Abstract
Histone tail acetylation and methylation are known to enhance accessibility of islet genes to transcription factors and the basal transcriptional machinery. In this brief report, we follow up on a recent study in which we identified the islet enriched factor Set7/9 as a potentially important histone methyltransferase in β cells (Deering, et al. Diabetes 2009; 58:185-93). We had suggested that the methylation of H3-Lys4 by Set7/9 enhances accessibility of the insulin gene to the basal transcriptional machinery. Consistent with this hypothesis, we show here that RNA polymerase II occupancy at the insulin and IAPP genes is considerably enhanced in β cells compared to α cells (or NIH3T3 cells), and that the converse is true for RNA polymerase II occupancy at the glucagon gene. The enrichment of Set7/9 in β cells appears to be dependent upon Pdx1, as knockdown of Pdx1 in INS-1 β cells using small hairpin RNAs almost completely abolishes Set7/9 expression. A LacZ expression vector driven by the -6.5 kilobase pair Set7/9 promoter that contains putative Pdx1 binding sites shows β cell-line-specific expression. Taken together, our data support further the hypothesis that Pdx1-dependent Set7/9 expression may be crucial to enhancing chromatin accessibility and transcription of β cell genes.
Results and Discussion
The expression of the insulin gene is a feature unique to pancreatic β cells, and many recent studies suggest that this property arises in part from the unique modifications of histone proteins surrounding the gene. In prior studies, we demonstrated that histone H3-Lys4 di-methylation and tri-methylation were strongly enriched at the insulin gene in βTC3 β cells, but not in non-β cell lines 1,2. Importantly, tri-methylation of H3-Lys4 is associated with actively transcribed loci and is typically found in coding regions of genes, whereas di-methylation is a marker of open (euchromatic) loci and is typically enriched in promoter regions of genes 3-6. To determine both the potential for active transcription and the openness of chromatin structure at the insulin gene, we assessed the occupancy of RNA polymerase II in different cell lines by chromatin immunoprecipitation using an antibody that recognizes all forms (phosphorylated and unphosphorylated) of RNA polymerase II (Covance). This antibody was selected so as to pick up both the initiation (i.e. promoter-bound) and elongation (coding region-bound) isoforms of RNA polymerase II. We found that RNA polymerase II occupied both the promoter and coding regions of the insulin gene in βTC3 cells, but not αTC1.6 α cells or NIH3T3 fibroblasts (Fig. 1). Conversely, RNA polymerase II occupancy was observed at the promoter and coding regions of the glucagon gene in αTC1.6 cells, but not βTC3 or NIH3T3 cells (Fig. 1). These studies demonstrate that the occupancy of islet genes by RNA polymerase II correlates directly to both their expression level and histone H3-Lys4 methylation status.
Figure 1. RNA polymerase II occupancy in islet and non-islet cell lines.
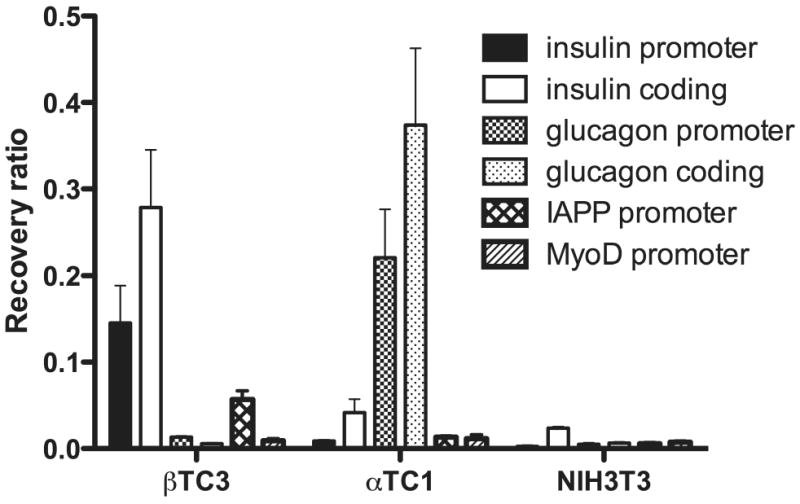
RNA polymerase II occupancy was evaluated in chromatin immunoprecipitation experiments. After fixation with 1% formaldehyde, βTC3, αTC1.6 and NIH3T3 cells were isolated and sonicated as described previously 13. DNA/protein complexes were immunoprecipitated by anti-RNA polymerase II antibody (C-terminal domain, Covance). Purified DNA was subjected to quantitative real-time PCR using primers to the indicated genes. Primer sequences and PCR conditions were described previously 13. RNA polymerase II occupancy is displayed as percent recovery of the indicated DNA fragment relative to input DNA.
