (PS) expressed on the outer surface of cell membranes. Annexin V is a member of an evolutionary conserved multigene family of Ca2+ and phospholipid-binding proteins (Gerke and Moss 2002). The protein harbors four domains, each of which is composed of five α-helixes. Annexin V binds to PS with Ca2+ as cofactor and forms a membrane-bound two-dimensional crystal lattice which may involve eight PS molecules per molecule annexin V (Meers and Mealy 1993). PS was originally expressed in the inner plasma membrane leaflet but is rapidly externalized on the outer leaflet of the plasma membrane in response to apoptosis-related signals (Comfurius et al. 1994; Chang et al. 1993; Zhao et al. 1998). Annexin V-based imaging provides a very specific, rapid and reliable technique to detect apoptosis. Molecular imaging of cell death using labeled-annexin V has been carried out in vitro (van Engeland et al. 1996; van den Eijnde et al. 1998) and in vivo in animal models (Blankenberg et al. 1999; Dumont et al. 2001; van den Eijnde et al. 1997) and in patients (Hofstra et al. 2000; Narula et al. 2001).
Recently, Guo et al. (2009) evaluated the imaging of in vivo 99mTc-HYNIC-annexin V and investigated its correlation with radiosensitivity. The imaging group of mice was injected with 4–8 MBq 99mTc-HYNIC-annexin V 24 h after irradiation and imaged 1-h post-injection. The 99mTc-HYNIC-annexin V uptake in E14 lymphoma significantly increased as the radiation dose escalated from 0 to 8 Gy, and significantly correlated with the number of TUNEL positive cells. Therefore, 99mTc-HYNIC-annexin V in vivo imaging is a feasible method to detect early radiation-induced apoptosis in different tumors, and might be predictive for radiation sensitivity.
A study was undertaken to evaluate the changes in relative 99mTc-HYNIC-annexin V tumor uptake over time in a total of 17 patients, who underwent chemotherapeutic treatment at baseline and at 5–7 and 40–44 h after treatment initiation (Rottey et al. 2006). The tumor response was evaluated with response evaluation criteria in solid tumors (RECIST) and related to the observed changes in the ratios of tumor activity to background activity for the largest known lesion. It was shown that patients who responded to treatment could be separated from non-responders by imaging with a 94% accuracy (16/17 patients). Therefore, sequential 99mTc-HYNIC-annexin V imaging allows for assessment of the response to chemotherapy.
Site-specific conjugation
One major challenge in protein chemistry is to achieve covalent coupling without affecting the biological activity. Conventional random conjugation techniques target lysine or cysteine residues on protein surfaces. These methods are unappealing because of their non-specific coupling feature, although they remain dominant in protein modifications. An on-going trend is to use site-specific labeling techniques, which permit conjugation take place in a stoichiometric fashion without compromising the protein activity. It is usually achieved by introducing a tag that is uniquely active toward the to-be-coupled agent at a site far from the active site. Several strategies are now available for such purpose, including the incorporation of unnatural amino acid (UAA), peptide tag, and intein tag, to name a few.
Introducing UAA to target protein
Usually, more than one residue of interests are present on the surface of one target protein. Random coupling may lead to heterogeneous products and is usually accompanied with activity loss. To overcome this limitation, one straightforward strategy is to introduce orthogonally reactive UAAs into proteins. Thereafter, the coupling will occur only between the UAA and the added reagent, allowing strict control over the site and stoichiometry of the reaction.
A method of encoding UAAs in bacteria, yeast and mammalian cells with diverse physicochemical and biological properties has been established by Xie and Schultz (2006). Using an engineered orthogonal tRNA/aminoacyl-tRNA synthetase pair, these UAAs can be introduced into proteins. So far, over 30 UAAs have been cotranslationally incorporated into proteins to facilitate site-specific labeling with high fidelity and efficiency.
Some of these UAAs are themselves of imaging functions. For instance, 7-hydroxycoumarin and dansyl side chains have been selectively incorporated into proteins (Wang et al. 2006; Summerer et al. 2006), both are fluorescently active. Also, the incorporated UAAs can be isotopically labeled, providing unique opportunities for site-specific labeling of proteins for NMR studies. Such opportunities, including new photocaged UAAs, outline usage of metal chelating and spin-labeled UAAs, were recently discussed by Jones et al. (2010).
The incorporation of UAA can induce single-molecule fluorescence resonance energy transfer (sm-FRET) that was found of use in studying protein conformational changes. For example, Schultz et al. selected a tRNA/aminoacyl-tRNA synthetase pair that encodes p-acetyl-phenylalanine, an unnatural ketone bearing amino acid. By incorporating p-acetylphenylalanine into proteins, hydroxylamine-containing fluorophore can be site-specifically coupled to protein molecules with high yield (>95%). As a proof-of-concept, T4L* variants were constructed, which possessed two unique binding sites: one cysteine and one p-acetylphenylalanine (the original T4L* is cysteine free). Two dyes, Alexa594-maleimide and Alexa488-alkoxyamine, were applied, each selectively coupled to one of the two binding sites to achieve dual-labeling of protein. The yielded protein, bearing two dyes, is ideal for sm-FRET studies, allowing the assessment of T4 lysozyme folding at single-molecule resolution (Brustad et al. 2008).
Along this line, Miyake-Stoner recently reported the site-specific incorporation of two kinds of UAAs, l-4-cyanophenylalanine (pCNPhe) and 4-ethynylphenylalanine (pENPhe), into protein molecules. Both UAAs have an emission spectrum that overlaps with the absorbance spectrum of Trp, making them good FRET donors of Trp (Miyake-Stoner et al. 2009). These two UAAs were separately introduced to T4 lysozyme, replacing a phenylalanine in the hydrophobic core of T4 lysozyme. In one demonstration, the urea-induced disruption of the hydrophobic core of the protein was successfully probed by the change in FRET efficiency between either pCNPhe or pENPhe and the Trp residues in T4 lysozyme.
Protein tags
Cysteine has been one of the most commonly used tags in site-specific protein conjugation. Cysteine harbors the specific thiol side chain among the normal amino acids, which can react with various thiol-specific coupling reagents without affecting other amino acid species, with the premise, of course, that no other cysteine is present on protein surface. A surface-accessible cysteine can be introduced into target protein by site-directed mutagenesis (Wingfield et al. 1989). For instance, scFvs have been cysteine modified with PEG to adjust the circulation half-life (Yang et al. 2003). Similarly, liposomes have been coupled via thiol chemistry to scFvs, yielding conjugates for tumor targeting and imaging (Weng et al. 2008). Recently, Sirk et al. (2008) demonstrated the utility of a C-terminal cysteine modification for site-specific labeling of two diabodies (CysDbs): one specific for CD20 and the other for HER2. Each CysDb was site-specifically conjugated to three different fluorophores for optical detection: the large fluorescent proteins phycoerythrin (PE) and allophycocyanin (APC), and the small fluorescent molecule AlexaFluor-488. Fluorophore-conjugated CysDbs bound specifically to their targets in both antigen systems and with all three fluorescent tags, as determined by flow cytometry. The success of this study confirms that relatively large functional groups can be linked to the C-termini of diabodies with unaffected binding and targeting capabilities.
One critical topic in protein-based PET/SPECT imaging is to site-specifically introduce chelators, the motifs which can bind efficiently with metal radioisotopes, into protein surface without affecting the protein functions. For this purpose, several short amino acid sequences (Mease and Lambert 2001) possessing coordinating nitrogen and sulfur atoms have been incorporated into the N- or C-terminus of bioactive molecules. Compared with random chemical conjugation, such an approach allows better adjustment of the charge and polarity of the final products, along with a good control over the chelation site. For instance, Berndorff et al. (2006) genetically inserted a (Gly)3-Cys-Ala sequence at the C-terminus of scFv L19, which specifically targets the ED-B sequence of fibronectin. The protein has good 99mTc binding capability, and the labeled conjugates showed favorable biodistribution properties, including high and fast tumor uptake (8.3% injected dose per gram at 3 h after injection), rapid blood clearance and renal excretion, all of which contributed to high signal-to-noise ratios (tumor-to-blood ratio of 6.4 at 3 h after injection). In another case, an endogenous Tc chelation tag (Ala-Gly-Gly-Cys-Gly-His) was added to the N-terminus of annexin V to create annexin V-128 (Tait et al. 2005). The comparison of 99mTc-annexin V-128 with 99mTc-HYNIC-annexin V showed that the protein labeled at the endogenous chelation site had the same or higher uptake in apoptotic tissues, while showing 88% lower renal uptake at 60 min after injection.
Protein labeling catalyzed by post-translational modification enzymes
Besides the above-mentioned conjugation techniques, which rely on chemical reactions to achieve coupling, a group of enzymes have also been studied to site-specifically introduce compound of interest onto target proteins at the post-translational stage (Sunbul and Yin 2009; Walsh et al. 2005). Based on the types of enzyme catalysis reactions during protein post-translational modifications, the present review sheds a light on some enzymatic tools which have been developed for efficient labeling of target proteins with chemical probes of diverse structures and functionalities.
