Abstract
The innate immune system senses pathogens by pattern recognition receptors (PRR) that signal to induce effector cytokines, such as type I interferons (IFNs). We characterized IFNε as a type I IFN because it signaled via the Ifnar1 and Ifnar2 receptors to induce IFN-regulated genes. In contrast to other type I IFNs, IFNε was not induced by known PRR pathways, but was instead constitutively expressed by epithelial cells of the female reproductive tract (FRT) and hormonally regulated. Ifnε-deficient mice had increased susceptibility to infection of the FRT by common sexually transmitted infections (STIs) Herpes Simplex Virus (HSV)-2 and Chlamydia muridarum. IFNε is thus a potent anti-pathogen and immunoregulatory cytokine that may be important in combating STIs which represent a major global health and socioeconomic burden.
Type I IFNs are crucial in host defence because of their antipathogen actions and ability to activate effector cells of the innate and adaptive immune responses (1, 2). The type I IFN locus contains genes encoding 13 IFNα subtypes, IFNβ and IFNω (3) whose promoters contain acute response elements (such as IRFs and NF-κB in IFNβ), which ensure their rapid induction by PRR pathways (4, 5). This locus also contains a gene, which we previously designated IFNε, but whose function has remained uncharacterized.
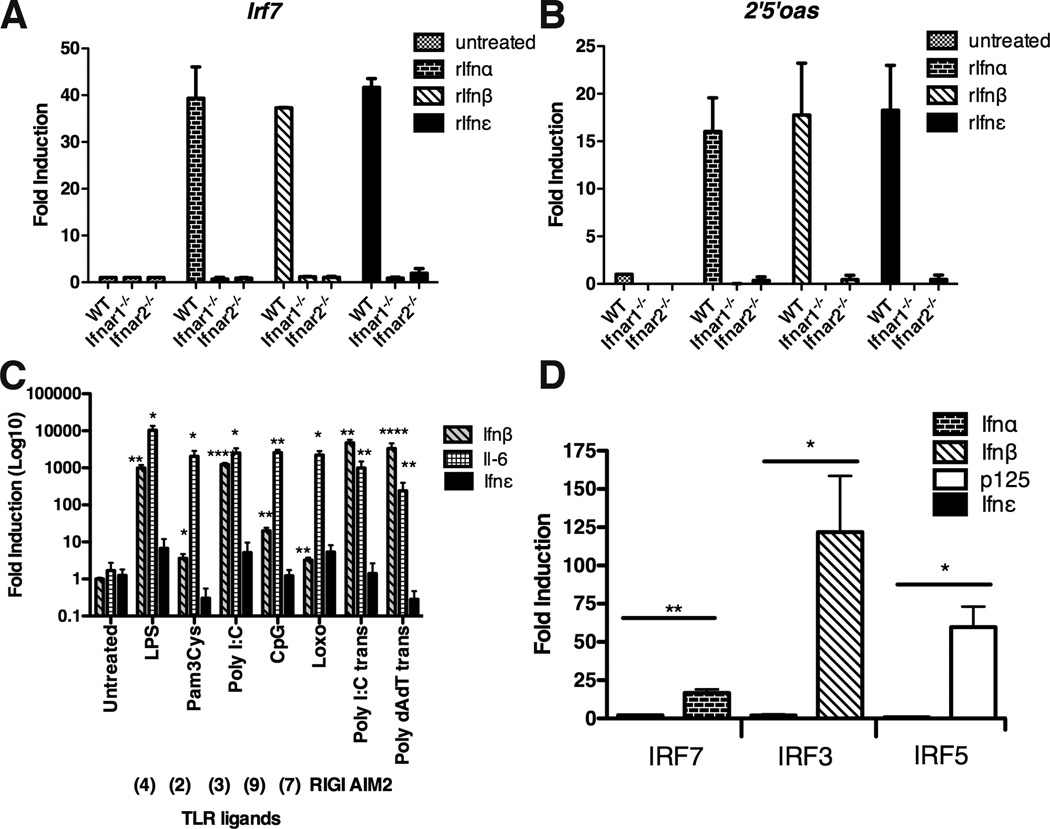
IFNε shares only 30% amino acid homology to a consensus IFNα sequence and to IFNβ. Therefore, we first demonstrated that IFNε was a type I IFN by showing that it transduced signals via the Ifnar1 and Ifnar2 receptors (6). Incubation of recombinant Ifnε with bone marrow derived macrophages (BMMs) from wild type (WT) mice induced IFN-regulated genes (IRGs) such as Irf7 and 2’5’oas (which encodes oligoadenylate synthetase) (Fig. 1A and 1B), whereas these IRGs were not induced in BMM from Ifnar1 or Ifnar2-deficient mice. Accordingly, Ifnε should be classed as a type I IFN.