In a recent studies 2,7, we showed that the islet-enriched histone methyltransferase Set7/9 may be the catalyst responsible for H3-Lys4 di-methylation at the insulin gene in β cells. Set7/9 has no sequence-specific DNA binding domain, and its actions at specific genetic loci are presumably dependent upon recruitment by sequence-specific transcription factors. Therefore, Set7/9 would be considered a transcriptional cofactor as opposed to a transcription factor. In glutathione S-transferase (GST) pull-down studies in vitro, we demonstrated direct and specific interactions between the respective N-termini of Pdx1 and Set7/9 2, a finding suggesting that this key islet β–cell enriched transcriptional activator serves as the “recruiter” for Set7/9. To determine if the two proteins actually interact in β cells, we performed co-immunoprecipitation studies in INS-1 β cells. As shown in Fig. 2A, Pdx1 and Set7/9 can be co-immunoprecipitated from an INS-1 extract, verifying that the two proteins form a complex. Importantly, Pdx1 is known to interact with a host of transcriptional cofactors, including p300/CBP, Bridge1, PCIF1, and histone deacetylases (HDACs) 8-12. In this setting, however, Set7/9 represents a very different type of interacting cofactor: unlike the Bridge1 and the ubiquitously-expressed acetyltransferase p300, it functions as a histone methyltransferase and displays islet-specificity within the pancreas, and unlike the co-repressors PCIF1 or HDACs, it augments Pdx1 action at target genes.
Figure 2. Pdx-1 forms a complex with Set7/9 and is required for Set7/9 expression in INS-1 cells.
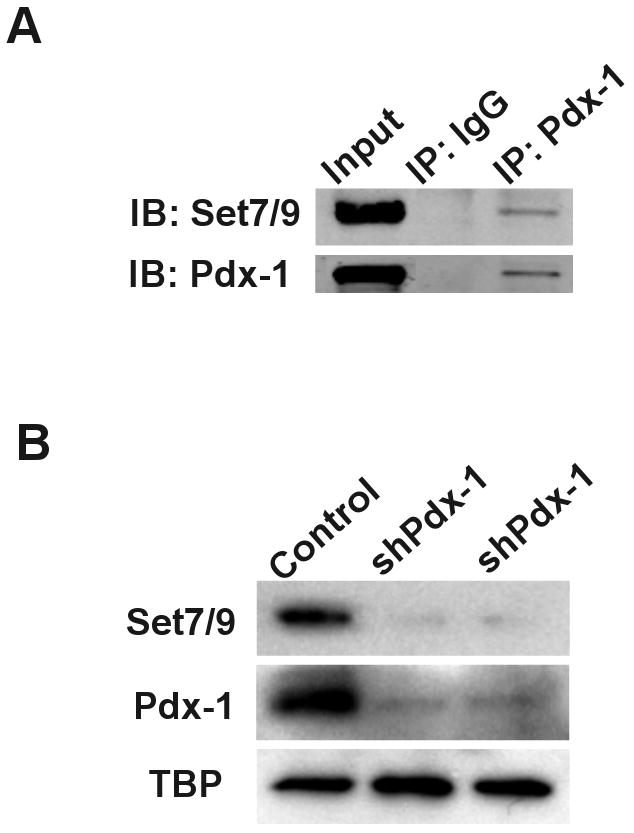
A, Protein extract from INS-1 cells was immunoprecipitated using control rabbit IgG or rabbit anti-Pdx-1 antibody (Millipore) as previously described 2, and subjected to immunoblot using mouse anti-Pdx-1 antibody (Developmental Studies Hybridoma Bank) or mouse anti-Set7/9 antibody (Abcam). Immunoblots were visualized using the Odyssey system (LI-COR Biosciences); B, INS-1 cells were treated with either a control adenovirus or an adenovirus encoding a small hairpin RNA against Pdx1 14 for 48 hours at a multiplicity of infection of 450 pfu/cell. Protein extract was then subjected to immunoblotting for the proteins shown (TBP, TATA binding protein). Representative samples from two independent shRNA infections are shown.