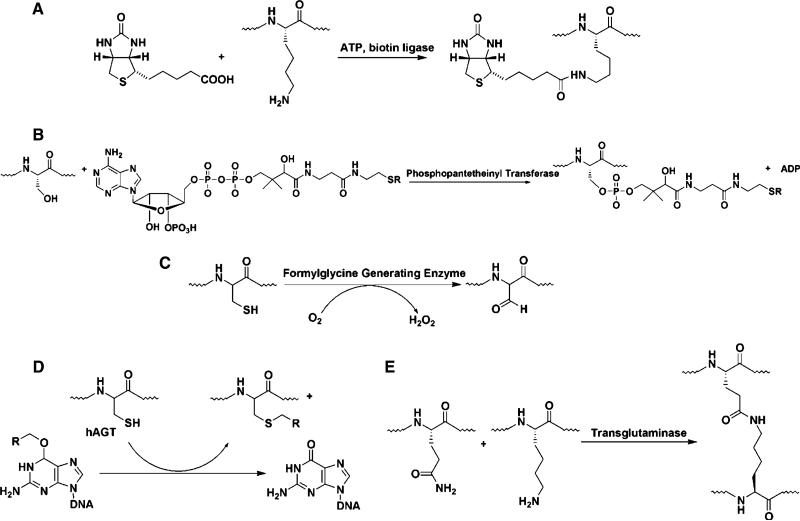
One most important application of protein chemistry in site-specific modification is biotinylation. The high affinity and specificity of biotin–avidin/streptavidin interaction have made it a reliable system for protein conjugation. One group of tags ranging from 123-amino acid to 15-amino acid sequence have been found to allow site-specific biotinylation when working along with E. coli enzyme biotin ligase (BirA) (Fig. 7a) (Beckett et al. 1999; Cull and Schatz 2000; Smith et al. 1998; Sibler et al. 1999). Although a 14-mer sequence was reported working well (Beckett et al. 1999), one 15-mer, termed AviTag (GLNDIFEAQKIEWHE), was considered a better option since its consistent biotinylation rate is slightly better than that of the natural substrate (Cull and Schatz 2000).
Fig. 7.
Schematic illustration of biotin ligase (a), phosphopantetheinyl transferase (PPTase) (b), formylglycine-generating enzyme (FGE) (c), human O6-alkylguanine-DNA alkyltransferases (hAGT) (d), and transglutaminase (TGase) (e) catalyzed modification
Recently, Wang et al. (2008) developed an Avi-tagged VEGF121 protein, which is site-specifically biotinylated in the presence of bacterial BirA biotin ligase. The resulting biotinylated VEGF121-Avi (VEGF121-Avib) forms a stable complex with streptavidin-IRDye800 (SA800) while retaining high affinity and targeting specificity to VEGFR in vitro. In contrast, chemical coupling of IRDye800 abrogated the VEGFR binding ability of the modified protein both in vitro and in vivo. This example clearly shows the advantage of site-specific biotinylation and makes VEGF121-Avib/SA800 complex (VEGF-Avib/SA800), a candidate agent for quantitative and repetitive evaluation of VEGFR expression level.
Another useful enzyme in site-specific labeling is phosphopantetheinyl transferation (PPTase), which transfers a 20Å long phosphopantetheinyl (Ppant) group derived from coenzyme A (CoA) to a conserved Ser residue in the acyl carrier proteins (ACP) of fatty acid synthase and polyketide synthase (PKS), and peptidyl carrier proteins (PCP) of nonribosomal peptide synthetase (NRPS) (Fig. 7b). The Ppant modification activates the carrier proteins for the attachment of biosynthetic intermediates during the enzymatic assembly of primary and secondary metabolites such as fatty acids, polyketides, and nonribosomal peptides. PPTase-catalyzed protein modification has been demonstrated as an efficient method for site-specific protein labeling with small molecules of diverse structures. For instance, Zou and Yin (2009) discovered that small molecules directly conjugated to the 5′-diphosphate moiety of ADP can serve as the substrates of a mutant Sfp PPTase, R4-4. Based on this, they synthesized ADP-conjugated small molecule probes by one-step coupling between phosphate-derivatized probes and morpholidate-activated AMP, and used R4-4 to transfer the small molecule labels to the carrier protein or peptide tags fused to the target protein.
Formylglycine-based modification that implicates sulfatase is also an important enzyme tool. The sulfatase family of enzymes catalyzes the hydrolysis of sulfate ester bonds of a wide variety of substrates. A catalytic cysteine residue, strictly conserved in prokaryotic and eukaryotic sulfatases, was found to be modified post-translationally into a formylglycine (Fig. 7c). All formylglycine-containing sulfatases possess a conserved pentapeptide motif, (C/S)X(P/A)XR, which directs the post-translational modification of the first cysteine (in eukaryotes and prokaryotes) or serine (in prokaryotes) residue to formylglycine. Later on, a new type of formylglycine-directing sulfatase signature, (C/S)XAXR, and a new type of anaerobic sulfatase-maturing enzyme were discovered (Berteau et al. 2006).
Carrico et al. described a method for the site-specific introduction of aldehyde groups into recombinant proteins using the 6-amino-acid consensus sequence recognized by the formylglycine-generating enzyme. This genetically encoded ‘aldehyde tag’ is no larger than a His6-tag and can be exploited for numerous protein labeling applications (Carrico et al. 2007). They found that the consensus sequence can be installed within heterologous proteins expressed in E. coli, where it is modified efficiently by a coexpressed bacterial formylglycine-generating enzyme. Recently, they further demonstrated that recombinant proteins expressed in mammalian cells can also be site-specifically modified using the aldehyde tag technology (Wu et al. 2009). Wu et al. engineered both Fc and intact IgG constructs bearing aldehyde tags at various sites. When expressed in CHO or HEK cells, the proteins were efficiently modified with aldehyde groups that were later exploited for site-specific modifications. They demonstrated that membrane-associated proteins can be labeled with aldehyde tags and then chemically derivatized on live cells. Also, they found that a cytosolic protein expressed in HEK cells can be modified by a coexpressed prokaryotic formylglycine-generating enzyme directed to that compartment.
Human O6-alkylguanine-DNA alkyltransferases (hAGT) is a DNA repair protein that plays an important role in protecting cells from the toxic effects of monofunctional alkylating agents and chloroethylating drugs. hAGT has the capability to transfer the alkyl group attached on the O″-position of guanine to a conserved cysteine located within the AGT active site (Fig. 7d) (Lindahl et al. 1988; Pegg et al. 1995). The resulting S-alkylcysteine in the protein cannot be converted reversibly to cysteine. It was demonstrated previously that hAGT have a very broad substrate specificity and can act at on a very wide range of benzylguanine groups (Margison and Santibanez-Koref 2002; Pegg 2000).
SNAP-tag, a 20 kDa mutant of hAGT as a fusion tag, has been developed to react specifically and rapidly with various benzylguanine derivatives, leading to the labeling of the fusion protein with a synthetic probe (Gronemeyer et al. 2006; Juillerat et al. 2003). Tirat et al. (2006) fused ubiquitin conjugating enzyme Rad6B with either N- or C-terminus of SNAP-tag using DNA recombinant technology. The fused protein was specifically labeled with SNAP-tag and was found fully retaining its activity. Recently, Kampmeier et al. (2009) site-specifically labeled a series of antibody fragments and protein ligands by fusing them to hAGT-based SNAP-tag. Substrates containing O6-benzylguanine were covalently bound to the fusion proteins via a stable thioether bond in a rapid and highly specific self-labeling reaction. They cloned different ligand SNAP-tag fusion proteins and expressed them in HEK 293T cells. The antibody/ligand fusions were characterized by labeling with different fluorophores, biotin, or fluorescent nanobeads, followed by analysis by flow cytometry and confocal microscopy. All ligands retained their original antigen-binding properties when fused to the SNAP-tag. Overall, the combination of recombinant protein with the SNAP-tag provides an efficient alternative to the existing coupling strategies. The major problem of this approach, however, is the rather long sequence of hAGT, which might have some effect on target protein conformation and function.
Transglutaminases (TGases) are widely distributed in most tissues and body fluids, including liver, hair follicles, epidermis, prostate and blood, and are involved in a variety of physiological functions (Fontana et al. 2008). TGase catalyzes the formation of a covalent bond between the protein-bound glutamine residue (Gln) and unbranched primary amines, including the amino group of lysine (Fig. 7e) (Folk 1983). One of the most studied TGases is the guinea pig liver transglutaminase (gpTGase), a 77-kDa monomeric protein that is expressed in the cytosol (Greenberg et al. 1991; Connellan et al. 1971). gpTGase exhibits high specificity for its glutamine-containing protein substrate and has wide tolerance for the structure of the amine-containing substrate (Coussons et al. 1992). Instead of lysine, amines as diverse as fluorescein cadaverine (Wolff and Lai 1988) and biotin cadaverine (Slaughter et al. 1992) can be utilized by gpTGase. gpTGase has been demonstrated in many cases to label various proteins fused with ‘Q-tag’, a 6- or 7-mer transglutaminase recognition sequence. Interleukin-2 and glutathione S-transferase have been attached with monodansyl cadaverine (Sato et al. 1996) and fluorescein cadaverine (Taki et al. 2004), respectively, by gpTGase. Lin and Ting (2006) demonstrated the labeling of Q-tagged proteins by gpTGase both in vitro and on the surface of living mammalian cells with biotin, fluorophores, and a benzophenone photoaffinity probe. To illustrate the utility of this labeling, they tagged the NF-kappaB p50 transcription factor with benzophenone, cross-linked with UV light, and observed increased levels of p50 homodimerization in the presence of DNA and the binding protein myotrophin.
Recently, Kamiya et al. (2009) reported that microbial TGases can accept diverse fluorophores (dansyl, fluorescein, and rhodamine derivatives) as substrates. The utility of the new fluorescent substrates was demonstrated by site-specific, covalent and quantitative labeling of a TGases-reactive Lys-containing peptide tag fused to the N-terminus of a recombinant bacterial alkaline phosphatase with retention of target protein functionality.
