Abstract
Herpes simplex virus type 1 (HSV-1) UL37 is a 1,123 amino acid tegument protein that self-associates and binds to the tegument protein UL36 (VP1/2). Studies were undertaken to identify regions of UL37 involved in these protein-protein interactions. Coimmunoprecipitation assays showed that residues within the carboxy-terminal half of UL37, amino acids 568–1123, are important for interaction with UL36. Coimmunoprecipitation assays also revealed that amino acids 1–300 and 568–1123 of UL37 are capable of self-association. UL37 appears to self-associate only under conditions when UL36 is not present or is present in low amounts, suggesting UL36 and UL37 may compete for binding. Transfection-infection experiments were performed to identify domains of UL37 that complement the UL37 deletion virus, KΔUL37. The carboxy terminal region of UL37 (residues 568–1123) partially rescues the KΔUL37 infection. These results suggest the C-terminus of UL37 may contribute to its essential functional role within the virus-infected cell.
Keywords: UL37, UL36, VP1/2, herpes, tegument, HSV-1, assembly, herpesvirus
INTRODUCTION
Herpesvirus virions are composed of three morphologically distinct structures: the capsid, the tegument and the envelope (reviewed in Roizman et al., 2007). The 150 kbp linear DNA genome is enclosed within the icosahedral capsid. The virion is bounded by a host-derived lipid envelope that contains approximately 12 virus-encoded glycoproteins (reviewed in Roizman et al., 2007). The proteinaceous region between the capsid and envelope is termed the tegument. Approximately 25 virus-encoded proteins, as well as trace amounts of host proteins, make up the tegument region (reviewed in Kelly et al., 2009; Mettenleiter et al., 2009).
During herpesvirus replication, the viral genome is packaged into assembled capsids within the nucleus of the infected cell. The capsid acquires an envelope by budding into the inner nuclear membrane and enters the perinuclear space. The capsid loses its primary envelope as it subsequently fuses with the outer nuclear membrane and the capsid is released into the cytoplasm (reviewed in Mettenleiter et al., 2009). It has also been reported that capsids may escape the nucleus by exiting through damaged nuclear pores (Leuzinger et al., 2005; Wild et al., 2005). Detectable quantities of HSV-1 tegument proteins UL36 (VP1/2) and UL37 (Bucks et al., 2007), as well as vhs (Read & Patterson, 2007), have been reported on capsids isolated from the nucleus. However, other groups failed to detect HSV-1 UL36 and UL37 associated with detergent-treated capsids isolated from the nucleus of infected cells (Newcomb & Brown, 2010, Radtke et al., 2010, Trus et al., 2007, Wolfstein et al., 2006). Other tegument proteins including HSV-1 VP16, HSV-1 VP22 and HSV and pseudorabies virus (PRV) US3 proteins have been shown to associate with capsids within the perinuclear space (Granzow et al., 2004; Naldinho-Souto et al., 2006; Padula et al., 2009; Reynolds et al., 2002) suggesting the earliest events in tegumentation occur within these sites. After exiting the nucleus the cytoplasmic capsids travel through the secretory pathway. It has been suggested that some tegument proteins, such as HSV-1 UL16, interact with the capsid as it travels in the cytoplasm on its journey toward the trans-Golgi network (TGN) (Meckes, Jr. & Wills, 2007). The capsid obtains a lipid envelope by budding into vesicles derived from the TGN. Many, if not all, tegument proteins are associated with TGN-derived vesicles through membrane binding domains or through interactions with membrane bound glycoproteins and/or membrane bound tegument proteins. The TGN-membrane-associated tegument proteins and glycoproteins are incorporated into the virion during final envelopment (Mettenleiter et al., 2009).
The morphogenesis and structure of the tegument appear to be complex, with many details only recently being uncovered. With the exception of the innermost layers, the tegument region is described as asymmetrical in nature (Grunewald et al., 2003; Zhou et al., 1999). Recent evidence suggests the tegument region may not be a static structure, but instead may undergo a time-dependent maturation process after release of the virion from the infected cell (Newcomb & Brown, 2009). The maturation of the virion results in changes in tegument structure including loss of protein symmetry surrounding the capsid and increased resistance to detergent extraction (Newcomb & Brown, 2009). There is also evidence that structural changes in the tegument occur upon binding of the virion to the host cellular receptor heparan (Meckes, Jr. & Wills, 2008). Virion binding to the heparan receptor causes the disassociation of at least one tegument protein, UL16, from the capsid (Meckes, Jr. & Wills, 2008).
A myriad of protein-protein interactions among tegument proteins has been described, and these interactions likely play important roles in facilitating the incorporation of tegument proteins into the assembling virion. Two proteins of interest in this study are tegument proteins encoded by the UL36 and UL37 genes. UL36, also known as VP1/2, binds UL37 (Klupp et al., 2002). UL36 is the largest known herpesvirus-encoded protein and the HSV-1-encoded UL36 protein is approximately 270 kDa (Heine et al., 1974; Honess & Roizman, 1973; Spear & Roizman, 1972). HSV-1 UL36 interacts with capsid proteins VP5 and UL25 (Coller et al., 2007; McNabb & Courtney, 1992; Pasdeloup et al., 2009), and is bound tightly to the capsid (Gibson & Roizman, 1972; Spear & Roizman, 1972). UL36, and its homologues in all herpesvirus subfamilies, functions as a deubiquitinating enzyme (Kattenhorn et al., 2005; Schlieker et al., 2005). The conserved UL36 tegument protein is important for productive infection, playing roles at early times, as well as late times during infection. During translocation of incoming capsids to the nucleus, UL36 associated with the incoming HSV-1 capsid is cleaved, resulting in changes to the capsid that allow the viral genome to be released into the nucleus (Jovasevic et al., 2008). Studies of HSV-1 and PRV UL36 deletion viruses indicate that in the absence of UL36, DNA filled capsids accumulate in the cytoplasm, are not associated with appreciable amounts of tegument and do not undergo secondary envelopment (Desai, 2000; Fuchs et al., 2004; Luxton et al., 2006; Roberts et al., 2009). Furthermore, Luxton and colleagues reported that a PRV UL36 deletion virus showed reduced microtubule-based transport of capsids in the cytoplasm (Luxton et al., 2006).
UL37 encodes a conserved 120 kDa phosphorylated tegument protein (Albright & Jenkins, 1993; McLauchlan et al., 1994; Schmitz et al., 1995; Shelton et al., 1990). UL37 is distributed throughout the cell and contains a nuclear export signal (McLauchlan et al., 1994; McLauchlan, 1997; Schmitz et al., 1995; Watanabe et al., 2000). HSV-1 UL37 functions to activate NF-κB signaling by binding TRAF6 (Liu et al., 2008). The amount of HSV-1 UL37 packaged into the virion is tightly controlled, as increasing expression of the protein does not increase the abundance of UL37 in virions (McLauchlan, 1997).
During virus assembly UL37 may have a function in the cytoplasm as well as the nucleus. In cells infected with a mutant virus containing a deletion of HSV-1 UL37, KΔUL37, DNA-filled capsids accumulate in the nucleus (Desai et al., 2001); however, in cells infected with another HSV-1 UL37 deletion virus, ΔUL37[86–1035], an accumulation of nuclear capsids was not observed (Leege et al., 2009). Roberts and colleagues constructed a third HSV-1 UL37 deletion virus, FRΔUL37. Infection with FRΔUL37 did not result in an accumulation of capsids in the nucleus but yielded an accumulation of large aggregates of cytoplasmic capsids, similar to capsid aggregates also observed in KΔUL37-infected cells (Desai et al., 2001; Roberts et al., 2009). None of the HSV-1 UL37 deletion viruses yield productive infections (Desai et al., 2001; Leege et al., 2009; Roberts et al., 2009). PRV UL37 deletion viruses produced titers approximately 100–1,000-fold lower than wild type and showed no accumulation of capsids within the nucleus (Klupp et al., 2001; Luxton et al., 2006). Secondary envelopment is inhibited in HSV-1 and PRV UL37 deletion viruses, resulting in an accumulation of capsids in the cytoplasm (Desai et al., 2001; Klupp et al., 2001; Leege et al., 2009; Luxton et al., 2006; Roberts et al., 2009).
Coimmunoprecipitation and yeast two-hybrid interaction studies have revealed that HSV-1 UL37 may interact with several other viral proteins. In a yeast two-hybrid analysis, HSV-1 UL37 interacts with the capsid proteins VP26 and VP19c (Lee et al., 2008). UL37 also participates in interactions with other tegument proteins, including UL46 and UL36 (Fossum et al., 2009; Klupp et al., 2002; Lee et al., 2008; Uetz et al., 2006; Vittone et al., 2005). Furthermore, HSV-1 UL37 interacts with itself, as indicated by yeast two-hybrid analysis and coimmunoprecipitation studies (Lee et al., 2008; Vittone et al., 2005).