FIGURE 1. Ifnε signals through the type I IFN receptor but is not induced by TLR ligands nor regulated by IRFs.
(A,B) BMMs from WT, Ifnar1−/− and Ifnar2−/− C57BL/6 mice were stimulated with recombinant mouse Ifnα1, Ifnβ or Ifnε (0.1µg/ml) for 3h. (A) Irf7 and (B) 2’5’ oas expression was measured by qRT-PCR. Data are expressed as mean + SEM. of at least three independent experiments. (C) BMMs from C57BL/6 WT mice were treated with a range of TLR ligands or transfected with Poly (I:C) and Poly (dA:dT) for 3h at 37°C. Ifnβ, Il-6 and Ifnε were measured by qRT-PCR. Data are expressed as mean + SEM. of at least three independent experiments. (D) Luciferase reporter plasmids containing Ifnα, Ifnβ, p125, or Ifnε were co-transfected with empty vector or IRF3, IRF7 or IRF5 expression vectors into HEK293 cells. Data are expressed as mean + SEM. All values are means of at least three independent experiments. *P<0.05, **P<0.01, ***P<0.001 (unpaired Student’s t-test).
We next determined whether IFNε was induced by PRR pathways. Primary BMMs, murine embryonic fibroblasts (MEFs) and the murine macrophage cell line, RAW264.7, treated with synthetic ligands of: TLRs 2, 3, 4, 7/8 and 9; cytosolic DNA sensors or AIM2 inflammasomes, potently induced known PRR response genes such as Ifnβ and/or Il-6 (7–9). In contrast, there was no significant change in the expression of Ifnε upon stimulation with these activators (Fig. 1C and fig. S1A and B). Because all PRRs induce type I IFN expression through the activation of the IRF family of transcription factors (5), we then examined whether IRFs could directly regulate the Ifnε promoter. IRF3, IRF7 and IRF5 induced promoter activity of Ifnβ, Ifnα and p125 (5) luciferase reporters in HEK293 cells (Fig. 1D). By contrast, no alteration of Ifnε promoter activity was observed (Fig. 1D). Semliki Forest Virus (SFV) infection of RAW264.7 cells stimulated the expression of the positive control antiviral response gene 2’5’oas, but not Ifnε expression (fig. S1C). Furthermore, Ifnε expression was not altered during in vivo infection with HSV-2 or Chlamydia muridarum (see below), nor by stimulation of human endometrial cell lines with PRR ligands (fig. S1D). This lack of regulation of Ifnε gene expression by conventional PRR pathways is consistent with the lack of response elements for these pathways (IRFs, NF-κB, STAT, ISRE) in the Ifnε proximal promoter compared to other type I IFN genes (fig. S1E).