Intein-mediated modification
Inteins are peptide sequences that mediate self-splicing when inserted within a larger precursor polypeptide (Xu and Evans 2005; Muralidharan and Muir 2006). During splicing, intein removes itself from the precursor sequence and joins the flanking N- and C-terminal parts into a new protein. In nature, inteins have been found to splice enzymes including polymerases, proteases, and ATPases. Intein can function also in split geometry, in which case the precursor protein comes from two genes. These unique features have made intein ligation a powerful method for the site-specific incorporation of a wide range of molecules (Muralidharan and Muir 2006), including proteins (Schwarzer and Cole 2005; Evans et al. 1999), peptides (Ayers et al. 1999), fluorescent labels (Muir 2003; Muralidharan et al. 2004), carbohydrates (Ma and Cooney 2004), oligonucleotides (Lovrinovic et al. 2003), affinity tags and metal chelators (Xu et al. 1999); and to prepare cyclic peptides and proteins (Iwai et al. 2001; Abel-Santos et al. 2003).
For example, Volkmann and Liu (2009) recently developed a new method of protein C-terminal labeling using a non-canonical split-intein, through an intein-catalyzed trans-splicing reaction between a protein and a small synthetic peptide carrying the desired labeling groups. As a demonstration, three different proteins were efficiently labeled at their C-termini with two different labels (fluorescein and biotin) either in solution or on a solid surface, and a transferrin receptor protein was labeled on the membrane surface of live mammalian cells.
Charalambous et al. (2009) described an intein-based method to site-specifically conjugate quantum dots (QDs) to target proteins within the cells of the developing embryo. They genetically fused a pleckstrinhomology (PH) domain with the IntN. The IntC was conjugated to QDs in vitro. IntC-QD's and RNA encoding PH-IntN were microinjected into Xenopus embryos. In vivo intein-splicing resulted in fully functional QD-PH conjugates that could be monitored in real time within live embryos. The use of near infrared (NIR)-emitting QDs allowed monitoring of QD conjugates within the embryo at depths. This novel in vivo strategy for the site-specific conjugation of QD's and other artificial structures to target proteins can be applied to different intracellular compartments and signaling complexes.
Conclusion
Protein-based molecular imaging has been an important modality in clinical oncology for monitoring the effects of conventional treatments, such as chemotherapy, external radiation therapy and surgery. Many antigens are more highly expressed in tumors compared to normal tissues and can be used as imaging targets. The advances in genetic engineering and in vitro selection technology have now made it possible to generate versatile protein-based probes with diversified functions and more favorable physiological properties. Such development, synergized with the improvement of molecular imaging instrumentation and software algorithms, has dramatically changed our way of detecting the progression of diseases. These techniques have great benefits in early stage diagnosis, therapeutic response monitoring and personalized treatment. The site-specifically labeled protein probes for molecular imaging can be prepared by diverse strategies, including UAAs incorporation, polypeptide tags, and enzymatic post-translational modification. Overall, protein based probes remain to be an important class of agents in molecular imaging. And with the on-going efforts in protein engineering and site-specific labeling, more exciting findings in this domain are expected to come.
References
- Abel-Santos E, Scott CP, Benkovic SJ. Use of inteins for the in vivo production of stable cyclic peptide libraries in E coli. Methods Mol Biol. 2003;205:281–294. doi: 10.1385/1-59259-301-1:281. [DOI] [PubMed] [Google Scholar]
- Adams GP, Schier R. Generating improved single-chain Fv molecules for tumor targeting. J Immunol Methods. 1999;231(1–2):249–260. doi: 10.1016/s0022-1759(99)00161-1. [DOI] [PubMed] [Google Scholar]
- Adams GP, McCartney JE, Tai MS, et al. Highly specific in vivo tumor targeting by monovalent and divalent forms of 741F8 anti-c-erbB-2 single-chain Fv. Cancer Res. 1993;53(17):4026–4034. [PubMed] [Google Scholar]
- Adams GP, Schier R, McCall AM, et al. Prolonged in vivo tumour retention of a human diabody targeting the extracellular domain of human HER2/neu. Br J Cancer. 1998;77(9):1405–1412. doi: 10.1038/bjc.1998.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan SM, Dean CJ, Eccles S, Sacks NP. Clinical radioimmunolocalization with a rat monoclonal antibody directed against c-erbB-2. Cell Biophys. 1994;24–25:93–98. doi: 10.1007/BF02789219. [DOI] [PubMed] [Google Scholar]
- Antoniw P, Farnsworth AP, Turner A, et al. Radioimmunotherapy of colorectal carcinoma xenografts in nude mice with yttrium-90 A33 IgG and Tri-Fab (TFM). Br J Cancer. 1996;74(4):513–524. doi: 10.1038/bjc.1996.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga C. Targeting HER1/EGFR: a molecular approach to cancer therapy. Semin Oncol. 2003;30(3 Suppl 7):3–14. [PubMed] [Google Scholar]
- Ayers B, Blaschke UK, Camarero JA, Cotton GJ, Holford M, Muir TW. Introduction of unnatural amino acids into proteins using expressed protein ligation. Biopolymers. 1999;51(5):343–354. doi: 10.1002/(SICI)1097-0282(1999)51:5<343::AID-BIP4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Backer MV, Levashova Z, Patel V, et al. Molecular imaging of VEGF receptors in angiogenic vasculature with single-chain VEGF-based probes. Nat Med. 2007;13(4):504–509. doi: 10.1038/nm1522. [DOI] [PubMed] [Google Scholar]
- Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8(4):921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman RA, Weiner LM, Davis HM. Antibody constructs in cancer therapy: protein engineering strategies to improve exposure in solid tumors. Cancer. 2007;109(2):170–179. doi: 10.1002/cncr.22402. [DOI] [PubMed] [Google Scholar]
- Behr TM, Goldenberg DM, Becker W. Reducing the renal uptake of radiolabeled antibody fragments and peptides for diagnosis and therapy: present status, future prospects and limitations. Eur J Nucl Med. 1998;25(2):201–212. doi: 10.1007/s002590050216. [DOI] [PubMed] [Google Scholar]
- Belardelli F, Ferrantini M. Cytokines as a link between innate and adaptive antitumor immunity. Trends Immunol. 2002;23(4):201–208. doi: 10.1016/s1471-4906(02)02195-6. [DOI] [PubMed] [Google Scholar]
- Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3(6):401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- Berndorff D, Borkowski S, Moosmayer D, et al. Imaging of tumor angiogenesis using 99mTc-labeled human recombinant anti-ED-B fibronectin antibody fragments. J Nucl Med. 2006;47(10):1707–1716. [PubMed] [Google Scholar]
- Berteau O, Guillot A, Benjdia A, Rabot S. A new type of bacterial sulfatase reveals a novel maturation pathway in prokaryotes. J Biol Chem. 2006;281(32):22464–22470. doi: 10.1074/jbc.M602504200. [DOI] [PubMed] [Google Scholar]
- Blankenberg FG, Katsikis PD, Tait JF, et al. Imaging of apoptosis (programmed cell death) with 99mTc annexin V. J Nucl Med. 1999;40(1):184–191. [PubMed] [Google Scholar]
- Boulianne GL, Hozumi N, Shulman MJ. Production of functional chimaeric mouse/human antibody. Nature. 1984;312(5995):643–646. doi: 10.1038/312643a0. [DOI] [PubMed] [Google Scholar]
- Brustad EM, Lemke EA, Schultz PG, Deniz AA. A general and efficient method for the site-specific dual-labeling of proteins for single molecule fluorescence resonance energy transfer. J Am Chem Soc. 2008;130(52):17664–17665. doi: 10.1021/ja807430h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Chen X. Anti-angiogenic cancer therapy based on integrin alphavbeta3 antagonism. Anticancer Agents Med Chem. 2006;6(5):407–428. doi: 10.2174/187152006778226530. [DOI] [PubMed] [Google Scholar]
- Cai W, Wu Y, Chen K, Cao Q, Tice DA, Chen X. In vitro and in vivo characterization of 64Cu-labeled Abegrin, a humanized monoclonal antibody against integrin alpha v beta 3. Cancer Res. 2006a;66(19):9673–9681. doi: 10.1158/0008-5472.CAN-06-1480. [DOI] [PubMed] [Google Scholar]
- Cai W, Chen K, Mohamedali KA, et al. PET of vascular endothelial growth factor receptor expression. J Nucl Med. 2006b;47(12):2048–2056. [PubMed] [Google Scholar]
- Cai W, Chen K, He L, Cao Q, Koong A, Chen X. Quantitative PET of EGFR expression in xenograft-bearing mice using 64Cu-labeled cetuximab, a chimeric anti-EGFR monoclonal antibody. Eur J Nucl Med Mol Imaging. 2007a;34(6):850–858. doi: 10.1007/s00259-006-0361-6. [DOI] [PubMed] [Google Scholar]