Based on its tight association with the capsid, it has been suggested that UL36 may be one of the first tegument proteins to associate with the capsid, and may act as a foundation to bind other tegument proteins to the capsid (Mettenleiter et al., 2009). Studies revealing the association of HSV-1 UL36 and UL37 with capsids isolated from the nuclear fraction also support this hypothesis (Bucks et al., 2007). The interaction of UL36 with the capsid may aid or enhance the incorporation of UL36 binding partners, such as UL37 or VP16 (Ko et al., 2010, Lee et al., 2008; Mettenleiter et al., 2009; Vittone et al., 2005). UL36 and UL37 physically interact, but the functional importance of the interaction is perhaps only beginning to be understood (Klupp et al., 2002). Desai and colleagues reported that the localization of HSV-1 UL37 in the Golgi complex is dependent on UL36 (Desai et al., 2008). Furthermore, based upon the work of Roberts and colleagues, HSV-1 UL36 and UL37 appear to rely on one another for incorporation into L particles (Roberts et al., 2009). Interestingly, the incorporation of UL37 and UL36 into L particles may be dependent on VP16, as L particles produced from a PRV VP16 deletion mutant, PRV-ΔUL48, do not contain these proteins (Fuchs et al., 2002). Consistent with this finding, HSV-1 UL36 has been reported to interact with VP16 in yeast two-hybrid and pulldown assays (Ko et al., 2010, Lee et al., 2008; Vittone et al., 2005).
Vittone and colleagues have determined that the region containing amino acids 512–767 of HSV-1 UL36 is necessary for binding UL37 (Vittone et al., 2005). Mijatov and colleagues extended these studies to identify residues F593 and E596 of HSV-1 UL36 as essential for the interaction (Mijatov et al., 2007). Furthermore, Fuchs and colleagues have reported that deletion of the UL37 binding domain of UL36 does not abolish PRV replication, suggesting that the essential function(s) of UL36 is not dependent on binding UL37 (Fuchs et al., 2004). The region of UL37 that participates in its interaction with UL36 as well as the region(s) of UL37 that participates in self-association have not been identified and are the focus of our report. These studies are important for several reasons. First, the interaction of UL36 and UL37 is conserved among the alpha-, beta-, and gammaherpesviruses (Bechtel & Shenk, 2002; Klupp et al., 2002; Lee et al., 2008; Rozen et al., 2008; Uetz et al., 2006; Vittone et al., 2005). Furthermore, UL36 plays an essential role in the assembly of HSV-1 and PRV (Desai, 2000; Fuchs et al., 2004; Luxton et al., 2006; Roberts et al., 2009). In addition, UL37 plays an essential role for assembly of HSV-1 and varicella zoster virus (VZV) and a critical role in the assembly of PRV (Desai et al., 2001; Klupp et al., 2001; Luxton et al., 2006; Pasdeloup et al., 2010, Roberts et al., 2009; Xia et al., 2003). A greater understanding of the interaction between UL36 and UL37 will help elucidate the role(s) that the UL36–UL37 interaction plays during assembly of herpesvirus virions. We have identified a region within the carboxy-terminal half of HSV-1 UL37 to be necessary for binding to UL36. Furthermore, we have determined that both carboxy and amino-terminal regions of UL37 are capable of self-association, and can interact with one another. In a trans-complementation assay, when plasmids encoding UL37 truncation mutants were transfected into cells followed by infection with a UL37 deletion virus, a mutant protein encompassing the carboxy-terminal half of UL37 was found to complement the infection with the UL37 deletion virus.
RESULTS
Identification of the region of UL37 involved in binding UL36
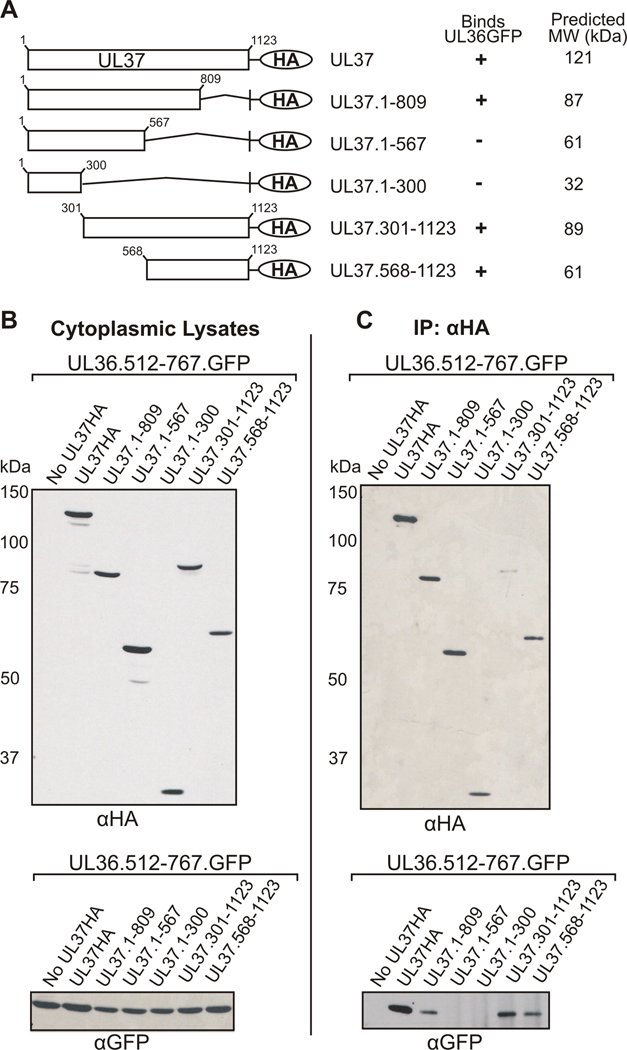
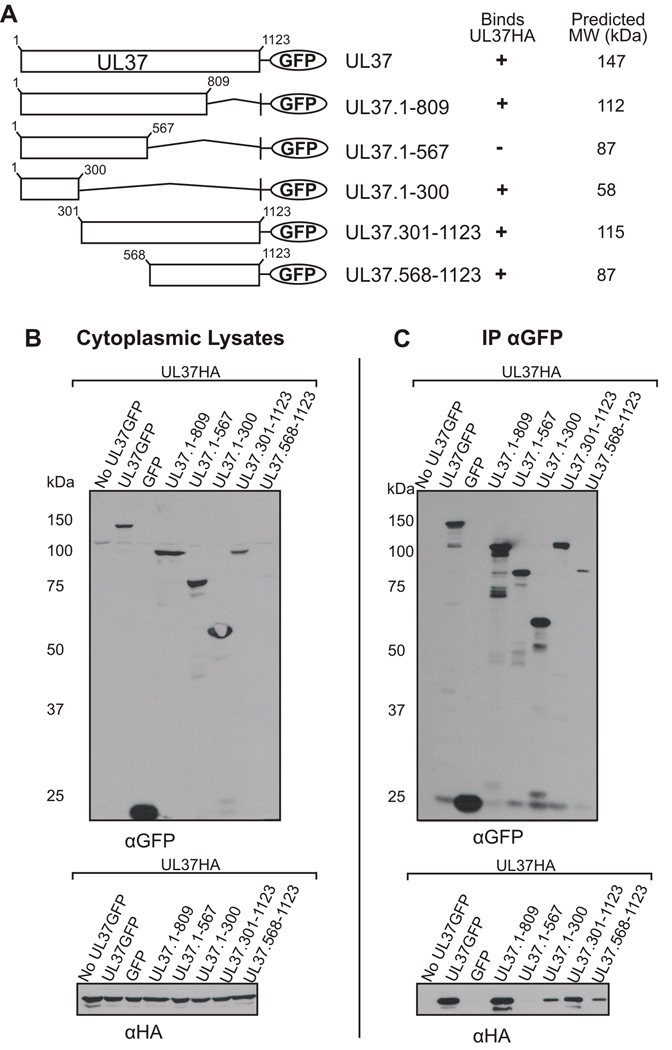
A series of hemagglutinin (HA) tagged UL37 truncation mutants was constructed and used in coimmunoprecipitation assays to identify the region of UL37 involved in binding UL36 (Fig. 1A). UL37 truncation mutants were designed to avoid major disruptions in secondary structure that may result from protein truncations in the middle of hydrophobic regions. A GFP expressing plasmid encoding the region of UL36 necessary for binding UL37, amino acids 512–767, was generated (UL36.512-767GFP). N-terminal and C-terminal truncation mutants of UL37, containing a hemagglutinin (HA) tag at the carboxy-terminus, were cotransfected into Vero cells with UL36.512-767GFP. Cells were lysed in NP40 lysis buffer and the nuclear fraction was discarded. The detergent lysis buffer presumably solubilizes cytoplasmic membranes, resulting in the disruption of cytoplasmic compartments and release of all cytoplasmic proteins to potentially interact in coimmunoprecipitation experiments. Anti-HA antiserum was used to immunoprecipitate the UL37HA mutant proteins. Immune complexes were captured by incubating the lysates with Protein G-agarose beads. SDS-PAGE and Western blotting were performed to verify expression of the cotransfected plasmids (Fig. 1B) and identify the UL37HA mutants capable of coimmunoprecipitating UL36.512-767GFP (Fig. 1C). As expected, full-length UL37HA coimmunoprecipitated UL36.512-767GFP. UL37 mutants encoding amino acids 1–300 or 1–567 of UL37 did not coimmunoprecipitate UL36. Extension of the amino-terminal mutants to include residues 1–809 restored binding to UL36. Furthermore, mutants UL37.568-1123HA and UL37.301-1123HA were also capable of binding UL36.512-767.GFP. These results suggest that residues 568–809 of UL37 are important for interaction with UL36.