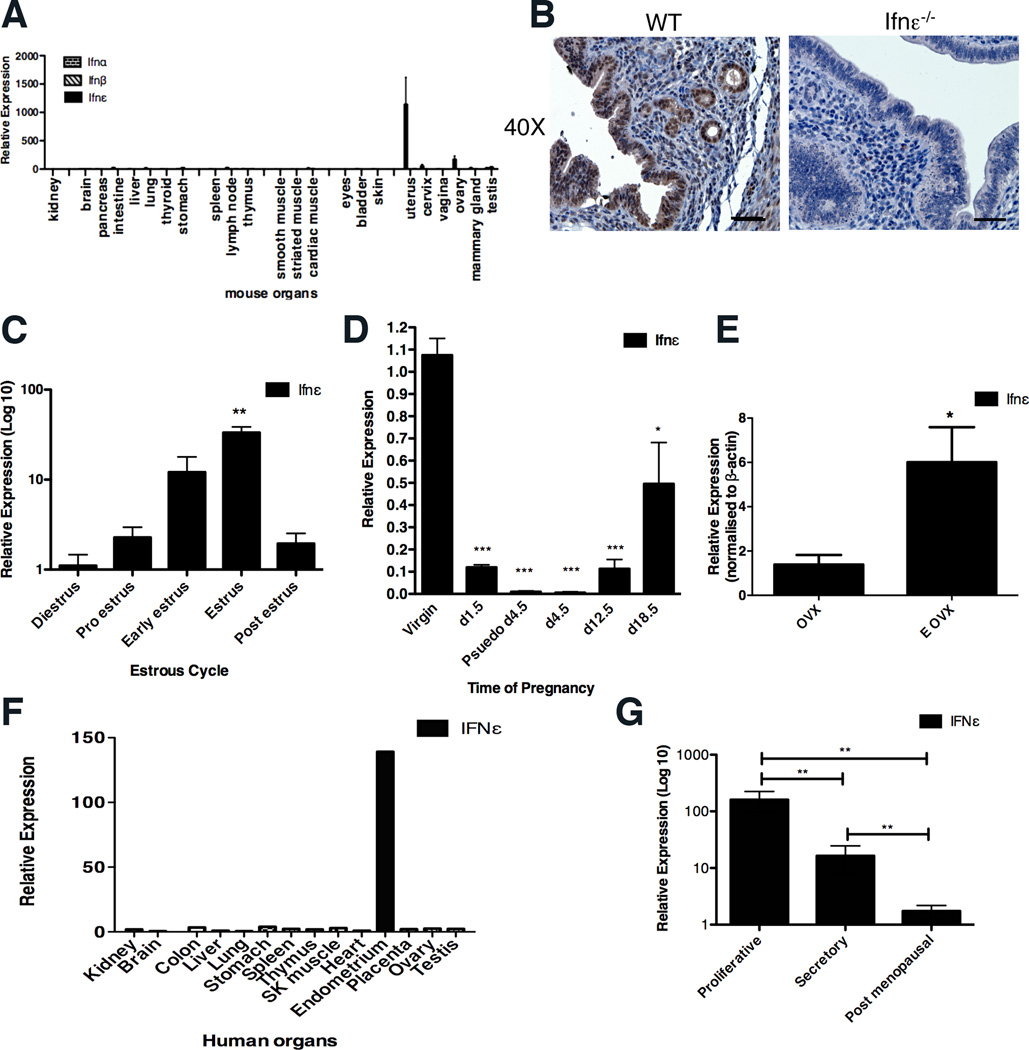
Because Ifnε was not regulated by PRR pathways, we examined its constitutive expression. The expression of Ifnα and β was undetectable in all organs (Fig. 2A). Similarly, the expression of Ifnε was not detectable at significant levels in any organ with the notable exception of the uterus, cervix, vagina and ovary (Fig. 2A). Immunohistochemistry demonstrated that Ifnε was expressed in the luminal and glandular epithelial cells of the endometrium (Fig. 2B). In support of these data, the uterine expression levels of Ifnε did not differ in NOD/SCID/IL-2rγ−/− mice, which are deficient in T, B and NK cells, relative to WT mice, indicating that the aforementioned cells do not express detectable levels, nor do they regulate this cytokine (fig. S1F). This contrasts with conventional type I IFNs, which are usually expressed in hemopoietic cells.
FIGURE 2. Ifnε is expressed in the female reproductive tract in both mice and humans.
(A) Mouse organs were harvested and Ifnε expression was measured by qRT-PCR, normalized to 18S RNA and presented relative to Ifnε expression in kidney. Data are expressed as the mean + SEM of at least three individual mice.. (B) Representative images showing Ifnε localization in uterine tissue (at oestrous stage) of WT and Ifnε−/− C57BL/6 mice by immunohistochemistry. Scale bar = 50µm. This is representative of at least five individual mice. (C, D) Ifnε expression was measured by qRT-PCR in mouse uterus at different stages of (C) estrous cycle and (D) pregnancy.. Data are expressed as mean + SEM of at least three separate experiments. (E) Ifnε expression was determined by qRT-PCR in ovariectomized (OVX) mice and OVX mice treated with estrogen (E OVX). Data are expressed as mean + SEM of at least six individual mice and are representative of at least two separate experiments. (F) A cDNA panel of human tissues was examined for IFNε expression by qRT-PCR and the results were expressed relative to IFNε expression in kidney. (G) Epithelial cells were isolated from endometrial samples of post-menopausal women or those at different stages of the menstrual cycle and IFNε expression was measured by qRT-PCR; values are presented relative to IFNε expression in human endometrial cell lines, ECC-1. Data are expressed as mean + SEM of six individual patient samples. *P<0.05, **P<0.01, ***P<0.001 (unpaired Student’s t-test).