- Cai W, Ebrahimnejad A, Chen K, et al. Quantitative radioimmunoPET imaging of EphA2 in tumor-bearing mice. Eur J Nucl Med Mol Imaging. 2007b;34(12):2024–2036. doi: 10.1007/s00259-007-0503-5. [DOI] [PubMed] [Google Scholar]
- Cai W, Olafsen T, Zhang X, et al. PET imaging of colorectal cancer in xenograft-bearing mice by use of an 18F-labeled T84.66 anti-carcinoembryonic antigen diabody. J Nucl Med. 2007c;48(2):304–310. [PubMed] [Google Scholar]
- Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438(7070):932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- Carrico IS, Carlson BL, Bertozzi CR. Introducing genetically encoded aldehydes into proteins. Nat Chem Biol. 2007;3(6):321–322. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]
- Casalini P, Iorio MV, Galmozzi E, Menard S. Role of HER receptors family in development and differentiation. J Cell Physiol. 2004;200(3):343–350. doi: 10.1002/jcp.20007. [DOI] [PubMed] [Google Scholar]
- Casey JL, King DJ, Chaplin LC, et al. Preparation, characterisation and tumour targeting of cross-linked divalent and trivalent anti-tumour Fab′ fragments. Br J Cancer. 1996;74(9):1397–1405. doi: 10.1038/bjc.1996.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JL, Napier MP, King DJ, et al. Tumour targeting of humanised cross-linked divalent-Fab′ antibody fragments: a clinical phase I/II study. Br J Cancer. 2002;86(9):1401–1410. doi: 10.1038/sj.bjc.6600198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chames P, Baty D. Antibody engineering and its applications in tumor targeting and intracellular immunization. FEMS Microbiol Lett. 2000;189(1):1–8. doi: 10.1111/j.1574-6968.2000.tb09197.x. [DOI] [PubMed] [Google Scholar]
- Chang CP, Zhao J, Wiedmer T, Sims PJ. Contribution of platelet microparticle formation and granule secretion to the transmembrane migration of phosphatidylserine. J Biol Chem. 1993;268(10):7171–7178. [PubMed] [Google Scholar]
- Charalambous A, Andreou M, Skourides PA. Intein-mediated site-specific conjugation of quantum dots to proteins in vivo. J Nanobiotechnol. 2009;7:9. doi: 10.1186/1477-3155-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Mrkobrada M, Vallis KA, et al. Comparative antiproliferative effects of (111)In-DTPA-hEGF, chemotherapeutic agents and gamma-radiation on EGFR-positive breast cancer cells. Nucl Med Biol. 2002;29(6):693–699. doi: 10.1016/s0969-8051(02)00325-6. [DOI] [PubMed] [Google Scholar]
- Cohen S. The epidermal growth factor (EGF). Cancer. 1983;51(10):1787–1791. doi: 10.1002/1097-0142(19830515)51:10<1787::aid-cncr2820511004>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Colcher D, Goel A, Pavlinkova G, Beresford G, Booth B, Batra SK. Effects of genetic engineering on the pharmacokinetics of antibodies. Q J Nucl Med. 1999;43(2):132–139. [PubMed] [Google Scholar]
- Collingridge DR, Carroll VA, Glaser M, et al. The development of [(124)I]iodinated-VG76e: a novel tracer for imaging vascular endothelial growth factor in vivo using positron emission tomography. Cancer Res. 2002;62(20):5912–5919. [PubMed] [Google Scholar]
- Colnot DR, Quak JJ, Roos JC, et al. Phase I therapy study of 186Re-labeled chimeric monoclonal antibody U36 in patients with squamous cell carcinoma of the head and neck. J Nucl Med. 2000;41(12):1999–2010. [PubMed] [Google Scholar]
- Comfurius P, Smeets EF, Willems GM, Bevers EM, Zwaal RF. Assembly of the prothrombinase complex on lipid vesicles depends on the stereochemical configuration of the polar head-group of phosphatidylserine. Biochemistry. 1994;33(34):10319–10324. doi: 10.1021/bi00200a012. [DOI] [PubMed] [Google Scholar]
- Connellan JM, Chung SI, Whetzel NK, Bradley LM, Folk JE. Structural properties of guinea pig liver transglutaminase. J Biol Chem. 1971;246(4):1093–1098. [PubMed] [Google Scholar]
- Coussons PJ, Price NC, Kelly SM, Smith B, Sawyer L. Factors that govern the specificity of transglutaminase-catalysed modification of proteins and peptides. Biochem J. 1992;282(Pt 3):929–930. doi: 10.1042/bj2820929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuartero-Plaza A, Martinez-Miralles E, Rosell R, Vadell-Nadal C, Farre M, Real FX. Radiolocalization of squamous lung carcinoma with 131I-labeled epidermal growth factor. Clin Cancer Res. 1996;2(1):13–20. [PubMed] [Google Scholar]
- Cull MG, Schatz PJ. Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol. 2000;326:430–440. doi: 10.1016/s0076-6879(00)26068-0. [DOI] [PubMed] [Google Scholar]
- De Bree R, Roos JC, Quak JJ, Den Hollander W, Snow GB, Van Dongen GA. Clinical screening of monoclonal antibodies 323/A3, cSF-25 and K928 for suitability of targeting tumours in the upper aerodigestive and respiratory tract. Nucl Med Commun. 1994;15(8):613–627. doi: 10.1097/00006231-199408000-00006. [DOI] [PubMed] [Google Scholar]
- Dean CJ, Eccles SA, Valeri M, et al. Rat MAbs to the product of the c-erbB-2 proto-oncogene for diagnosis and therapy in breast cancer. Cell Biophys. 1993;22(1–3):111–127. doi: 10.1007/BF03033870. [DOI] [PubMed] [Google Scholar]
- Dijkers EC, Kosterink JG, Rademaker AP, et al. Development and characterization of clinical-grade 89Zr-trastuzumab for HER2/neu immunoPET imaging. J Nucl Med. 2009;50(6):974–981. doi: 10.2967/jnumed.108.060392. [DOI] [PubMed] [Google Scholar]
- Downward J, Yarden Y, Mayes E, et al. Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature. 1984;307(5951):521–527. doi: 10.1038/307521a0. [DOI] [PubMed] [Google Scholar]
- Dumont EA, Reutelingsperger CP, Smits JF, et al. Real-time imaging of apoptotic cell-membrane changes at the single-cell level in the beating murine heart. Nat Med. 2001;7(12):1352–1355. doi: 10.1038/nm1201-1352. [DOI] [PubMed] [Google Scholar]
- Engfeldt T, Renberg B, Brumer H, Nygren PA, Karlstrom AE. Chemical synthesis of triple-labelled three-helix bundle binding proteins for specific fluorescent detection of unlabelled protein. Chembiochem. 2005;6(6):1043–1050. doi: 10.1002/cbic.200400388. [DOI] [PubMed] [Google Scholar]
- Engfeldt T, Orlova A, Tran T, et al. Imaging of HER2-expressing tumours using a synthetic Affibody molecule containing the 99mTc-chelating mercaptoacetyl-glycyl-glycyl-glycyl (MAG3) sequence. Eur J Nucl Med Mol Imaging. 2007;34(5):722–733. doi: 10.1007/s00259-006-0266-4. [DOI] [PubMed] [Google Scholar]
- Evans TC, Jr, Benner J, Xu MQ. The in vitro ligation of bacterially expressed proteins using an intein from Methanobacterium thermoautotrophicum. J Biol Chem. 1999;274(7):3923–3926. doi: 10.1074/jbc.274.7.3923. [DOI] [PubMed] [Google Scholar]
- Fernandez A, Spitzer E, Perez R, et al. A new monoclonal antibody for detection of EGF-receptors in western blots and paraffin-embedded tissue sections. J Cell Biochem. 1992;49(2):157–165. doi: 10.1002/jcb.240490208. [DOI] [PubMed] [Google Scholar]
- Ferrara N. VEGF and the quest for tumour angiogenesis factors. Nat Rev Cancer. 2002;2(10):795–803. doi: 10.1038/nrc909. [DOI] [PubMed] [Google Scholar]
- Ferrara N. Vascular endothelial growth factor: basic science and clinical progress. Endocr Rev. 2004;25(4):581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- Ferrara N. The role of VEGF in the regulation of physiological and pathological angiogenesis. EXS. 2005;(94):209–231. doi: 10.1007/3-7643-7311-3_15. [DOI] [PubMed] [Google Scholar]
- Fields GB, Noble RL. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int J Pept Protein Res. 1990;35(3):161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- Fishwild DM, O'Donnell SL, Bengoechea T, et al. High-avidity human IgG kappa monoclonal antibodies from a novel strain of minilocus transgenic mice. Nat Biotechnol. 1996;14(7):845–851. doi: 10.1038/nbt0796-845. [DOI] [PubMed] [Google Scholar]
- Folk JE. Mechanism and basis for specificity of transglutaminase-catalyzed epsilon-(gamma-glutamyl) lysine bond formation. Adv Enzymol Relat Areas Mol Biol. 1983;54:1–56. doi: 10.1002/9780470122990.ch1. [DOI] [PubMed] [Google Scholar]
- Folkman J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat Med. 1995;1(1):27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- Fontana A, Spolaore B, Mero A, Veronese FM. Site-specific modification and PEGylation of pharmaceutical proteins mediated by transglutaminase. Adv Drug Deliv Rev. 2008;60(1):13–28. doi: 10.1016/j.addr.2007.06.015. [DOI] [PubMed] [Google Scholar]
- Francis RJ, Sharma SK, Springer C, et al. A phase I trial of antibody directed enzyme prodrug therapy (ADEPT) in patients with advanced colorectal carcinoma or other CEA producing tumours. Br J Cancer. 2002;87(6):600–607. doi: 10.1038/sj.bjc.6600517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman M, Stahl S. Engineered affinity proteins for tumour-targeting applications. Biotechnol Appl Biochem. 2009;53(Pt 1):1–29. doi: 10.1042/BA20080287. [DOI] [PubMed] [Google Scholar]