Figure 1. Coimmunoprecipitation of UL36.512-767GFP with N-Terminal and C-Terminal UL37HA Truncation Mutants.
(A) A schematic representation of HA tagged full length, N-terminal and C-terminal UL37 truncation mutants. Additionally, a tabular summary of results indicate the UL37HA truncation mutants that bind (+) or do not bind (−) UL36.512-767GFP (UL36GFP) in coimmunoprecipitation assays. Molecular weight values were predicted by Statistical Analysis of Protein Sequence accessed through SDSC Biology Workbench. (B) Expression of UL37HA truncation mutants and UL36.512-767GFP. Vero cells were cotransfected with UL36.512-767GFP and the indicated UL37HA constructs, harvested at 24 h post-transfection and lysed in NP40 lysis buffer. Each cytoplasmic lysate was analyzed by Western blotting using rabbit HA antibody (αHA, top panel) or goat GFP antibody (αGFP, bottom panel). (C) Coimmunoprecipitation of UL36.512-767GFP with UL37 HA constructs. Cotransfected cytoplasmic lysates were incubated with an anti-HA rabbit antibody and then bound to protein-G agarose beads. Immunoprecipitated material was separated on an SDS-PAGE gel and transferred to nitrocellulose. Proteins were detected by Western blotting using a goat anti-HA antibody (αHA, top panel) or goat anti-GFP antibody (αGFP, bottom panel).
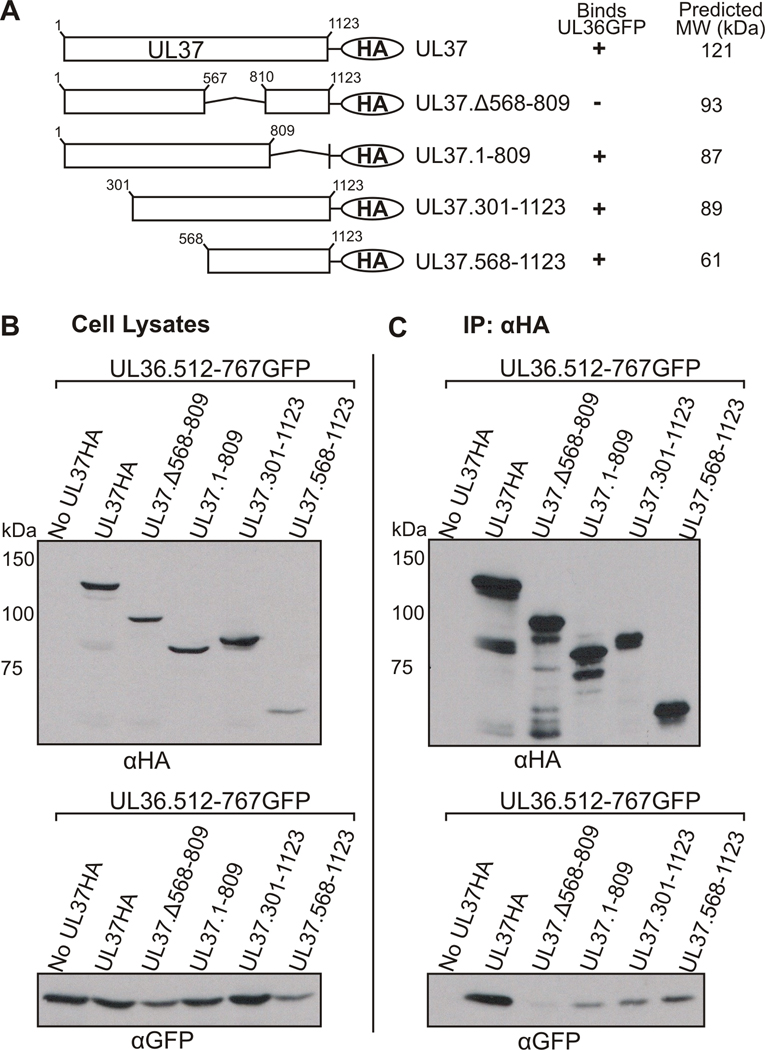
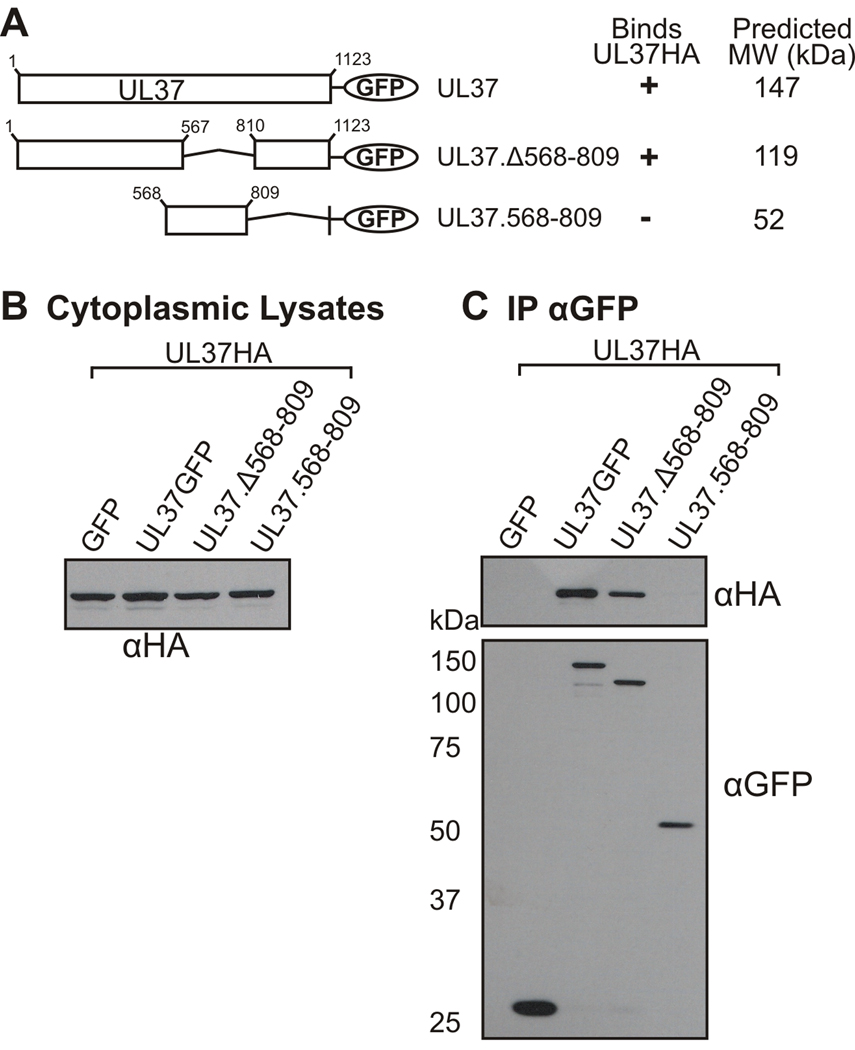
To further address the role that amino acids 568–809 of UL37 play in the interaction with UL36, a UL37 mutant containing an internal deletion of these amino acids, UL37.Δ568-809HA, was constructed (Fig. 2A). Vero cells were cotransfected with UL37HA constructs and UL36.512-767GFP, subsequently lysed for coimmunoprecipitation with anti-HA antiserum, and protein interactions visualized by SDS-PAGE and Western blotting. Expression of the transfected plasmids in cell lysates is shown in Fig. 2B. Immunoprecipitated material is presented in Fig. 2C. As also seen in Figure 1, truncation mutants UL37.1-809HA, UL37.568-1123HA and UL37.301-1123HA coimmunoprecipitated UL36.512-767GFP. When analyzed in coimmunoprecipitation assays, deletion of amino acids 568–809 within full-length UL37 decreased binding to UL36 to nearly background levels (Fig. 2C). This result suggests that amino acids 568–809 of UL37 are functionally important for the interaction of UL36 and UL37, or alternatively this region plays an integral role in the structure of UL37 that is necessary for its interaction with UL36.
Figure 2. Analysis of the Ability of UL36.512-767GFP to Interact with a UL37HA Mutant Lacking Residues 568–809.
(A) A schematic representation of the UL37HA mutants. UL37HA mutants were cotransfected into Vero cells with UL36.512-767GFP. A tabular summary of results indicate the UL37HA truncation mutants that bind (+) or do not bind (−) UL36.512-767GFP (UL36GFP) in coimmunoprecipitation assays. Molecular weight values were predicted by Statistical Analysis of Protein Sequence accessed through SDSC Biology Workbench. Immunoprecipitation and Western blot analysis was performed as described in the legend to Fig. 1. (B) Expression of the GFP and HA fusion proteins in whole cell lysates was analyzed by Western blotting with rabbit anti-HA (αHA, top panel) and rabbit anti-GFP (αGFP, bottom panel) antibodies. (C) The ability of the UL37HA mutants to bind UL36.512-767GFP was analyzed by Western blotting membranes of the coimmunoprecipitated material with goat anti-GFP antibody (αGFP, bottom panel). Rabbit anti-HA antibody and rabbit TrueBlot HRP-conjugated anti-rabbit secondary antibody was used to verify immunoprecipitated UL37HA constructs (αHA, top panel).