Ifnε expression was found to vary approximately 30-fold at different stages of the estrous cycle, with lowest levels during diestrus and highest at estrus (Fig. 2C). During pregnancy, uterine Ifnε expression was dramatically reduced at day 1.5 post coitus (p.c.) and lowest at day 4.5, coincident with the time of embryo implantation (Fig. 2D). Ifnε expression was also reduced in pseudo-pregnant mice 4.5 days p.c. after mating with vasectomised males (Fig. 2D), which suggests that maternal hormones, not the embryo or its products, were required for the reduction in Ifnε. In addition, there was a slight increase in expression of Ifnε (1.8–1.9-fold) 8h p.c., which had returned to normal levels by 16h, showing that neither seminal fluid nor sperm directly suppress Ifnε expression (fig. S1G). Because changes in expression occur after mating with vasectomised or intact males, they are likely to be secondary to the physiological and hormonal changes, which are known to be comparable at day 4.5 p.c. whether or not conception occurs. Together these data are consistent with Ifnε expression being hormonally regulated. To evaluate this, we then ovariectomized female mice, and administered ovarian sex steroid hormones. Estrogen administration induced Ifnε expression over 6-fold (Fig. 2E). Such hormonal regulation was not observed for Ifnα or β expression (10).
Expression analysis of a panel of tissues confirmed the lack of basal expression of IFNε in all organs in women with the exception of endometrium (Fig. 2F). In order to determine whether human IFNε was also regulated in different hormonal states, we tested epithelial cells isolated from uterine endometrium from six women in secretory or proliferative stages of the menstrual cycle or post-menopause. IFNε expression was highest in the proliferative phase when estrogen levels are high and was approximately 10-fold lower in the secretory phase when estrogen levels are low and progesterone is high. IFNε levels were virtually undetectable in samples from post-menopausal women (Fig. 2G) (11). Consistent with the epithelial cell origin of this cytokine, several endometrial cancer-derived cell lines were shown to express IFNε (fig S1H).
We next generated Ifnε−/− mice to characterize its pathophysiological functions (fig. S2 A–E) (Table S1). Male and female fertility was normal (fig. S3A) as were the reproductive organs from male and female mice (fig. S3B) and immune organs characterized by immunophenotyping (fig. S3C–H).
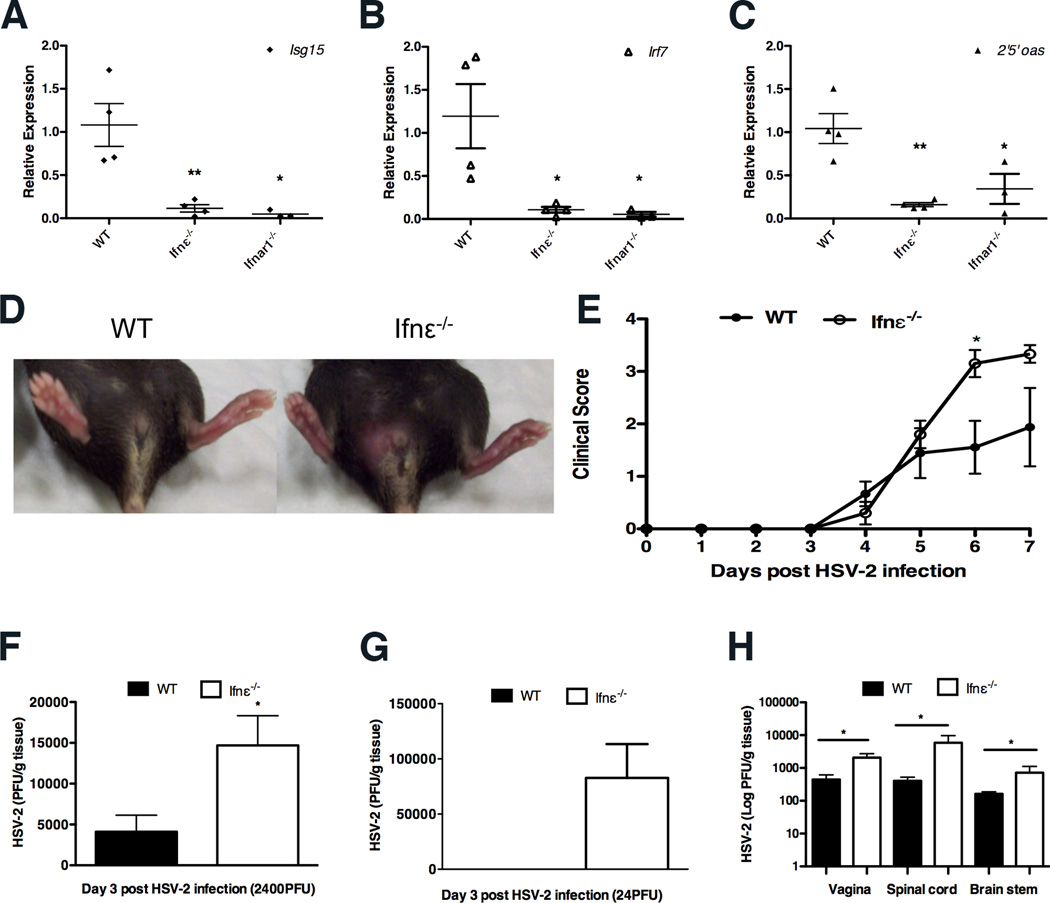
The basal levels of 2’5’oas, Irf7, and Isg15 were significantly reduced in uteri from Ifnε−/− mice, similar to the very low levels observed in Ifnar1−/− mice (Fig. 3A) indicating that Ifnε did signal in vivo. IRG levels in other organs were the same between WT and Ifnε−/− mice (fig. S3I). Furthermore, this difference in IRG levels resulting from constitutive Ifnε expression was similar in magnitude to the induction of these IRGs in wild type mice administered intravaginal Ifns α, β or ε (fig. S4), and to the degree of altered expression observed after Chlamydia or HSV-2 infection (see below). These data demonstrate that expression of IFNε in the FRT is required for maintaining basal levels of IRGs, have important in innate immunity.