- Friedman M, Nordberg E, Hoiden-Guthenberg I, et al. Phage display selection of Affibody molecules with specific binding to the extracellular domain of the epidermal growth factor receptor. Protein Eng Des Sel. 2007;20(4):189–199. doi: 10.1093/protein/gzm011. [DOI] [PubMed] [Google Scholar]
- Friedman M, Orlova A, Johansson E, et al. Directed evolution to low nanomolar affinity of a tumor-targeting epidermal growth factor receptor-binding affibody molecule. J Mol Biol. 2008;376(5):1388–1402. doi: 10.1016/j.jmb.2007.12.060. [DOI] [PubMed] [Google Scholar]
- Funaro A, Horenstein AL, Santoro P, Cinti C, Gregorini A, Malavasi F. Monoclonal antibodies and therapy of human cancers. Biotechnol Adv. 2000;18(5):385–401. doi: 10.1016/s0734-9750(00)00043-4. [DOI] [PubMed] [Google Scholar]
- Gerke V, Moss SE. Annexins: from structure to function. Physiol Rev. 2002;82(2):331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- Goldenberg A, Masui H, Divgi C, Kamrath H, Pentlow K, Mendelsohn J. Imaging of human tumor xenografts with an indium-111-labeled anti-epidermal growth factor receptor monoclonal antibody. J Natl Cancer Inst. 1989;81(21):1616–1625. doi: 10.1093/jnci/81.21.1616. [DOI] [PubMed] [Google Scholar]
- Graslund S, Eklund M, Falk R, Uhlen M, Nygren PA, Stahl S. A novel affinity gene fusion system allowing protein A-based recovery of non-immunoglobulin gene products. J Biotechnol. 2002;99(1):41–50. doi: 10.1016/s0168-1656(02)00158-x. [DOI] [PubMed] [Google Scholar]
- Greenberg CS, Birckbichler PJ, Rice RH. Transglutaminases: multifunctional cross-linking enzymes that stabilize tissues. FASEB J. 1991;5(15):3071–3077. doi: 10.1096/fasebj.5.15.1683845. [DOI] [PubMed] [Google Scholar]
- Gronemeyer T, Chidley C, Juillerat A, Heinis C, Johnsson K. Directed evolution of O6-alkylguanine-DNA alkyltransferase for applications in protein labeling. Protein Eng Des Sel. 2006;19(7):309–316. doi: 10.1093/protein/gzl014. [DOI] [PubMed] [Google Scholar]
- Gronwall C, Jonsson A, Lindstrom S, Gunneriusson E, Stahl S, Herne N. Selection and characterization of Affibody ligands binding to Alzheimer amyloid beta peptides. J Biotechnol. 2007a;128(1):162–183. doi: 10.1016/j.jbiotec.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Gronwall C, Sjoberg A, Ramstrom M, et al. Affibody-mediated transferrin depletion for proteomics applications. Biotechnol J. 2007b;2(11):1389–1398. doi: 10.1002/biot.200700053. [DOI] [PubMed] [Google Scholar]
- Gronwall C, Snelders E, Palm AJ, Eriksson F, Herne N, Stahl S. Generation of Affibody ligands binding interleukin-2 receptor alpha/CD25. Biotechnol Appl Biochem. 2008;50(Pt 2):97–112. doi: 10.1042/BA20070261. [DOI] [PubMed] [Google Scholar]
- Gross ME, Shazer RL, Agus DB. Targeting the HER-kinase axis in cancer. Semin Oncol. 2004;31(1 Suppl 3):9–20. doi: 10.1053/j.seminoncol.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Gunneriusson E, Nord K, Uhlen M, Nygren P. Affinity maturation of a Taq DNA polymerase specific affibody by helix shuffling. Protein Eng. 1999;12(10):873–878. doi: 10.1093/protein/12.10.873. [DOI] [PubMed] [Google Scholar]
- Guo MF, Zhao Y, Tian R, et al. In vivo99mTc-HYNIC-annexin V imaging of early tumor apoptosis in mice after single dose irradiation. J Exp Clin Cancer Res. 2009;28:136. doi: 10.1186/1756-9966-28-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning P, Magnusson MK, Gunneriusson E, et al. Genetic modification of adenovirus 5 tropism by a novel class of ligands based on a three-helix bundle scaffold derived from staphylococcal protein A. Hum Gene Ther. 2002;13(12):1427–1439. doi: 10.1089/10430340260185067. [DOI] [PubMed] [Google Scholar]
- Herberman RB. Cancer immunotherapy with natural killer cells. Semin Oncol. 2002;29(3 Suppl 7):27–30. doi: 10.1053/sonc.2002.33079. [DOI] [PubMed] [Google Scholar]
- Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23(5):1011–1027. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]
- Hofer A, Saez JC, Chang CC, Trosko JE, Spray DC, Dermietzel R. C-erbB2/neu transfection induces gap junctional communication incompetence in glial cells. J Neurosci. 1996;16(14):4311–4321. doi: 10.1523/JNEUROSCI.16-14-04311.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstra L, Liem IH, Dumont EA, et al. Visualisation of cell death in vivo in patients with acute myocardial infarction. Lancet. 2000;356(9225):209–212. doi: 10.1016/s0140-6736(00)02482-x. [DOI] [PubMed] [Google Scholar]
- Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23(9):1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2(2):91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- Hsu AR, Cai W, Veeravagu A, et al. Multimodality molecular imaging of glioblastoma growth inhibition with vasculature-targeting fusion toxin VEGF121/rGel. J Nucl Med. 2007;48(3):445–454. [PubMed] [Google Scholar]
- Hu S, Shively L, Raubitschek A, et al. Minibody: a novel engineered anti-carcinoembryonic antigen antibody fragment (single-chain Fv-CH3) which exhibits rapid, high-level targeting of xenografts. Cancer Res. 1996;56(13):3055–3061. [PubMed] [Google Scholar]
- Hudson PJ. Recombinant antibody constructs in cancer therapy. Curr Opin Immunol. 1999;11(5):548–557. doi: 10.1016/s0952-7915(99)00013-8. [DOI] [PubMed] [Google Scholar]
- Hudson PJ. Recombinant antibodies: a novel approach to cancer diagnosis and therapy. Expert Opin Invest Drugs. 2000;9(6):1231–1242. doi: 10.1517/13543784.9.6.1231. [DOI] [PubMed] [Google Scholar]
- Hudson PJ, Kortt AA. High avidity scFv multimers; diabodies and triabodies. J Immunol Methods. 1999;231(1–2):177–189. doi: 10.1016/s0022-1759(99)00157-x. [DOI] [PubMed] [Google Scholar]
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110(6):673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Iwai H, Lingel A, Pluckthun A. Cyclic green fluorescent protein produced in vivo using an artificially split PI-PfuI intein from Pyrococcus furiosus. J Biol Chem. 2001;276(19):16548–16554. doi: 10.1074/jbc.M011639200. [DOI] [PubMed] [Google Scholar]
- Jayson GC, Zweit J, Jackson A, et al. Molecular imaging and biological evaluation of HuMV833 anti-VEGF antibody: implications for trial design of antiangiogenic antibodies. J Natl Cancer Inst. 2002;94(19):1484–1493. doi: 10.1093/jnci/94.19.1484. [DOI] [PubMed] [Google Scholar]
- Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature. 1986;321(6069):522–525. doi: 10.1038/321522a0. [DOI] [PubMed] [Google Scholar]
- Jones DH, Cellitti SE, Hao X, et al. Site-specific labeling of proteins with NMR-active unnatural amino acids. J Biomol NMR. 2010;46(1):89–100. doi: 10.1007/s10858-009-9365-4. [DOI] [PubMed] [Google Scholar]
- Juillerat A, Gronemeyer T, Keppler A, et al. Directed evolution of O6-alkylguanine-DNA alkyltransferase for efficient labeling of fusion proteins with small molecules in vivo. Chem Biol. 2003;10(4):313–317. doi: 10.1016/s1074-5521(03)00068-1. [DOI] [PubMed] [Google Scholar]
- Kamiya N, Abe H, Goto M, Tsuji Y, Jikuya H. Fluorescent substrates for covalent protein labeling catalyzed by microbial transglutaminase. Org Biomol Chem. 2009;7(17):3407–3412. doi: 10.1039/b904046c. [DOI] [PubMed] [Google Scholar]
- Kampmeier F, Ribbert M, Nachreiner T, et al. Site-specific, covalent labeling of recombinant antibody fragments via fusion to an engineered version of 6-O-alkylguanine DNA alkyltransferase. Bioconjug Chem. 2009;20(5):1010–1015. doi: 10.1021/bc9000257. [DOI] [PubMed] [Google Scholar]
- Karlstrom A, Nygren PA. Dual labeling of a binding protein allows for specific fluorescence detection of native protein. Anal Biochem. 2001;295(1):22–30. doi: 10.1006/abio.2001.5186. [DOI] [PubMed] [Google Scholar]
- Keyt BA, Nguyen HV, Berleau LT, et al. Identification of vascular endothelial growth factor determinants for binding KDR and FLT-1 receptors Generation of receptor-selective VEGF variants by site-directed mutagenesis. J Biol Chem. 1996;271(10):5638–5646. doi: 10.1074/jbc.271.10.5638. [DOI] [PubMed] [Google Scholar]
- Koyama Y, Barrett T, Hama Y, Ravizzini G, Choyke PL, Kobayashi H. In vivo molecular imaging to diagnose and subtype tumors through receptor-targeted optically labeled monoclonal antibodies. Neoplasia. 2007;9(12):1021–1029. doi: 10.1593/neo.07787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer-Marek G, Kiesewetter DO, Capala J. Changes in HER2 expression in breast cancer xenografts after therapy can be quantified using PET and (18)F-labeled affibody molecules. J Nucl Med. 2009;50(7):1131–1139. doi: 10.2967/jnumed.108.057695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronqvist N, Lofblom J, Jonsson A, Wernerus H, Stahl S. A novel affinity protein selection system based on staphylococcal cell surface display and flow cytometry. Protein Eng Des Sel. 2008;21(4):247–255. doi: 10.1093/protein/gzm090. [DOI] [PubMed] [Google Scholar]