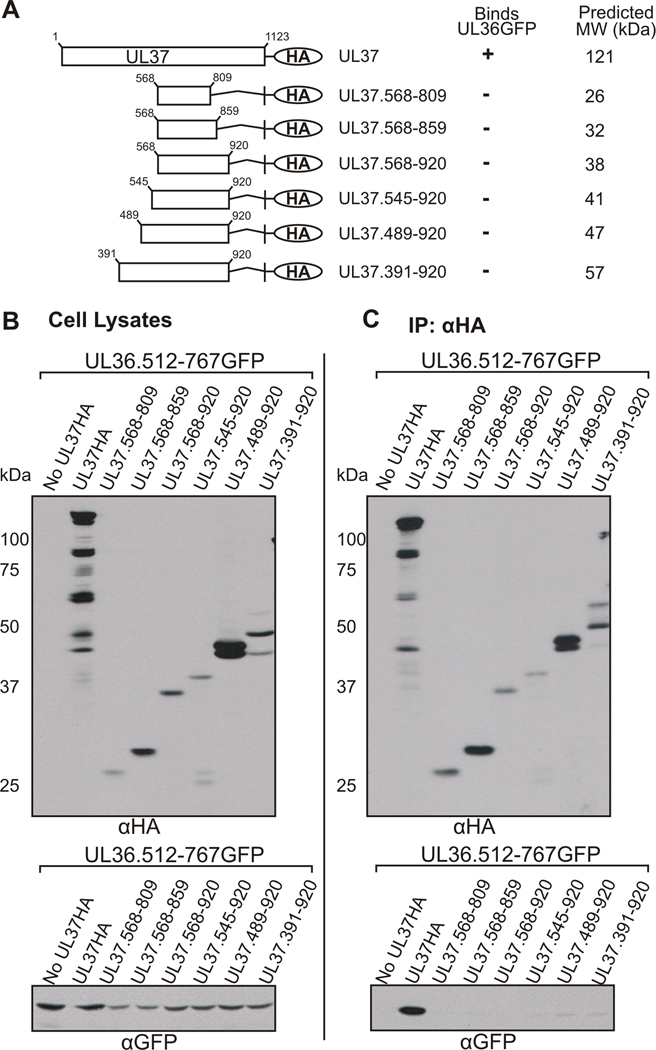
Results shown in Figures 1 and 2 suggest that amino acids 568–809 of UL37 are necessary for the interaction with UL36. Experiments were performed to determine if these amino acids are sufficient to enable UL36 binding. HA tagged UL37 truncation mutants containing amino acids 568–809, or amino acids extending outward from that region were generated (Fig. 3A). Plasmids encoding the UL37HA truncation mutants and UL36.512-767GFP were cotransfected into cells and analyzed by coimmunoprecipitation and Western blotting. Several of the internal UL37 mutants reproducibly exhibited relatively low protein expression as compared to the full length protein (Fig. 3B). When the internal UL37 mutants were immunoprecipitated with anti-HA antisera, Western blotting with anti-GFP antisera revealed that UL36.512-767GFP failed to coimmunoprecipitate with any of the internal UL37HA mutant proteins (Fig. 3C). These results suggest that although amino acids 568–809 of UL37 are necessary for interaction with UL36, they are not sufficient for this interaction. The failure of the UL37HA truncation mutants to bind UL36 may suggest that amino acids outside the region of 568–809 are involved in binding to UL36 and/or maintaining the proper conformation of UL37 necessary for its interaction with UL36.
Figure 3. Coimmunoprecipitation of UL36.512-767GFP with Internal UL37HA Mutants.
(A) A schematic representation of the internal UL37HA truncation mutants. Plasmids encoding internal domains of UL37HA were cotransfected into Vero cells with UL36.512-767GFP. Immunoprecipitation and Western blot analysis was performed as described in the legend to Fig. 1. A tabular summary of results indicate the UL37HA truncation mutants that bind (+) or do not bind (−) UL36.512-767GFP (UL36GFP) in coimmunoprecipitation assays. Molecular weight values were predicted by Statistical Analysis of Protein Sequence accessed through SDSC Biology Workbench. (B) Expression of the GFP and HA fusion proteins in whole cell lysates was analyzed by Western blotting with rabbit anti-HA (αHA, top panel) and rabbit anti-GFP (αGFP, bottom panel) antibodies. (C) The ability of the internal UL37HA mutants to bind UL36.512-767GFP was analyzed by Western blotting membranes of the coimmunoprecipitated material with goat anti-GFP antibody (αGFP, bottom panel). Rabbit anti-HA antibody and rabbit TrueBlot HRP-conjugated anti-rabbit secondary antibody was used to verify immunoprecipitated UL37HA constructs (αHA, top panel).
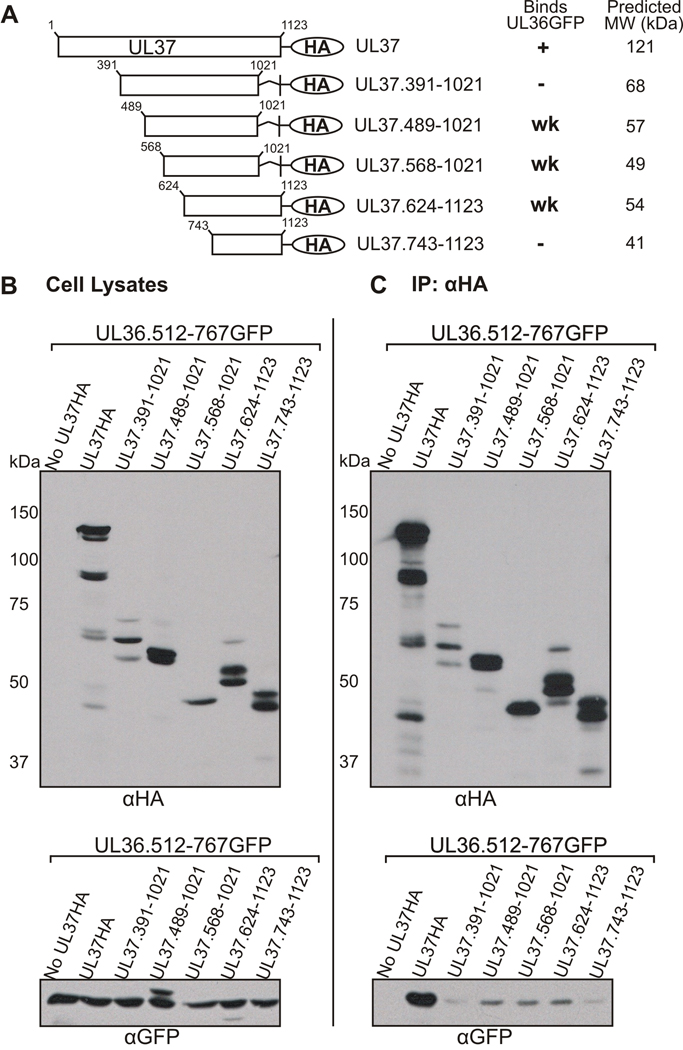
As shown in Figures 1 and 2, the C-terminal half of UL37, containing residues 568–1123, is sufficient for binding UL36.512-767GFP. In an effort to define a smaller region of UL37 that is sufficient for UL36 binding, additional HA tagged plasmids encoding internal regions of UL37 were constructed (Fig. 4A). UL37HA truncation mutants and UL36.512-767GFP were cotransfected into cells to be used in coimmunoprecipitation assays. Protein expression (Fig. 4B) and protein interactions (Fig. 4C) were visualized by SDS-PAGE and Western blotting. The internal UL37HA truncation mutants reproducibly displayed multiple protein bands, suggestive of protein degradation. The GFP Western blot of cell lysates (Fig. 4B, bottom panel) depicts multiple protein bands in the UL37.489-1021 sample due to incomplete stripping of anti-HA antibody from the membrane before the GFP Western blot was performed. Coimmunoprecipitation with anti-HA antibody showed that UL37 truncation mutants encoding amino acids 391–1021 or 743–1123 do not reproducibly bind UL36.512-767GFP above background levels. However, UL37 truncation mutants containing amino acids 489–1021, 568–1021, or 624–1123 coimmunoprecipitated UL36.512-767GFP; although binding is markedly lower than binding with full length UL37. The smallest truncation mutant of UL37 that facilitates an interaction with UL36 is UL37.568-1021HA. Together, these results suggest that amino acids 624–1021 would also be sufficient for interaction with UL36. To address this issue a UL37 truncation mutant expressing amino acids 624–1021 was constructed and analyzed in coimmunoprecipitation assays as described above. Surprisingly, UL37.624-1021HA was not able to interact with UL36, although protein expression was considerably less than full length UL37 (data not shown). Collectively, these coimmunoprecipitation studies indicate that residues 568–809 of UL37 are necessary for interaction with UL36. Amino acids 568–809 may directly interact with UL36 or, alternatively, deletion of this region may cause major disruptions in the protein structure, thus inhibiting the interaction with UL36. The carboxy-terminal half of UL37 is sufficient for interaction with UL36, but further truncation of this region results in little, if any, binding to UL36. These results suggest that UL37 may be conformationally-sensitive to truncations within the carboxy-terminus.
Figure 4. Coimmunoprecipitation of UL36.512-767GFP with Additional Internal UL37HA Mutants.