FIGURE 3. Ifnε−/− mice are more susceptible to HSV-2 vaginal infection.
(A) Isg15, Irf7 and 2’5’ oas expression between WT and Ifnε−/− C57BL/6 mice was determined by qRT-PCR. The values represent means + SEM of four individual mice (B–C, E) Mice pretreated with Depo-ralovera at day -5 were infected with HSV-2 (B, E) 2400 PFU/mouse or (C) 24 PFU/mouse on day 0. (B) Representative images demonstrating overt genital lesions, redness and swelling in HSV-2 infected Ifnε−/− mice at day 7 p.i., but absent in C57BL/6 WT mice. Clinical scores of WT and Ifnε−/− C57BL/6 mice during the 7 day course of infection. Data are means + SEM of 5 individual mice and are representative of at least three separate experiments. (C–D) HSV-2 titres (PFU) from vaginal tissue of WT and Ifnε−/− C57BL/6 mice infected with (C) 2400 and (D) 24 pfu, respectively at day 3 p.i. were determined by titration of clarified vaginal tissue samples on Vero cell monolayers by plaque assay. Data are expressed as mean + SEM of five individual mice. E) HSV-2 titres from homogenates of vaginal tissue, spinal cord and brain stem of infected WT and Ifnε−/− C57BL/6 mice at day 7 p.i. were determined as in (B). Data are expressed as mean + SEM of five individual mice. *P<0.05 (unpaired Student’s t-test).
To determine whether Ifnε is important in protecting the FRT from viral infection, we examined the effect of genital HSV-2 infection in Ifnε−/− mice. Following a sublethal dose of a clinical isolate of HSV-2 strain 186 (12), Ifnε−/− mice had significantly more severe clinical scores of disease (day 6 and 7 post infection [p.i.]) with severe epidermal lesions evident compared to WT mice (Fig. 3B). These effects were observed at virus doses of 24 and 2400 pfu/mouse (Fig. 3C and D), and were consistent with elevated viral titres in infected vaginal tissues of Ifnε−/− mice at day 3 p.i., compared with WT animals. At the low dose of 24pfu, Ifnε was protective as virus was only detectable in the null mice and not wild type. In addition, Ifnε−/− mice had significantly higher viral titres in the spinal cord and brain stem 7 days post infection, consistent with either increased replication or retrograde transport of virus (Fig. 3E). Notably, there was no significant change in the expression of Ifnε in the first three days following viral infection, consistent with our in vitro data that this gene is not pathogen induced (fig. S5A). The susceptibility of Ifnε−/− was less than that of Ifnar1−/− mice which cannot respond to Ifns α, β nor ε (fig. S5B). However, since Ifnβ and IRGs were not induced less in Ifnε−/− mice, the protective effects of Ifnε in this model of a prevalent STI were independent of other type I IFNs (fig. S5C–F).