In primary mouse tissues, we showed that Set7/9 exhibits a restricted expression pattern that includes brain, muscle, liver, and pancreatic islets, a pattern consistent with regulation at the transcriptional level 7. We identified a highly conserved β cell-specific gene enhancer between base pairs −5768 and −6030 of the Set7/9 promoter that appeared to be dependent upon Pdx1 for activation 7. The dependency of Set7/9 expression on Pdx1 was further examined in Fig. 2B, where short hairpin RNA-mediated knockdown in INS-1 cells resulted in a rapid and near-complete loss of Set7/9 protein. To show that the promoter fragment containing this Pdx1-dependent enhancer exhibited β cell-specific expression, we constructed a LacZ reporter plasmid driven by the −6.5 kilo-base pair Set7/9 promoter (Fig. 3A). As shown in Fig. 3B and 3C, β-galactosidase expression was observed only in βTC3 cells, and not NIH3T3 cells (by immunoblot, Fig. 3B, and by immunocytochemistry, Fig. 3C). These data further support that enrichment of Set7/9 in islet β cells is mediated by transcriptional enhancer elements driven by Pdx1.
Figure 3. -6.5 kilobase pair Set7/9 promoter is sufficient for β cell-line-specific transcription.
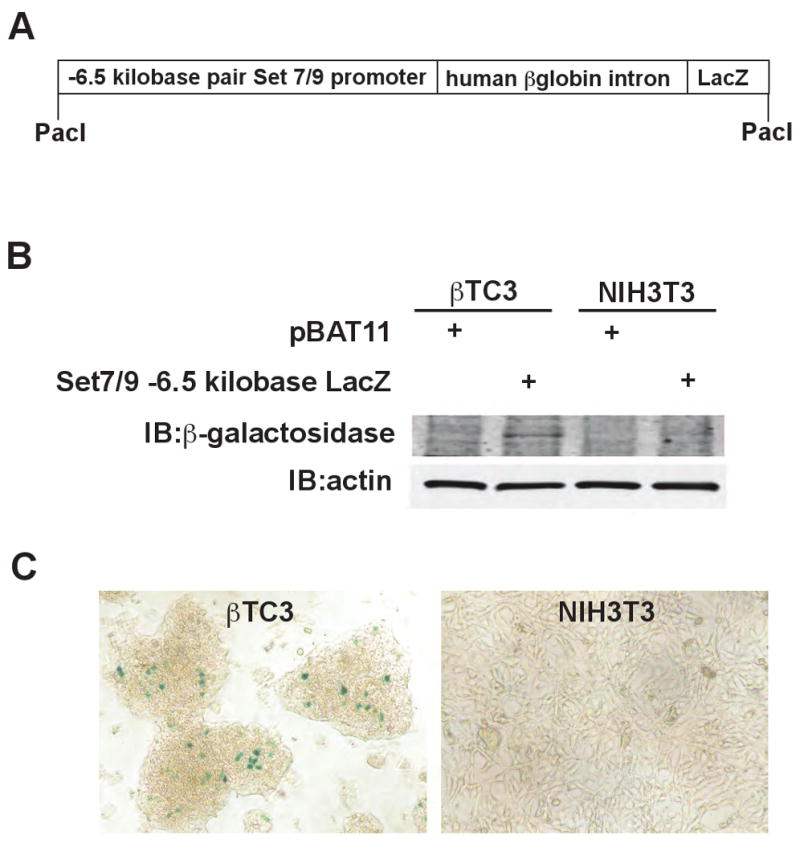
A, schematic representation of the Set7/9 promoter (-6584 bp to +316 bp)-LacZ expression construct (in vector pBAT11); B, the expression construct in A or pBAT11 (control vector) was transfected into βTC3 and NIH3T3 cells using Metafectene Pro (Biontex) according to manufacturer's protocol. Total cellular protein was isolated and subjected to immunoblotting using anti-β-galactosidase (Abcam) and anti-actin (clone C4, MP Biomedicals) antibodies, and visualized with the Odyssey system (LICOR Biosciences); C, enzymatic staining for β-galactosidase was performed on βTC3 and NIH3T3 cells transfected with the expression construct shown in A according to a protocol by Open Wet Ware (http://openwetware.org). Blue staining indicates positive β-galactosidase activity.
Taken together, our data support the hypothesis that Set7/9 is a chromatin-modifying cofactor that functions as an effector of Pdx1 action in the developing and mature islet. The model in Fig. 4 proposes that the action of Set7/9 is closely linked to Pdx1, such that Pdx1 activates Set7/9 expression and subsequently recruits it to specific target genes. Set7/9 then effects histone H3-Lys4 methylation, upon which nucleosomes are “remodeled” resulting in greater gene accessibility to the RNA polymerase II transcriptional machinery. We emphasize that although physical and functional interactions between Set7/9 and Pdx1 is supported by our preliminary data, not all actions of Pdx1 and Set7/9 will be linked and that further refinement of this model will undoubtedly occur.