(A) A schematic representation of the internal UL37 truncation mutants. Plasmids encoding internal domains of UL37HA were cotransfected into Vero cells with UL36.512-767GFP. Immunoprecipitation and Western blot analysis was performed as described in the legend to Fig. 1. A tabular summary of results indicate the UL37HA truncation mutants that bind (+), do not bind (−) or reproducibly bind UL36.512-767GFP (UL36GFP) weakly (wk) in coimmunoprecipitation assays. Molecular weight values were predicted by Statistical Analysis of Protein Sequence accessed through SDSC Biology Workbench. (B) Expression of the GFP and HA fusion proteins in whole cell lysates was analyzed by Western blotting using rabbit anti-HA (αHA, top panel) and rabbit anti-GFP (αGFP, bottom panel) antibodies. (C) The ability of the internal UL37HA mutants to bind UL36.512-767GFP was analyzed by Western blotting the immunoprecipitated material with goat anti-GFP antibody. Rabbit anti-HA antibody and rabbit TrueBlot HRP-conjugated anti-rabbit secondary antibody was used to verify immunoprecipitated UL37HA constructs.
Identification of domains involved in self-association of UL37
UL37 has been shown by yeast two-hybrid and coimmunoprecipitation assays to self associate (Lee et al., 2008; Vittone et al., 2005). To ascertain the region(s) of UL37 involved in self-association of the protein, GFP-tagged UL37 truncation mutants were generated (Fig. 5A). A plasmid encoding full length UL37HA was cotransfected into cells with plasmids encoding UL37GFP truncation mutants. Protein interactions were assayed by coimmunoprecipitation with anti-GFP antisera. Western blotting with rabbit antibodies to GFP and HA were used to verify protein expression (Fig. 5B). Anti-HA immunoblots show that truncation mutants encoding either N-terminal or C-terminal regions of UL37 were capable of interacting with full length UL37HA (Fig. 5C). UL37 truncation mutants containing amino acids 1–809, 1–300, 301–1123 or 589–1123 were able to coimmunoprecipitate full length UL37HA (Fig. 5C). A construct encoding amino acids 1–567 repeatedly did not interact with UL37HA in binding assays; this is an intriguing observation because a smaller truncation mutant encoding amino acids 1–300 does bind full length UL37HA. This finding suggests that removal of the C-terminal half of UL37GFP may cause the protein to misfold, thus blocking its ability to bind UL37HA. However, further truncation of the UL37GFP mutant from the C-terminus, resulting in UL37.1-300GFP, restores binding to UL37HA, possibly by exposing an amino-terminal UL37 self-association domain that was presumably masked or misfolded in UL37.1-567GFP.
Figure 5. Coimmunoprecipitation of UL37HA with GFP tagged N-Terminal and C-Terminal UL37 Truncation Mutants.
(A) A schematic representation of full length, N-terminal and C-terminal UL37 truncation mutants fused to GFP. A tabular summary of results indicate the UL37GFP truncation mutants that bind (+) or do not bind (−) UL37HA in coimmunoprecipitation assays. Molecular weight values were predicted by Statistical Analysis of Protein Sequence accessed through SDSC Biology Workbench. (B) Expression of the UL37 constructs. Vero cells cotransfected with the indicated constructs were harvested approximately 24 h post-transfection and lysed in NP40 lysis buffer. A fraction of the cytoplasmic lysates was analyzed using rabbit antibodies to GFP (αGFP, top panel) and HA (αHA, bottom panel). (C) Coimmunopreciptitation of UL37GFP constructs with UL37HA. Transfected cell lysates were incubated with goat anti-GFP antibody and then bound to protein-G agarose beads. Immunoprecipitated material was analyzed on a SDS-PAGE gel and transferred to nitrocellulose. Proteins were detected by Western blotting using rabbit antibodies to GFP (αGFP, top panel) and HA (αHA, bottom panel).
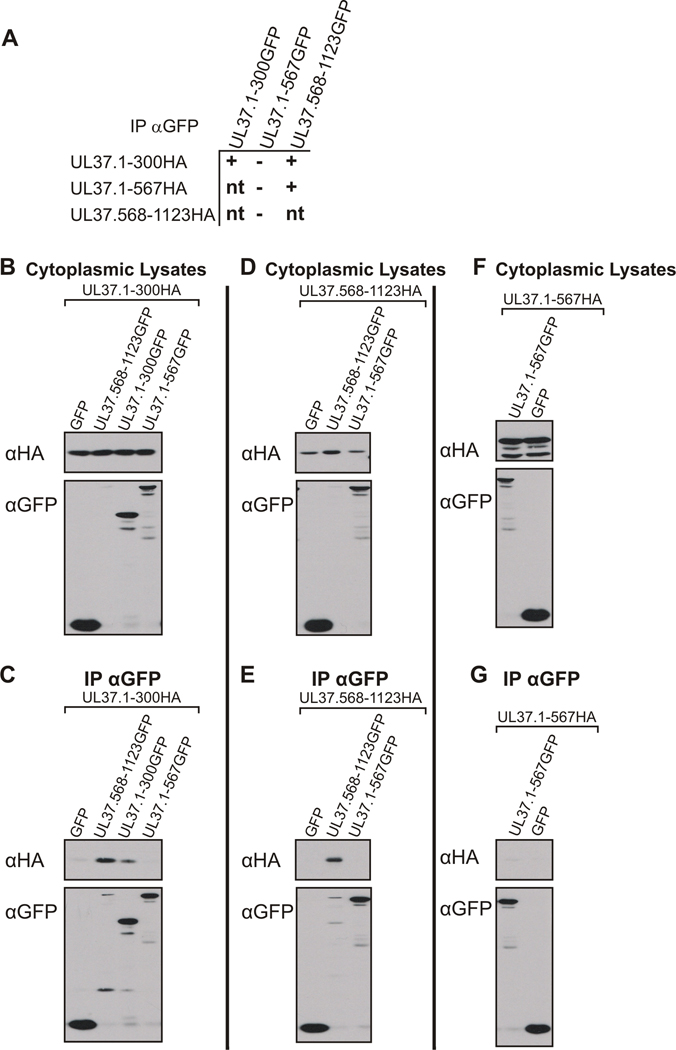
UL37 appears to contain two domains involved in interaction with full length UL37, residues 1–300 and 568–1123. Cells were cotransfected with HA and GFP tagged UL37 constructs and coimmunoprecipitation assays were used to determine if the amino and carboxy-terminal interaction domains bind to one another and if the mutant UL37 domains self interact (Fig. 6A). In UL37 binding assays, UL37.1-567GFP, which does not bind full length UL37, was used as a negative control. Protein expression was assayed by Western blotting cytoplasmic lysates with anti-HA (top panels) and anti-GFP (bottom panels) antibodies (Figs. 6B, D and F). Immunoprecipitation with anti-GFP antisera and Western blotting with anti-HA antibody revealed that UL37.1-300HA and UL37.568-1123GFP interact in cotransfected cells (Fig. 6C). Binding assays also showed that UL37.1-300GFP is capable of binding UL37.1-300HA and that UL37.568-1123GFP is capable of interacting with UL37.568-1123HA (Figs. 6C and E, respectively). Fig. 6G shows that HA and GFP-tagged truncation mutants containing amino acids 1–567 are not capable of self-association, further suggesting that this mutant protein may lack proper structure that enables self-interaction. These results suggest that UL37 contains two domains involved in self-association of the protein. Mutants encoding either of the interaction domains are capable of self-association with full length UL37 and the two interaction domains can be coimmunoprecipitated in a complex. Furthermore, HA-tagged mutants encoding residues 1–300 or 568–1123 can self-interact with GFP-tagged mutants encoding residues 1–300 or 568–1123, respectively.
Figure 6. Interactions of Amino- and Carboxy-Terminal UL37 Self-Association Domains Analyzed by Coimmunoprecipitation-Western Blot Analysis.
(A) A schematic representation of the UL37 N-terminal and C-terminal HA and GFP fusion constructs used in the coimmunoprecipitation assays. A tabular summary of results indicate the UL37GFP truncation mutants that bind (+) or do not bind (−) UL37HA truncation mutants in coimmunoprecipitation assays. Possible combinations of coimmunoprecipitations that were not tested are indicated by “nt.” (B, D, F) Expression of cotransfected UL37HA and UL37 GFP mutants. Vero cells were cotransfected with the indicated plasmids, harvested at approximately 24 h post-transfection and lysed in NP40 lysis buffer. A portion of each cytoplasmic lysate was analyzed for protein expression using rabbit antibodies to HA (αHA, top panels) and GFP (αGFP, bottom panels). (C, E, G) Coimmunoprecipitation of UL37 HA-tagged and UL37 GFP-tagged mutant proteins. Coimmunoprecipitations were performed and analyzed as described in the legend to Fig. 5. Western blotting with rabbit anti-HA (αHA, top panels) and anti-GFP (αGFP, bottom panels) antibodies was used to determine if the UL37HA mutants coimmunoprecipitated with the GFP fusion constructs.