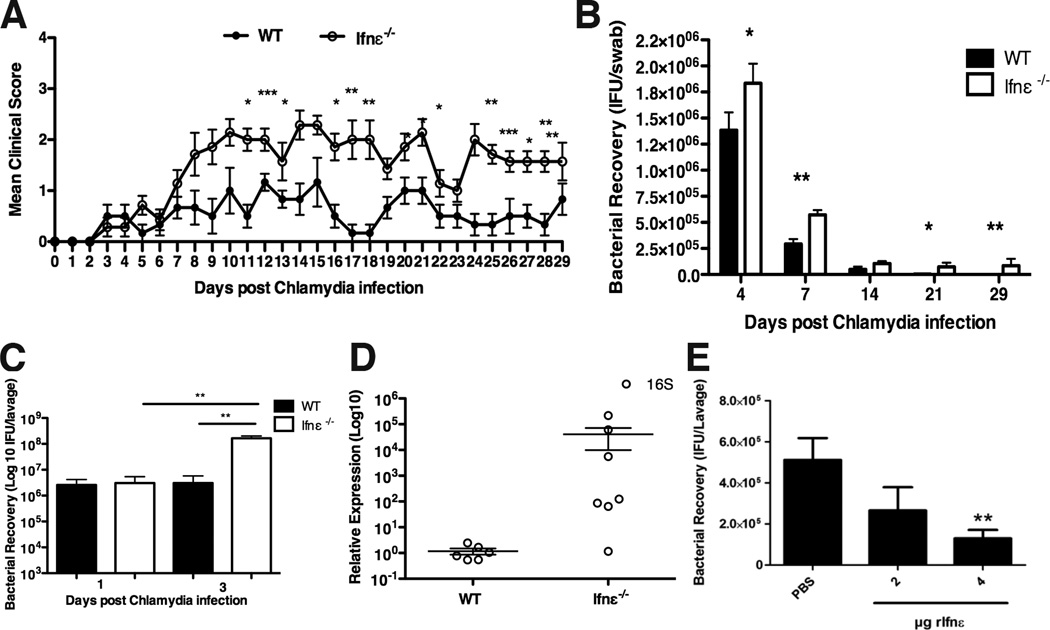
We next investigated the role of Ifnε in a murine model of FRT infection by Chlamydia -the most prevalent bacterial STI (13, 14). Following a sublethal, intravaginal infection of WT and Ifnε−/− mice with C. muridarum (15), Ifnε−/− mice displayed more severe clinical signs of disease from 7 until 30 days p.i. (Fig. 4A). More bacteria were detected in vaginal swabs of Ifnε−/− mice throughout the course of infection (Fig. 4B). C. muridarum recovery from vaginal lavage 3 days p.i. in WT mice had not increased from day 1 inoculum levels, but there was a 40-fold increase in the levels of bacteria in Ifnε−/− mice (Fig. 4C). We also observed significantly increased levels of Chlamydia at 30 days p.i., indicative of increased chlamydial growth in the upper FRT (uterine horns) of Ifnε−/− mice compared to very low levels in WT mice (Fig. 4D). This finding in particular indicates that Ifnε−/− mice are substantially more susceptible to (and less able to clear), an ascending infection in the FRT than WT mice. Because NK cells have a protective role against this infection (16), we measured their levels at 3 days p.i. Notably, both the percentage and total numbers of these cells were decreased in the uteri of Ifnε−/− mice (fig. S6A and B). Importantly, there were no changes in Ifnε RNA expression at the early or late in the infection (fig. S6C), consistent with our in vitro data that Ifnε is not regulated by PRR pathways. Furthermore, production of Ifnβ and IRGs was higher than the levels in wild type mice (fig. S7A–D), indicating that the protective effects of Ifnε were not solely due to priming for the production of other type I IFNs. To demonstrate that Ifnε could directly mediate protection against infection, we observed a dose-dependent reduction in bacteria (Fig. 4E), demonstrating that “reconstitution” of (progesterone) lowered Ifnε levels protected against this bacterial infection.