Figure 4. Expression and function of Set7/9 in β cells.

We propose that Pdx1 participates in the β cell-specific activation of the Set7/9 gene in β cells (top). Set7/9 protein is then recruited to target genes (e.g. insulin) by Pdx1. Subsequent methylation of histone H3-Lys4 by Set7/9 leads to the remodeling of nucleosomes to create a more open platform that permits the recruitment of other transcription factors and the basal transcriptional machinery. Small circles represent nucleosomes (containing 146 bp of DNA wrapped around histone octamers). “Me”, represents methylated-H3-Lys4.
Footnotes
This work was supported by grants from the National Institutes of Health (R01 DK60581 to RGM; R01 DK50203 to RWS; 5T32 DK007061 to NLV) and by a Juvenile Diabetes Research Foundation Grant (to RGM)
References
- 1.Chakrabarti SK, Francis J, Ziesmann SM, Garmey JC, Mirmira RG. Covalent histone modifications underlie the developmental regulation of insulin gene transcription in pancreatic beta cells. J Biol Chem. 2003;278:23617–23. doi: 10.1074/jbc.M303423200. [DOI] [PubMed] [Google Scholar]
- 2.Francis J, Chakrabarti SK, Garmey JC, Mirmira RG. Pdx-1 links histone H3-Lys-4 methylation to RNA polymerase II Elongation during activation of insulin transcription. J Biol Chem. 2005;280:36244–53. doi: 10.1074/jbc.M505741200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–5. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 4.Noma K, Grewal SI. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16438–45. doi: 10.1073/pnas.182436399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 6.Musri MM, Corominola H, Casamitjana R, Gomis R, Parrizas M. Histone H3 lysine 4 dimethylation signals the transcriptional competence of the adiponectin promoter in preadipocytes. J Biol Chem. 2006;281:17180–8. doi: 10.1074/jbc.M601295200. [DOI] [PubMed] [Google Scholar]
- 7.Deering TG, Ogihara T, Trace AP, Maier B, Mirmira RG. Methyltransferase Set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes. 2009;58:185–93. doi: 10.2337/db08-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosley AL, Corbett JA, Ozcan S. Glucose regulation of insulin gene expression requires the recruitment of p300 by the beta-cell-specific transcription factor Pdx-1. Mol Endocrinol. 2004;18:2279–90. doi: 10.1210/me.2003-0463. [DOI] [PubMed] [Google Scholar]
- 9.Mosley AL, Ozcan S. The pancreatic duodenal homeobox-1 protein (Pdx-1) interacts with histone deacetylases Hdac-1 and Hdac-2 on low levels of glucose. J Biol Chem. 2004;279:54241–7. doi: 10.1074/jbc.M410379200. [DOI] [PubMed] [Google Scholar]
- 10.Volinic JL, Lee JH, Eto K, Kaur V, Thomas MK. Overexpression of the coactivator bridge-1 results in insulin deficiency and diabetes. Mol Endocrinol. 2006;20:167–82. doi: 10.1210/me.2005-0127. [DOI] [PubMed] [Google Scholar]
- 11.Liu A, Desai BM, Stoffers DA. Identification of PCIF1, a POZ domain protein that inhibits PDX-1 (MODY4) transcriptional activity. Mol Cell Biol. 2004;24:4372–83. doi: 10.1128/MCB.24.10.4372-4383.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu Y, Guo M, Huang S, Stein R. Insulin Gene Transcription Is Mediated by Interactions between the p300 Coactivator and PDX-1, BETA2, and E47. Mol Cell Biol. 2002;22:412–20. doi: 10.1128/MCB.22.2.412-420.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakrabarti SK, James JC, Mirmira RG. Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1. Importance of chromatin structure in directing promoter binding. J Biol Chem. 2002;277:13286–93. doi: 10.1074/jbc.M111857200. [DOI] [PubMed] [Google Scholar]
- 14.Iype T, Francis J, Garmey JC, Schisler JC, Nesher R, Weir GC, Becker TC, Newgard CB, Griffen SC, Mirmira RG. Mechanism of insulin gene regulation by the pancreatic transcription factor Pdx-1: application of pre-mRNA analysis and chromatin immunoprecipitation to assess formation of functional transcriptional complexes. J Biol Chem. 2005;280:16798–807. doi: 10.1074/jbc.M414381200. [DOI] [PubMed] [Google Scholar]