- Langone JJ. Protein A of Staphylococcus aureus and related immunoglobulin receptors produced by streptococci and pneumonococci. Adv Immunol. 1982;32:157–252. [PubMed] [Google Scholar]
- Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nat Rev Drug Discov. 2008;7(1):21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- Li PC, Wang CR, Shieh DB, et al. In vivo photoacoustic molecular imaging with simultaneous multiple selective targeting using antibody-conjugated gold nanorods. Opt Express. 2008;16(23):18605–18615. doi: 10.1364/oe.16.018605. [DOI] [PubMed] [Google Scholar]
- Lin CW, Ting AY. Transglutaminase-catalyzed site-specific conjugation of small-molecule probes to proteins in vitro and on the surface of living cells. J Am Chem Soc. 2006;128(14):4542–4543. doi: 10.1021/ja0604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CR, Chen WS, Kruiger W, et al. Expression cloning of human EGF receptor complementary DNA: gene amplification and three related messenger RNA products in A431 cells. Science. 1984;224(4651):843–848. doi: 10.1126/science.6326261. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Lobato MN, Rabbitts TH. Intracellular antibodies as specific reagents for functional ablation: future therapeutic molecules. Curr Mol Med. 2004;4(5):519–528. doi: 10.2174/1566524043360384. [DOI] [PubMed] [Google Scholar]
- Lovrinovic M, Seidel R, Wacker R, et al. Synthesis of protein-nucleic acid conjugates by expressed protein ligation. Chem Commun (Camb) 2003;(7):822–823. doi: 10.1039/b212294d. [DOI] [PubMed] [Google Scholar]
- Lundberg E, Hoiden-Guthenberg I, Larsson B, Uhlen M, Graslund T. Site-specifically conjugated anti-HER2 Affibody molecules as one-step reagents for target expression analyses on cells and xenograft samples. J Immunol Methods. 2007;319(1–2):53–63. doi: 10.1016/j.jim.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Lyakhov I, Zielinski R, Kuban M, et al. HER2- and EGFR-specific affiprobes: novel recombinant optical probes for cell imaging. Chembiochem. 2010;11(3):345–350. doi: 10.1002/cbic.200900532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Cooney CL. Application of vortex flow adsorption technology to intein-mediated recovery of recombinant human alpha1-antitrypsin. Biotechnol Prog. 2004;20(1):269–276. doi: 10.1021/bp0341803. [DOI] [PubMed] [Google Scholar]
- Mankoff DA. A definition of molecular imaging. J Nucl Med. 2007;48(6):18N–21N. [PubMed] [Google Scholar]
- Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- Margison GP, Santibanez-Koref MF. O6-alkylguanine-DNA alkyltransferase: role in carcinogenesis and chemotherapy. Bioessays. 2002;24(3):255–266. doi: 10.1002/bies.10063. [DOI] [PubMed] [Google Scholar]
- Mass RD. The HER receptor family: a rich target for therapeutic development. Int J Radiat Oncol Biol Phys. 2004;58(3):932–940. doi: 10.1016/j.ijrobp.2003.09.093. [DOI] [PubMed] [Google Scholar]
- Mayer A, Tsiompanou E, O'Malley D, et al. Radioimmunoguided surgery in colorectal cancer using a genetically engineered anti-CEA single-chain Fv antibody. Clin Cancer Res. 2000;6(5):1711–1719. [PubMed] [Google Scholar]
- Mease RC, Lambert C. Newer methods of labeling diagnostic agents with Tc-99m. Semin Nucl Med. 2001;31(4):278–285. doi: 10.1053/snuc.2001.26182. [DOI] [PubMed] [Google Scholar]
- Meers P, Mealy T. Calcium-dependent annexin V binding to phospholipids: stoichiometry, specificity, and the role of negative charge. Biochemistry. 1993;32(43):11711–11721. doi: 10.1021/bi00094a030. [DOI] [PubMed] [Google Scholar]
- Miyake-Stoner SJ, Miller AM, Hammill JT, et al. Probing protein folding using site-specifically encoded unnatural amino acids as FRET donors with tryptophan. Biochemistry. 2009;48(25):5953–5962. doi: 10.1021/bi900426d. [DOI] [PubMed] [Google Scholar]
- Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- Mume E, Orlova A, Larsson B, et al. Evaluation of ((4-hydroxyphenyl)ethyl)maleimide for site-specific radiobromination of anti-HER2 affibody. Bioconjug Chem. 2005;16(6):1547–1555. doi: 10.1021/bc050056o. [DOI] [PubMed] [Google Scholar]
- Muralidharan V, Muir TW. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat Methods. 2006;3(6):429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- Muralidharan V, Cho J, Trester-Zedlitz M, et al. Domain-specific incorporation of noninvasive optical probes into recombinant proteins. J Am Chem Soc. 2004;126(43):14004–14012. doi: 10.1021/ja0466199. [DOI] [PubMed] [Google Scholar]
- Myhre S, Henning P, Granio O, et al. Decreased immune reactivity towards a knobless, affibody-targeted adenovirus type 5 vector. Gene Ther. 2007;14(4):376–381. doi: 10.1038/sj.gt.3302875. [DOI] [PubMed] [Google Scholar]
- Nagengast WB, de Vries EG, Hospers GA, et al. In vivo VEGF imaging with radiolabeled bevacizumab in a human ovarian tumor xenograft. J Nucl Med. 2007;48(8):1313–1319. doi: 10.2967/jnumed.107.041301. [DOI] [PubMed] [Google Scholar]
- Narula J, Acio ER, Narula N, et al. Annexin-V imaging for noninvasive detection of cardiac allograft rejection. Nat Med. 2001;7(12):1347–1352. doi: 10.1038/nm1201-1347. [DOI] [PubMed] [Google Scholar]
- Niu G, Li Z, Cao Q, Chen X. Monitoring therapeutic response of human ovarian cancer to 17-DMAG by noninvasive PET imaging with (64)Cu-DOTA-trastuzumab. Eur J Nucl Med Mol Imaging. 2009;36(9):1510–1519. doi: 10.1007/s00259-009-1158-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord K, Gunneriusson E, Ringdahl J, Stahl S, Uhlen M, Nygren PA. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat Biotechnol. 1997;15(8):772–777. doi: 10.1038/nbt0897-772. [DOI] [PubMed] [Google Scholar]
- Nord K, Gunneriusson E, Uhlen M, Nygren PA. Ligands selected from combinatorial libraries of protein A for use in affinity capture of apolipoprotein A-1M and taq DNA polymerase. J Biotechnol. 2000;80(1):45–54. doi: 10.1016/s0168-1656(00)00232-7. [DOI] [PubMed] [Google Scholar]
- Nord K, Nord O, Uhlen M, Kelley B, Ljungqvist C, Nygren PA. Recombinant human factor VIII-specific affinity ligands selected from phage-displayed combinatorial libraries of protein A. Eur J Biochem. 2001;268(15):4269–4277. doi: 10.1046/j.1432-1327.2001.02344.x. [DOI] [PubMed] [Google Scholar]
- Nord O, Gustrin A, Nygren PA. Fluorescent detection of beta-lactamase activity in living Escherichia coli cells via esterase supplementation. FEMS Microbiol Lett. 2005;242(1):73–79. doi: 10.1016/j.femsle.2004.10.047. [DOI] [PubMed] [Google Scholar]
- Normanno N, De Luca A, Bianco C, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366(1):2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Kosaka N, Longmire MR, Urano Y, Choyke PL, Kobayashi H. Fluorophore–quencher based activatable targeted optical probes for detecting in vivo cancer metastases. Mol Pharm. 2009;6(2):386–395. doi: 10.1021/mp800115t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova A, Nilsson FY, Wikman M, et al. Comparative in vivo evaluation of technetium and iodine labels on an anti-HER2 affibody for single-photon imaging of HER2 expression in tumors. J Nucl Med. 2006a;47(3):512–519. [PubMed] [Google Scholar]
- Orlova A, Magnusson M, Eriksson TL, et al. Tumor imaging using a picomolar affinity HER2 binding affibody molecule. Cancer Res. 2006b;66(8):4339–4348. doi: 10.1158/0008-5472.CAN-05-3521. [DOI] [PubMed] [Google Scholar]
- Orlova A, Tolmachev V, Pehrson R, et al. Synthetic affibody molecules: a novel class of affinity ligands for molecular imaging of HER2-expressing malignant tumors. Cancer Res. 2007;67(5):2178–2186. doi: 10.1158/0008-5472.CAN-06-2887. [DOI] [PubMed] [Google Scholar]
- Park H, Lee S, Chen L, et al. SERS imaging of HER2-overexpressed MCF7 cells using antibody-conjugated gold nanorods. Phys Chem Chem Phys. 2009;11(34):7444–7449. doi: 10.1039/b904592a. [DOI] [PubMed] [Google Scholar]
- Pateisky N, Schatten C, Vavra N, et al. Lymphoscintigraphy using epidermal growth factor as tumour-seeking agent in uterine cervical cancer. Wien Klin Wochenschr. 1991;103(21):654–656. [PubMed] [Google Scholar]
- Pegg AE. Repair of O(6)-alkylguanine by alkyltransferases. Mutat Res. 2000;462(2–3):83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- Pegg AE, Dolan ME, Moschel RC. Structure, function, and inhibition of O6-alkylguanine-DNA alkyltransferase. Prog Nucleic Acid Res Mol Biol. 1995;51:167–223. doi: 10.1016/s0079-6603(08)60879-x. [DOI] [PubMed] [Google Scholar]