Results from UL36–UL37 binding assays indicated that amino acids 568–809 of UL37 are necessary for the interaction with UL36 (Fig. 1 and 2). It is possible that deletion of amino acids 568–809 of UL37 causes the protein to misfold, thus inhibiting protein-protein interactions. To determine if an internal deletion of this region of UL37 negatively affects self-association of UL37, we generated a UL37.Δ568-809GFP mutant to be used in coimmunoprecipitation studies. Plasmids encoding UL37.Δ568-809GFP and UL37HA were cotransfected into cells, lysed and analyzed by coimmunoprecipitation with anti-GFP antisera. Immunoblots with anti-HA antisera indicate that residues 568–809 of UL37 are not sufficient for self-association of UL37. UL37.Δ568-809GFP coimmunoprecipitated UL37GFP, indicating that amino acids 568–809 of UL37 are not necessary for the self-interaction (Fig. 7C). These results suggest that this region, amino acids 568–809, is integral for the interaction with UL36 but not for self-association of UL37.
Figure 7. Coimmunoprecipitation of UL37HA and UL37.Δ568-809GFP.
(A) A schematic representation of the UL37GFP constructs cotransfected with full length UL37HA construct. A tabular summary of results indicate the UL37GFP truncation mutants that bind (+) or do not bind (−) UL37HA in coimmunoprecipitation assays. Molecular weight values were predicted by Statistical Analysis of Protein Sequence accessed through SDSC Biology Workbench. (B) Expression of UL37HA. Vero cells cotransfected with the indicated constructs were harvested at approximately 24 h post-transfection and lysed in NP40 lysis buffer. A portion of the cytoplasmic lysates was analyzed for protein expression using a rabbit anti-HA antibody. (C) Coimmunoprecipitation of UL37.Δ568-809GFP with UL37HA. Coimmunoprecipitations were performed and analyzed as described in the legend to Fig. 5. Coimmunoprecipitated proteins were detected by Western blotting with rabbit anti-GFP (αGFP, bottom panel) and rabbit anti-HA (αHA, top panel) antibodies.
Do UL37 and UL36 compete for binding to UL37?
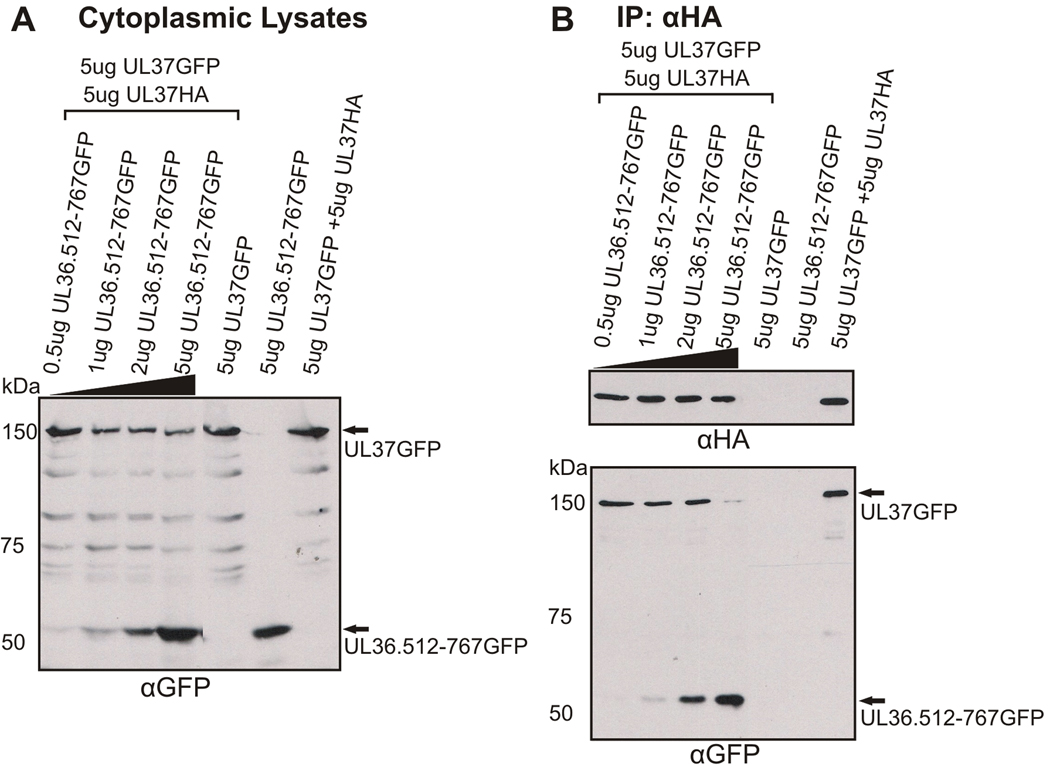
UL37 contains two self-association domains, amino acids 1–300 and 568–1123. The UL36 binding domain of UL37 overlaps the C-terminal UL37 self-association domain. In the infected cell, UL37 and UL36 are present in the cytoplasm and nucleus at approximately the same times. These observations raise the question whether UL36 and UL37 compete for binding to UL37. Coimmunoprecipitation studies were used to determine if UL37 is capable of binding UL37 and UL36 simultaneously or if one binding reaction inhibits the other. Cells were cotransfected with UL37GFP, UL37HA and increasing amounts of UL36.512-767GFP plasmids. Cells were lysed, proteins were coimmunoprecipitated with anti-HA antisera and immune complexes were visualized by SDS-PAGE and Western blotting. Western blotting of cytoplasmic lysates with anti-GFP antibody verified protein expression of both GFP-expressing plasmids (Fig. 8A). Western blotting of the immunoprecipitated material with anti-GFP antisera was used to determine if UL37GFP and/or UL36.512-767GFP coimmunoprecipitated with UL37HA. When 0.5 ug of plasmid encoding UL36.512-767GFP was cotransfected into cells with plasmids encoding UL37HA and UL3 GFP, anti-GFP Western blots revealed that both UL37GFP and UL36.512-767GFP coimmunoprecipitated with UL37HA (Fig. 8B). Thus, under relatively low levels of expression of UL36.512-767GFP, UL37GFP bound UL37HA (Fig. 8B). However, as the expression of UL36.512-767GFP protein increased, the amount of UL37GFP that coimmunoprecipitated with UL37HA decreased. The self-interaction of UL37HA and UL37GFP appeared to occur only when UL36.512-767GFP is not present in high quantities. Similar results from seven independent experiments indicated that appreciable expression of UL36.512-767GFP inhibits self-association of UL37HA and UL37GFP in coimmunoprecipitation experiments. Furthermore, expression of UL37HA was increased by transfecting greater amounts of DNA, but the increased expression of UL37HA failed to restore UL37GFP interaction with UL37HA in the presence of UL36.512-767GFP (data not shown). Based on these results it appears that UL37GFP and UL36.512-767GFP compete for binding to UL37HA. This suggests that UL37HA does not bind UL37GFP and UL36.512-767GFP simultaneously.
Figure 8. Coimmunoprecipitation Analysis of UL37HA when Coexpressed with UL37GFP and UL36.512-767GFP.
(A) Vero cells were cotransfected with the indicated amounts UL36.512-767GFP, UL37HA and UL37GFP plasmids. At approximately 24 h post-transfection, cells were harvested, lysed in NP40 lysis buffer and a portion of the cytoplasmic lysate was analyzed for protein expression by Western blotting using goat anti-GFP antibodies. Arrows indicate the positions of UL37GFP and UL36.512-767GFP. (B) Coimmunoprecipitation of UL37GFP and/or UL36.512-767GFP with UL37HA. Coimmunoprecipitation with anti-HA antisera was performed as described in the legend to Fig. 1. Immunoprecipitated material was analyzed by Western blot analysis using a goat anti-HA antibody (αHA, top panel) and goat anti-GFP antibody (αGFP, bottom panel). Arrows indicate the positions of UL37GFP and UL36.512-767GFP in the bottom panel.