FIGURE 4. Ifnε−/− mice are more susceptible to Chlamydia muridarum vaginal infection.
(A–D) Mice were pretreated with progesterone at day -7 and infected intra-vaginally with 5×104 IFU C. muridarum. (A) Clinical scores were recorded daily for 30 days. Data are means + SEM of at least six individual mice. (B) Bacterial recovery from vaginal swabs of WT and Ifnε−/− C57BL/6 mice at different time points, determined by qRT-PCR for bacterial MOMP. Data are means + SEM of at least six individual mice. (C) Bacterial recovery, measured by qRT-PCR from vaginal lavage at day 1 and 3 p.i. Data are means + SEM of at least six individual mice. (D) Bacterial 16S RNA from the uterine horns of WT and Ifnε−/− C57BL/6 mice at 30 days p.i. was examined by qRT-PCR. Data are means ± SEM of at least six individual mice (E) WT C57BL/6 mice were pretreated with progesterone at day -7 and treated intra-vaginally with rIfnε (2 or 4 µg) 6h prior to C. muridarum infection. Bacterial recovery from the vaginal lavage at day 3 p.i. was measured by qRT-PCR. Data are means + SEM of at least six individual mice. *P<0.05, **P<0.01, ***P<0.001 (unpaired Student’s t-test).
The distinct properties of IFNε compared to other type I IFNs (table S2) make it the only one that protects against Chlamydia, whereas others exacerbate disease (17–20). All type I IFNs protect against HSV2 infection (21, 22), with IFNε likely contributing because its constitutive expression by epithelial cells afford it immediate efficacy at the site of first contact of mucosal pathogens. Interestingly, the increased susceptibility to FRT infections of women on progestagen-containing contraception (23, 24) may be explained by the lowering of Ifnε levels (fig. S8A) progestin pretreatment that is required for all FRT infection models (25, 26). The local effect of IFNε is supported by our observation that IFNε makes no difference in a systemic model (fig. S8B–D). Consistent with the importance of IFNε in FRT immunity, it is evolutionarily conserved in eutherian mammals, particularly in residues predicted to contact the two receptor components (fig. S9) (27). Since STIs are major global health and socioeconomic problems, the distinctive regulatory and protective properties of this new IFNε may facilitate the development of new strategies for preventing and treating STIs, and perhaps other diseases.
Supplementary Material
Acknowledgements
The authors wish to acknowledge the contributions of A. Mansell, R. Ferrero and L. Salamonsen, N. Burke and S. Forster for helpful discussions and reading of the manuscript, K. Fitzgerald for reagents and C. Berry for assistance with viral plaque assays. The data presented in this paper are tabulated in the main paper and the supplementary materials. This work was supported by funding from Australian National Health and Medical Research Council (PJH, NEM, PMH, JR), the Australian Research Council (PJH, NEM, JR) and the Victorian Government's Operational Infrastructure Support Program.