- Pluckthun A, Pack P. New protein engineering approaches to multivalent and bispecific antibody fragments. Immunotechnology. 1997;3(2):83–105. doi: 10.1016/s1380-2933(97)00067-5. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B, Boeggeman E, Manzoni M, et al. Multiple site-specific in vitro labeling of single-chain antibody. Bioconjug Chem. 2009;20(7):1383–1389. doi: 10.1021/bc900149r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Suzarte M, Rodriguez N, Oliva JP, et al. 99mTc-labeled antihuman epidermal growth factor receptor antibody in patients with tumors of epithelial origin. Part III. Clinical trials safety and diagnostic efficacy. J Nucl Med. 1999;40(5):768–775. [PubMed] [Google Scholar]
- Reilly RM, Kiarash R, Cameron RG, et al. 111In-labeled EGF is selectively radiotoxic to human breast cancer cells over-expressing EGFR. J Nucl Med. 2000;41(3):429–438. [PubMed] [Google Scholar]
- Ren G, Zhang R, Liu Z, et al. A 2-helix small protein labeled with 68Ga for PET imaging of HER2 expression. J Nucl Med. 2009;50(9):1492–1499. doi: 10.2967/jnumed.109.064287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnmark J, Hansson M, Nguyen T, et al. Construction and characterization of affibody-Fc chimeras produced in Escherichia coli. J Immunol Methods. 2002;261(1–2):199–211. doi: 10.1016/s0022-1759(01)00563-4. [DOI] [PubMed] [Google Scholar]
- Roopenian DC, Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nat Rev Immunol. 2007;7(9):715–725. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- Rottey S, Slegers G, Van Belle S, Goethals I, Van de Wiele C. Sequential 99mTc-hydrazinonicotinamide-annexin V imaging for predicting response to chemotherapy. J Nucl Med. 2006;47(11):1813–1818. [PubMed] [Google Scholar]
- Saga T, Endo K, Akiyama T, et al. Scintigraphic detection of overexpressed c-erbB-2 protooncogene products by a class-switched murine anti-c-erbB-2 protein monoclonal antibody. Cancer Res. 1991;51(3):990–994. [PubMed] [Google Scholar]
- Sampath L, Kwon S, Ke S, et al. Dual-labeled trastuzumab-based imaging agent for the detection of human epidermal growth factor receptor 2 overexpression in breast cancer. J Nucl Med. 2007;48(9):1501–1510. doi: 10.2967/jnumed.107.042234. [DOI] [PubMed] [Google Scholar]
- Sandstrom K, Xu Z, Forsberg G, Nygren PA. Inhibition of the CD28–CD80 co-stimulation signal by a CD28-binding affibody ligand developed by combinatorial protein engineering. Protein Eng. 2003;16(9):691–697. doi: 10.1093/protein/gzg086. [DOI] [PubMed] [Google Scholar]
- Sato H, Ikeda M, Suzuki K, Hirayama K. Site-specific modification of interleukin-2 by the combined use of genetic engineering techniques and transglutaminase. Biochemistry. 1996;35(40):13072–13080. doi: 10.1021/bi952616k. [DOI] [PubMed] [Google Scholar]
- Schatten C, Pateisky N, Vavra N, et al. Lymphoscintigraphy with 123I-labelled epidermal growth factor. Lancet. 1991;337(8738):395–396. doi: 10.1016/0140-6736(91)91169-u. [DOI] [PubMed] [Google Scholar]
- Schechter NR, Yang DJ, Azhdarinia A, et al. Assessment of epidermal growth factor receptor with 99mTc-ethylenedicysteine-C225 monoclonal antibody. Anticancer Drugs. 2003;14(1):49–56. doi: 10.1097/00001813-200301000-00007. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103(2):211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Schneider PM, Hung MC, Chiocca SM, et al. Differential expression of the c-erbB-2 gene in human small cell and non-small cell lung cancer. Cancer Res. 1989;49(18):4968–4971. [PubMed] [Google Scholar]
- Schuhmacher J, Kaul S, Klivenyi G, et al. Immunoscintigraphy with positron emission tomography: gallium-68 chelate imaging of breast cancer pretargeted with bispecific anti-MUC1/anti-Ga chelate antibodies. Cancer Res. 2001;61(9):3712–3717. [PubMed] [Google Scholar]
- Schwarzer D, Cole PA. Protein semisynthesis and expressed protein ligation: chasing a protein's tail. Curr Opin Chem Biol. 2005;9(6):561–569. doi: 10.1016/j.cbpa.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Scopinaro F, De Vincentis G, Banci M, et al. In vivo study of a technetium labelled anti-EGFr MoAB. Anticancer Res. 1997;17(3B):1761–1765. [PubMed] [Google Scholar]
- Sebastian S, Settleman J, Reshkin SJ, Azzariti A, Bellizzi A, Paradiso A. The complexity of targeting EGFR signalling in cancer: from expression to turnover. Biochim Biophys Acta. 2006;1766(1):120–139. doi: 10.1016/j.bbcan.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Sibler AP, Kempf E, Glacet A, Orfanoudakis G, Bourel D, Weiss E. In vivo biotinylated recombinant antibodies: high efficiency of labelling and application to the cloning of active anti-human IgG1 Fab fragments. J Immunol Methods. 1999;224(1–2):129–140. doi: 10.1016/s0022-1759(99)00016-2. [DOI] [PubMed] [Google Scholar]
- Signore A, Annovazzi A, Corsetti F, et al. Biological imaging for the diagnosis of inflammatory conditions. BioDrugs. 2002;16(4):241–259. doi: 10.2165/00063030-200216040-00002. [DOI] [PubMed] [Google Scholar]
- Signore A, Chianelli M, Bei R, Oyen W, Modesti A. Targeting cytokine/chemokine receptors: a challenge for molecular nuclear medicine. Eur J Nucl Med Mol Imaging. 2003a;30(1):149–156. doi: 10.1007/s00259-002-0941-z. [DOI] [PubMed] [Google Scholar]
- Signore A, Capriotti G, Scopinaro F, Bonanno E, Modesti A. Radiolabelled lymphokines and growth factors for in vivo imaging of inflammation, infection and cancer. Trends Immunol. 2003b;24(7):395–402. doi: 10.1016/s1471-4906(03)00174-1. [DOI] [PubMed] [Google Scholar]
- Simon M, Rockl W, Hornig C, et al. Receptors of vascular endothelial growth factor/vascular permeability factor (VEGF/VPF) in fetal and adult human kidney: localization and [125I]VEGF binding sites. J Am Soc Nephrol. 1998;9(6):1032–1044. doi: 10.1681/ASN.V961032. [DOI] [PubMed] [Google Scholar]
- Sirk SJ, Olafsen T, Barat B, Bauer KB, Wu AM. Site-specific, thiol-mediated conjugation of fluorescent probes to cysteinemodified diabodies targeting CD20 or HER2. Bioconjug Chem. 2008;19(12):2527–2534. doi: 10.1021/bc800113v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- Slaughter TF, Achyuthan KE, Lai TS, Greenberg CS. A microtiter plate transglutaminase assay utilizing 5-(biotinamido)pentylamine as substrate. Anal Biochem. 1992;205(1):166–171. doi: 10.1016/0003-2697(92)90594-w. [DOI] [PubMed] [Google Scholar]
- Smith PA, Tripp BC, DiBlasio-Smith EA, Lu Z, LaVallie ER, McCoy JM. A plasmid expression system for quantitative in vivo biotinylation of thioredoxin fusion proteins in Escherichia coli. Nucleic Acids Res. 1998;26(6):1414–1420. doi: 10.1093/nar/26.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen AC, Wikman M, Tolmachev V, et al. In vitro characterization of a bivalent anti-HER-2 affibody with potential for radionuclide-based diagnostics. Cancer Biother Radiopharm. 2005;20(3):239–248. doi: 10.1089/cbr.2005.20.239. [DOI] [PubMed] [Google Scholar]
- Styles JM, Harrison S, Gusterson BA, Dean CJ. Rat monoclonal antibodies to the external domain of the product of the C-erbB-2 proto-oncogene. Int J Cancer. 1990;45(2):320–324. doi: 10.1002/ijc.2910450219. [DOI] [PubMed] [Google Scholar]
- Summerer D, Chen S, Wu N, Deiters A, Chin JW, Schultz PG. A genetically encoded fluorescent amino acid. Proc Natl Acad Sci USA. 2006;103(26):9785–9789. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunbul M, Yin J. Site specific protein labeling by enzymatic posttranslational modification. Org Biomol Chem. 2009;7(17):3361–3371. doi: 10.1039/b908687k. [DOI] [PubMed] [Google Scholar]
- Sundaresan G, Yazaki PJ, Shively JE, et al. 124I-labeled engineered anti-CEA minibodies and diabodies allow high-contrast, antigen-specific small-animal PET imaging of xenografts in athymic mice. J Nucl Med. 2003;44(12):1962–1969. [PMC free article] [PubMed] [Google Scholar]
- Tait JF, Smith C, Blankenberg FG. Structural requirements for in vivo detection of cell death with 99mTc-annexin V. J Nucl Med. 2005;46(5):807–815. [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Nakada T, Nakaki M, Wands JR. Inhibition of hepatic metastases of human colon cancer in nude mice by a chimeric SF-25 monoclonal antibody. Gastroenterology. 1995;108(1):172–182. doi: 10.1016/0016-5085(95)90022-5. [DOI] [PubMed] [Google Scholar]
- Takasu S, Takahashi T, Okamoto S, et al. Radioimmunoscintigraphy of intracranial glioma xenograft with a technetium-99m-labeled mouse monoclonal antibody specifically recognizing type III mutant epidermal growth factor receptor. J Neurooncol. 2003;63(3):247–256. doi: 10.1023/a:1024320516341. [DOI] [PubMed] [Google Scholar]
- Taki M, Shiota M, Taira K. Transglutaminase-mediated N- and C-terminal fluorescein labeling of a protein can support the native activity of the modified protein. Protein Eng Des Sel. 2004;17(2):119–126. doi: 10.1093/protein/gzh015. [DOI] [PubMed] [Google Scholar]