Identification of domains of UL37 that complement the UL37 deletion virus, KΔUL37
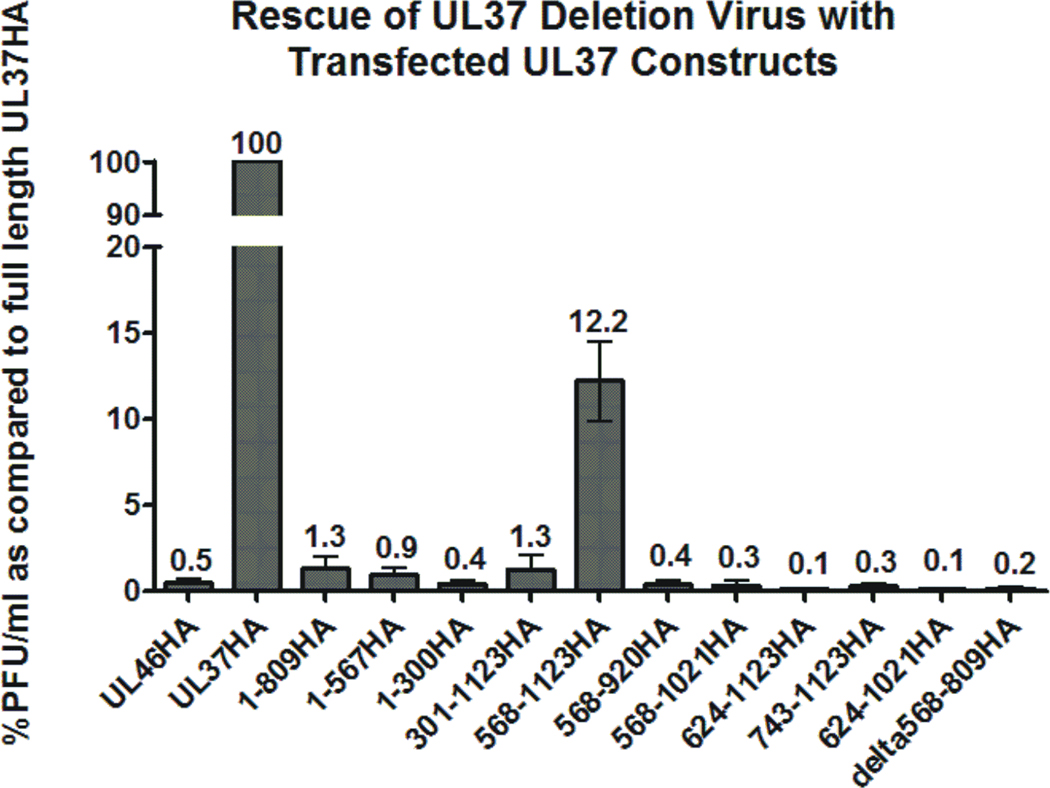
UL37 is necessary for assembly of HSV-1 virions; in the absence of UL37 proper tegumentation and secondary envelopment do not occur (Desai et al., 2001; Roberts et al., 2009). A trans-complementation assay was designed to determine the regions of UL37 that rescue the replication deficient KΔUL37 virus to result in the production of infectious virions. Cells were transfected with the indicated mutant UL37HA plasmids and 24 h later infected with KΔUL37. UL46HA was transfected as a negative control. At 48 h after infection, cells and media were collected and subjected to three rounds of freeze/thaw, cellular debris was pelleted and infectious virions were quantified by plaque assay on BD45 cells (Vero cells that stably express UL37). Western blotting of the transfected-infected cell lysates was performed to verify expression of each of the transfected plasmids. All plasmids used in this study contained the immediate early CMV promoter and produced robust protein expression of all transfected plasmids in transfected-infected cells (data not shown). The amounts of infectious virions released from cells transfected with UL37HA mutants were determined and are represented as the percent of PFU/ml as compared to virions released from cells transfected with full length UL37HA (Fig. 9). Plaque assays revealed that a mutant protein encoding the C-terminal half of UL37, residues 568–1123, was capable of rescuing the KΔUL37 virus to approximately 12% of the levels, or approximately one log less than the amount released when a plasmid encoding full length UL37HA is transfected (Fig. 9). The other tested UL37HA mutants yielded viral titers comparable to the UL46HA control sample. These results indicate that the carboxy-terminal half of UL37 confers partial complementation to the replication defective KΔUL37 virus. This result suggests that the region of UL37, which is involved in binding UL36 and self-association, may play an essential functional role during virus infection.
Figure 9. Transfection-Infection Experiments to Assess the Ability of UL37HA Mutants to Complement the KΔUL37 Virus.
Cells were transfected with the indicated UL37HA constructs, and approximately 24 h later infected with KΔUL37 virus with an MOI of 1. Approximately 48 h after infection, cells and media were harvested and lysed by three cycles of freeze/thaw. Cellular debris was pelleted and the supernatant was titered on UL37 expressing cells (BD45) to quantitate the amount of infectious virus released. Titrations for each UL37 construct represent the average of a minimum of three independent experiments, error bars represent the calculated standard error of measurement, (SEM). For each independent experiment, the titers were compared by determining the percent of virus released as compared to UL37HA (%PFU/ml for the UL37HA sample equals 100% for each experiment).
Does UL37.568-1123HA interact with UL36 in infected cells?
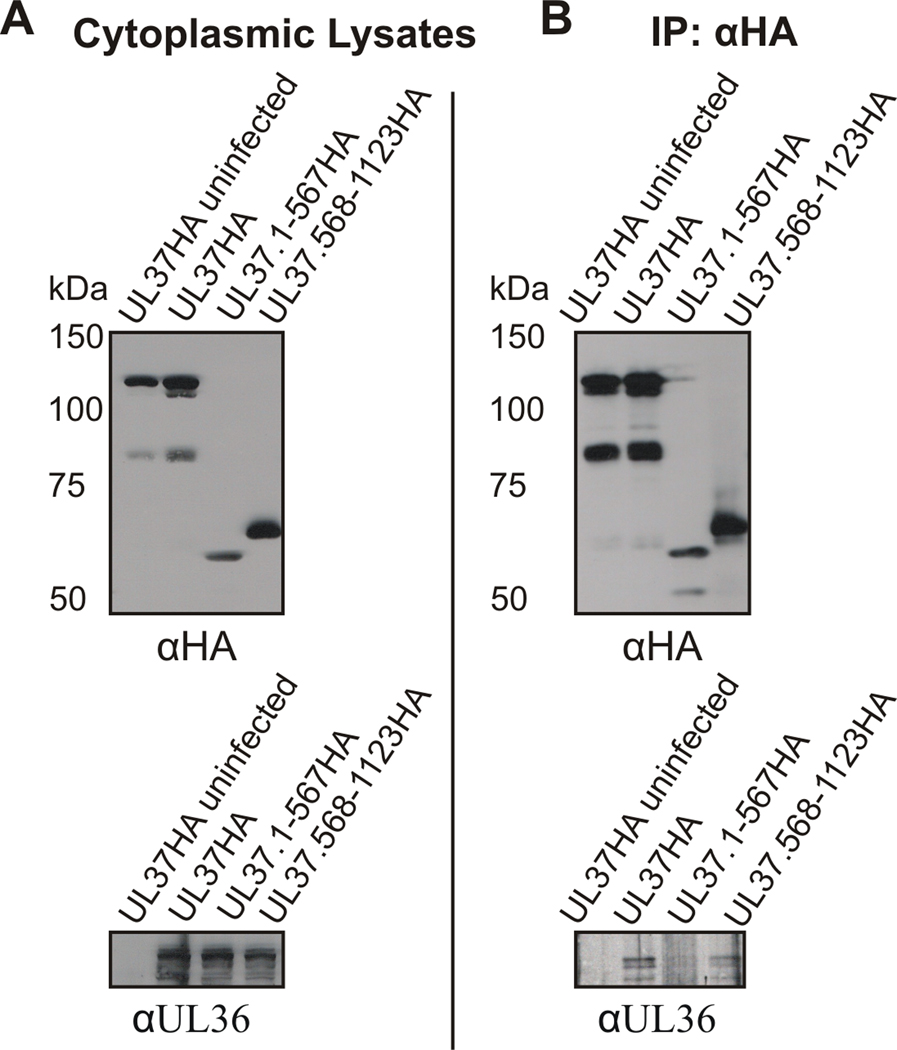
Results shown in figure 9 indicate that UL37.568-1123HA partially rescues the KΔUL37 virus. UL37.568-1123HA interacts with UL36.512-767GFP in cotransfected cells (Figs. 1 and 2). To determine if the ability of UL37.568-1123HA to rescue the KΔUL37 virus was possibly due to its ability to interact with UL36, it was necessary to demonstrate that UL37.568-1123HA interacts with virus-expressed full-length UL36. A coimmunoprecipitation assay from transfected-infected cells was utilized to address this question. To minimize the risk of coimmunoprecipitating UL37.568-1123HA and UL36 bound to a large capsid complex, transfected cells were infected with K23Z HSV-1 (Desai et al., 1993), in which capsid formation is abrogated. Therefore, K23Z-infected cells allowed the examination of the UL36 interaction with UL37.568-1123HA in the absence of capsid formation. Cells were transfected with the indicated UL37HA plasmids and 24 h later infected with K23Z HSV-1. Approximately 24 h after infection, cells were lysed, proteins were coimmunoprecipitated with anti-HA antisera and immune complexes were visualized by SDS-PAGE and Western blotting. Western blotting using anti-HA antiserum and anti-UL36 antiserum verified expression of the transfected plasmids and viral UL36 respectively (Fig. 10A). Immunoblots with anti-UL36 antiserum indicate that K23Z-expressed UL36 coimmunoprecipitated with full length UL37HA (Fig. 10B). As a negative control, cells were transfected with UL37.1-567HA which does not interact with UL36.512-767GFP. As expected, K23Z-expressed UL36 did not coimmunoprecipitate with UL37.1-567HA. In contrast, K23Z-expressed UL36 coimmunoprecipitated with UL37.568-1123HA. These results indicate that UL37.568-1123HA interacts with full-length UL36 in infected cells.
Figure 10. Coimmunoprecipitation of virus-expressed full-length UL36 and UL37.568-1123HA.
Vero cells were transfected with the indicated UL37HA plasmids and 24 h later infected with K23Z HSV-1 at an MOI of 10. Approximately 24 h post-infection cells were harvested and immunoprecipitation was performed as described in the legend to Fig. 1. (A) Each cytoplasmic lysate was analyzed by Western blotting using rabbit HA antibody (αHA, top panel) or rabbit UL36 antibody (αUL36, bottom panel). (B) The ability of K23Z-expressed full-length UL36 to bind UL37HA constructs was analyzed by Western blotting the membranes of the coimmunoprecipitated material with rabbit anti-UL36 antibody (αUL36, bottom panel) and rabbit TrueBlot HRP-conjugated anti-rabbit secondary antibody. Western blotting with rabbit anti-HA antibody and rabbit TrueBlot HRP-conjugated anti-rabbit secondary antibody verified imunoprecipitated UL37HA complexes.