Footnotes
Additional data and Materials and Methods and associated additional references (28–33) can be found in the Supporting Online Material
REFERENCES
- 1.Hervas-Stubbs S, et al. Clin Cancer Res. 2011 May 1;17:2619. doi: 10.1158/1078-0432.CCR-10-1114. [DOI] [PubMed] [Google Scholar]
- 2.Isaacs A, Lindenmann J. Proc R Soc Lond B Biol Sci. 1957 Sep 12;147:258. [PubMed] [Google Scholar]
- 3.Hardy MP, Owczarek CM, Jermiin LS, Ejdeback M, Hertzog PJ. Genomics. 2004 Aug;84:331. doi: 10.1016/j.ygeno.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Honda K, Takaoka A, Taniguchi T. Immunity. 2006 Sep;25:349. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Sato M, et al. Immunity. 2000 Oct;13:539. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 6.de Weerd NA, Samarajiwa SA, Hertzog PJ. J Biol Chem. 2007 Jul 13;282:20053. doi: 10.1074/jbc.R700006200. [DOI] [PubMed] [Google Scholar]
- 7.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Nature. 2001 Oct 18;413:732. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 8.Doyle SE, et al. J Immunol. 2003 Apr 1;170:3565. doi: 10.4049/jimmunol.170.7.3565. [DOI] [PubMed] [Google Scholar]
- 9.Hornung V, Latz E. Nat Rev Immunol. 2010 Feb;10:123. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 10.Patel MV, Ghosh M, Fahey JV, Wira CR. PLoS One. 2012;7:e35654. doi: 10.1371/journal.pone.0035654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salamonsen LA. In: The endometrium. Aplin ATF John D, Glasser Stanley R, Giudice Linda C., editors. United Kingdom: informa healthcare; 2008. pp. 25–45. [Google Scholar]
- 12.Thapa M, Carr DJ. J Virol. 2009 Sep;83:9486. doi: 10.1128/JVI.00854-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beagley KW, Huston WM, Hansbro PM, Timms P. Crit Rev Immunol. 2009;29:275. doi: 10.1615/critrevimmunol.v29.i4.10. [DOI] [PubMed] [Google Scholar]
- 14.WHO. Geneva: 2001. [Google Scholar]
- 15.Asquith KL, et al. PLoS Pathog. 2011 May;7:e1001339. doi: 10.1371/journal.ppat.1001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tseng CT, Rank RG. Infect Immun. 1998 Dec;66:5867. doi: 10.1128/iai.66.12.5867-5875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devitt A, Lund PA, Morris AG, Pearce JH. Infect Immun. 1996 Oct;64:3951. doi: 10.1128/iai.64.10.3951-3956.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lad SP, Fukuda EY, Li J, de la Maza LM, Li E. J Immunol. 2005 Jun 1;174:7186. doi: 10.4049/jimmunol.174.11.7186. [DOI] [PubMed] [Google Scholar]
- 19.Nagarajan UM, Ojcius DM, Stahl L, Rank RG, Darville T. J Immunol. 2005 Jul 1;175:450. doi: 10.4049/jimmunol.175.1.450. [DOI] [PubMed] [Google Scholar]
- 20.Nagarajan UM, et al. Infect Immun. 2008 Oct;76:4642. doi: 10.1128/IAI.00629-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Austin BA, James CM, Harle P, Carr DJ. Biol Proced Online. 2006;8:55. doi: 10.1251/bpo118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conrady CD, Halford WP, Carr DJ. J Virol. 2011 Feb;85:1625. doi: 10.1128/JVI.01715-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baeten JM, et al. Am J Obstet Gynecol. 2001 Aug;185:380. doi: 10.1067/mob.2001.115862. [DOI] [PubMed] [Google Scholar]
- 24.Wang CC, Reilly M, Kreiss JK. J Acquir Immune Defic Syndr. 1999 May 1;21:51. doi: 10.1097/00126334-199905010-00007. [DOI] [PubMed] [Google Scholar]
- 25.Morrison RP, Caldwell HD. Infect Immun. 2002 Jun;70:2741. doi: 10.1128/IAI.70.6.2741-2751.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parr MB, et al. Lab Invest. 1994 Mar;70:369. [PubMed] [Google Scholar]
- 27.Thomas C, et al. Cell. 2011 Aug 19;146:621. [Google Scholar]
- 28.Robertson SA, Mayrhofer G, Seamark RF. Biol Reprod. 1996 Jan;54:183. doi: 10.1095/biolreprod54.1.183. [DOI] [PubMed] [Google Scholar]
- 29.Hertzog PJ. Methods in Molecular Biology. In: Martin IK, Tymms J, editors. Gene Knockout Protocols. vol. 158. Australia: 2001. [Google Scholar]
- 30.Gargett CE, Schwab KE, Zillwood RM, Nguyen HP, Wu D. Biol Reprod. 2009 Jun;80:1136. doi: 10.1095/biolreprod.108.075226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenhill CJ, et al. Immunol Cell Biol. 2012 May;90:559. doi: 10.1038/icb.2011.56. [DOI] [PubMed] [Google Scholar]
- 32.Bidwell BN, et al. Nat Med. 2012 Jul 22; [Google Scholar]
- 33.Byers SL, Wiles MV, Dunn SL, Taft RA. PLoS One. 2012;7:e35538. doi: 10.1371/journal.pone.0035538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.