- Tang Y, Wang J, Scollard DA, et al. Imaging of HER2/neu-positive BT-474 human breast cancer xenografts in athymic mice using (111)In-trastuzumab (Herceptin) Fab fragments. Nucl Med Biol. 2005;32(1):51–58. doi: 10.1016/j.nucmedbio.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Tirat A, Freuler F, Stettler T, Mayr LM, Leder L. Evaluation of two novel tag-based labelling technologies for site-specific modification of proteins. Int J Biol Macromol. 2006;39(1–3):66–76. doi: 10.1016/j.ijbiomac.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Todorovska A, Roovers RC, Dolezal O, Kortt AA, Hoogenboom HR, Hudson PJ. Design and application of diabodies, triabodies and tetrabodies for cancer targeting. J Immunol Methods. 2001;248(1–2):47–66. doi: 10.1016/s0022-1759(00)00342-2. [DOI] [PubMed] [Google Scholar]
- Tolmachev V, Nilsson FY, Widstrom C, et al. 111In-benzyl-DTPA-ZHER2:342, an affibody-based conjugate for in vivo imaging of HER2 expression in malignant tumors. J Nucl Med. 2006;47(5):846–853. [PubMed] [Google Scholar]
- Tolmachev V, Orlova A, Nilsson FY, Feldwisch J, Wennborg A, Abrahmsen L. Affibody molecules: potential for in vivo imaging of molecular targets for cancer therapy. Expert Opin Biol Ther. 2007;7(4):555–568. doi: 10.1517/14712598.7.4.555. [DOI] [PubMed] [Google Scholar]
- Tran T, Engfeldt T, Orlova A, et al. In vivo evaluation of cysteine-based chelators for attachment of 99mTc to tumor-targeting affibody molecules. Bioconjug Chem. 2007;18(2):549–558. doi: 10.1021/bc060291m. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Nilsson B, Guss B, Lindberg M, Gatenbeck S, Philipson L. Gene fusion vectors based on the gene for staphylococcal protein A. Gene. 1983;23(3):369–378. doi: 10.1016/0378-1119(83)90025-2. [DOI] [PubMed] [Google Scholar]
- Underiner TL, Ruggeri B, Gingrich DE. Development of vascular endothelial growth factor receptor (VEGFR) kinase inhibitors as anti-angiogenic agents in cancer therapy. Curr Med Chem. 2004;11(6):731–745. doi: 10.2174/0929867043455756. [DOI] [PubMed] [Google Scholar]
- Van den Eijnde SM, Boshart L, Reutelingsperger CP, De Zeeuw CI, Vermeij-Keers C. Phosphatidylserine plasma membrane asymmetry in vivo: a pancellular phenomenon which alters during apoptosis. Cell Death Differ. 1997;4(4):311–316. doi: 10.1038/sj.cdd.4400241. [DOI] [PubMed] [Google Scholar]
- van den Eijnde SM, Boshart L, Baehrecke EH, De Zeeuw CI, Reutelingsperger CP, Vermeij-Keers C. Cell surface exposure of phosphatidylserine during apoptosis is phylogenetically conserved. Apoptosis. 1998;3(1):9–16. doi: 10.1023/a:1009650917818. [DOI] [PubMed] [Google Scholar]
- van Engeland M, Ramaekers FC, Schutte B, Reutelingsperger CP. A novel assay to measure loss of plasma membrane asymmetry during apoptosis of adherent cells in culture. Cytometry. 1996;24(2):131–139. doi: 10.1002/(SICI)1097-0320(19960601)24:2<131::AID-CYTO5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Velikyan I, Sundberg AL, Lindhe O, et al. Preparation and evaluation of (68)Ga-DOTA-hEGF for visualization of EGFR expression in malignant tumors. J Nucl Med. 2005;46(11):1881–1888. [PubMed] [Google Scholar]
- Viti F, Tarli L, Giovannoni L, Zardi L, Neri D. Increased binding affinity and valence of recombinant antibody fragments lead to improved targeting of tumoral angiogenesis. Cancer Res. 1999;59(2):347–352. [PubMed] [Google Scholar]
- Volkmann G, Liu XQ. Protein C-terminal labeling and biotinylation using synthetic peptide and split-intein. PLoS One. 2009;4(12):e8381. doi: 10.1371/journal.pone.0008381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorzanger-Rousselot N, Garnero P. Biochemical markers in oncology. Part I. Molecular basis. Part II. Clinical uses. Cancer Treat Rev. 2007;33(3):230–283. doi: 10.1016/j.ctrv.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44(45):7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- Wang J, Xie J, Schultz PG. A genetically encoded fluorescent amino acid. J Am Chem Soc. 2006;128(27):8738–8739. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- Wang H, Cai W, Chen K, et al. A new PET tracer specific for vascular endothelial growth factor receptor 2. Eur J Nucl Med Mol Imaging. 2007;34(12):2001–2010. doi: 10.1007/s00259-007-0524-0. [DOI] [PubMed] [Google Scholar]
- Wang H, Chen K, Niu G, Chen X. Site-specifically biotinylated VEGF121 for near-infrared fluorescence imaging of tumor angiogenesis. Mol Pharm. 2008;6(1):285–294. doi: 10.1021/mp800185h. [DOI] [PubMed] [Google Scholar]
- Ward ES, Ghetie V. The effector functions of immunoglobulins: implications for therapy. Ther Immunol. 1995;2(2):77–94. [PubMed] [Google Scholar]
- Weir AN, Nesbitt A, Chapman AP, Popplewell AG, Antoniw P, Lawson AD. Formatting antibody fragments to mediate specific therapeutic functions. Biochem Soc Trans. 2002;30(4):512–516. doi: 10.1042/bst0300512. [DOI] [PubMed] [Google Scholar]
- Weng KC, Noble CO, Papahadjopoulos-Sternberg B, et al. Targeted tumor cell internalization and imaging of multifunctional quantum dot-conjugated immunoliposomes in vitro and in vivo. Nano Lett. 2008;8(9):2851–2857. doi: 10.1021/nl801488u. [DOI] [PubMed] [Google Scholar]
- Wikman M, Steffen AC, Gunneriusson E, et al. Selection and characterization of HER2/neu-binding affibody ligands. Protein Eng Des Sel. 2004;17(5):455–462. doi: 10.1093/protein/gzh053. [DOI] [PubMed] [Google Scholar]
- Wikman M, Rowcliffe E, Friedman M, et al. Selection and characterization of an HIV-1 gp120-binding affibody ligand. Biotechnol Appl Biochem. 2006;45(Pt 2):93–105. doi: 10.1042/BA20060016. [DOI] [PubMed] [Google Scholar]
- Wingfield P, Graber P, Shaw AR, Gronenborn AM, Clore GM, MacDonald HR. Preparation, characterization and application of interleukin-1 beta mutant proteins with surface-accessible cysteine residues. Eur J Biochem. 1989;179(3):565–571. doi: 10.1111/j.1432-1033.1989.tb14584.x. [DOI] [PubMed] [Google Scholar]
- Wolff C, Lai CS. Evidence that the two amino termini of plasma fibronectin are in close proximity: a fluorescence energy transfer study. Biochemistry. 1988;27(9):3483–3487. doi: 10.1021/bi00409a053. [DOI] [PubMed] [Google Scholar]
- Wolfgang WJ, Miller TW, Webster JM, et al. Suppression of Huntington's disease pathology in Drosophila by human single-chain Fv antibodies. Proc Natl Acad Sci USA. 2005;102(32):11563–11568. doi: 10.1073/pnas.0505321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JY, Chu DZ, Williams LE, et al. A phase I trial of (90)Y-DOTA-anti-CEA chimeric T84.66 (cT84.66) radioimmunotherapy in patients with metastatic CEA-producing malignancies. Cancer Biother Radiopharm. 2006;21(2):88–100. doi: 10.1089/cbr.2006.21.88. [DOI] [PubMed] [Google Scholar]
- Wu AM, Chen W, Raubitschek A, et al. Tumor localization of anti-CEA single-chain Fvs: improved targeting by non-covalent dimers. Immunotechnology. 1996;2(1):21–36. doi: 10.1016/1380-2933(95)00027-5. [DOI] [PubMed] [Google Scholar]
- Wu P, Shui W, Carlson BL, et al. Site-specific chemical modification of recombinant proteins produced in mammalian cells by using the genetically encoded aldehyde tag. Proc Natl Acad Sci USA. 2009;106(9):3000–3005. doi: 10.1073/pnas.0807820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Gao X, Taratula O, et al. Anti-HER2 IgY antibody-functionalized single-walled carbon nanotubes for detection and selective destruction of breast cancer cells. BMC Cancer. 2009;9:351. doi: 10.1186/1471-2407-9-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Schultz PG. A chemical toolkit for proteins—an expanded genetic code. Nat Rev Mol Cell Biol. 2006;7(10):775–782. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Zhang R, et al. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296(5565):151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- Xu MQ, Evans TC., Jr Recent advances in protein splicing: manipulating proteins in vitro and in vivo. Curr Opin Biotechnol. 2005;16(4):440–446. doi: 10.1016/j.copbio.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Xu R, Ayers B, Cowburn D, Muir TW. Chemical ligation of folded recombinant proteins: segmental isotopic labeling of domains for NMR studies. Proc Natl Acad Sci USA. 1999;96(2):388–393. doi: 10.1073/pnas.96.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Basu A, Wang M, et al. Tailoring structure-function and pharmacokinetic properties of single-chain Fv proteins by site-specific PEGylation. Protein Eng. 2003;16(10):761–770. doi: 10.1093/protein/gzg093. [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhou Q, Wiedmer T, Sims PJ. Level of expression of phospholipid scramblase regulates induced movement of phosphatidylserine to the cell surface. J Biol Chem. 1998;273(12):6603–6606. doi: 10.1074/jbc.273.12.6603. [DOI] [PubMed] [Google Scholar]
- Zou Y, Yin J. Phosphopantetheinyl transferase catalyzed site-specific protein labeling with ADP conjugated chemical probes. J Am Chem Soc. 2009;131(22):7548–7549. doi: 10.1021/ja902464v. [DOI] [PubMed] [Google Scholar]