DISCUSSION
The studies described within this report were designed to identify domains of the UL37 tegument protein involved in its self-association, as well as its interaction with UL36. To our knowledge, these are the first studies identifying domains of UL37 involved in these interactions. These studies will help elucidate the role of the interaction between UL36 and UL37 and the role that self-association of UL37 play during viral assembly. A schematic summary of domains and epitopes of UL37 identified by this report and others is presented in Figure 11.
Figure 11. Schematic Representation of Identified Domains and Epitopes of UL37.
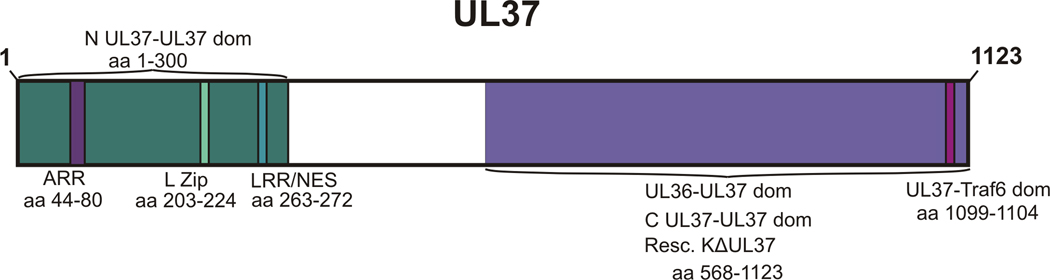
Amino acids 1–300 function as a UL37 multimerization domain (N UL37-UL37 dom), an alanine-rich region encompasses residues 44–80 (ARR), a leucine zipper motif spans residues 203–224 (L Zip) and a leucine-rich region at amino acids 263–272 functions as a nuclear export signal (LRR/NES) (Watanabe et al., 2000). The C-terminal half of UL37, amino acids 568–1123, functions as a UL37 self-association domain (C UL37-UL37 dom), is involved in binding UL36 (UL36–UL37 dom) and this region partially complements the KΔUL37 virus in transfected cells (Resc. KΔUL37). UL37 activates NFκ-B through binding TRAF6 at residues 1099–1104 (UL37-TRAF6 dom) (Liu et al., 2008). Throughout UL37 are 17 dileucine motifs (LL or LI) and 3 tyrosine-based motifs (YXXΦ, where Φ represents a large hydrophobic residue) which may facilitate interactions with clathrin adaptor proteins to promote vesicular trafficking (Boll et al., 1996, Kirchhausen et al., 1997; Kirchhausen, 1999; Wan et al., 1998). Conserved residues span the length of UL37, with the exception of approximately 100 unconserved amino acids at the amino and carboxy-termini.
Several observations throughout these studies suggested that the conformation of UL37 may be sensitive to mutations. First, UL37.1-567HA was the only truncation mutant that did not interact with full length UL37. Although UL37.1-567HA contains the amino-terminal self-association sequence (amino acids 1–300); UL37.1-567HA did not interact with either of the self-association domains (regions 1–300 and 568–1123). These results suggest that due to truncation, the structural conformation of UL37.1-567HA may be altered in such a way that inhibits the self-interaction of UL37. Results from mapping the domain of UL37 involved in UL36 binding also suggest that the conformation of UL37 is sensitive to truncations. The carboxy-terminal half of UL37, encompassing amino acids 568–1123, is sufficient for interaction with UL36. However, deletion of 102 amino acids from the unconserved C-terminus of UL37 yields a truncation mutant, UL37.568-1021HA, that binds UL36 at low levels. Furthermore, UL37.624-1123HA showed low levels of binding to UL36 (Fig. 4). Collectively, these results suggested that a smaller construct encoding amino acids 624–1021 would also bind UL36. However, when a plasmid encoding this region was constructed and analyzed in coimmunoprecipitation assays, UL37.624-1021HA failed to bind UL36 (data not shown), suggesting that amino acids surrounding this region within the carboxy-terminus are important in maintaining the conformation of UL37 needed to bind UL36. Interestingly, Luxton and colleagues reported that a GFP tag on the amino-terminus of PRV UL37 was deleterious for viral growth, perhaps suggesting that GFP at this position disrupts the conformation of UL37 (Luxton et al., 2005). Furthermore, Antinone and Smith also reported that a GFP tag on the amino-terminus of HSV-1 UL37 decreased viral titers by nearly one order of magnitude (Antinone & Smith, 2010).
The studies presented in this manuscript do not investigate the possibility that cellular and/or viral proteins may bridge the interaction of UL36 and UL37. Previous studies, have reported the HSV-1 UL36 and UL37 interaction, as well as HSV-1 UL37 self-association, in yeast two-hybrid and coimmunoprecipitation studies in the absence of other viral proteins (Lee et al., 2008, Vittone et al., 2005). Furthermore, coimmunoprecipitation results presented in this manuscript demonstrate that additional viral proteins are not necessary to facilitate or bridge the interaction of UL36 and UL37 minimal binding domains or UL37 self-association. The possibility that cellular proteins bind UL36 and/or UL37 to facilitate the above interactions cannot be dismissed entirely. Based upon the fact that the minimal binding domains of UL36 and UL37 interact in cotransfected cells, it seems unlikely that cellular factors are necessary for the interaction of UL36 and UL37. In addition, relatively small self-association domains of UL37 coimmunoprecipitate in transfected cells, also suggesting that cellular proteins are not necessary for interaction. Although we suspect that cellular proteins are not necessary for the UL36–UL37 interaction or UL37 self-association, it is possible that cellular and/or viral proteins may enhance or facilitate these interactions in a variety of ways.
The UL37 domains involved in interaction with UL36, as well as its self-association, are located in the carboxy-terminal region of UL37. We were curious if UL37 binds to molecules of UL36 and UL37 at the same time, forming a complex. From cells cotransfected with UL37HA, UL37GFP and UL36.512-767GFP, we observed that as the amount of UL36.512-767GFP protein increased, the amount of UL37GFP that coimmunoprecipitated with UL37HA decreased (Fig. 8). Thus, it appears that UL36.512-767GFP competes with UL37GFP for binding to UL37HA. It may be possible that binding of UL36 to UL37 alters the conformation of UL37, resulting in steric hindrance that prevents the self-association of UL37. The role that self-association of UL37 may play during infection is unclear. When UL37HA and UL37GFP are cotransfected into cells that are subsequently infected with HSV-1, the self-association of UL37HA and UL37GFP is readily detected in coimmunoprecipitation assays (data not shown). Although UL37 is overexpressed through transient transfection in this experimental design, it appears that UL37 does self-associate within the infected cell. Fuchs and colleagues reported that deletion of the domain of UL36 necessary for binding UL37 does not abrogate PRV virus production, but reduces viral titers by 50-fold and produces capsids within the cytoplasm in large ordered structures (Fuchs et al., 2004). Therefore, it appears that in PRV, UL36 is not required to interact with UL37 in order to execute its essential function. UL37 plays a critical role for assembly of HSV-1, but is not strictly essential for PRV replication. The function and/or necessity of the UL36–UL37 interaction may differ among HSV-1 and PRV. It is not yet understood if UL37 is dependent on the UL36–UL37 interaction to perform its essential function during HSV-1 infection. Similarly, it is unknown if the self-interaction of UL37 plays a vital role in virus replication. Exactly how the competition between UL36 and UL37 to bind UL37 may impact virus assembly is unclear.
The amount of HSV-1 UL37 packaged into virions is tightly controlled (McLauchlan, 1997). Furthermore, relatively few copies of HSV-1 UL36 are packaged into virions; an estimated 100–150 copies per virion (Heine et al., 1974). Based upon the incorporation of UL36 and UL37, it is possible that the protein levels of UL37 and UL36 within the infected cell may also be tightly controlled. Protein levels of the UL36 homologue of human cytomegalovirus have been reported to be dependent on the presence of the UL37 homologue (Bechtel & Shenk, 2002). Therefore, protein levels of UL36 and UL37 within the infected cell may also regulate the competition between UL36 and UL37 for binding with other molecules of UL37. One may speculate that the self-association of UL37 and the interaction of UL37 with UL36 may play roles at different sites of the infected cell. Although UL37 and UL36 are binding partners, it is likely that they are not always in abundance at the exact same locations within the cell, thus allowing UL37 to self associate in some locations and bind UL36 in others.
In an effort to identify regions of UL37 that perform necessary functions for assembly of infectious particles, plasmids encoding regions of UL37 were transfected into cells that were subsequently infected with the UL37 deletion virus, KΔUL37. Cells and media were harvested and analyzed by plaque assays to identify the regions of UL37 that rescue the replication defective virus. Surprisingly, the only region of UL37 that provided partial complementation was the region encoding amino acids 568–1123. The domain involved in UL36 binding and a self-association domain are located within this region. However, other truncation mutants that are capable of UL36 binding, such as UL37.301-1123HA and UL37.1-809HA were not able to rescue the mutant virus. Similarly, other UL37 truncation mutants capable of self-association were not able to complement KΔUL37. It is difficult to interpret what these results teach us about the essential function of UL37. It is possible that UL37 mutant proteins may perform a dominant-negative function within the infected cell, interacting with binding partners, but in a manner that disrupts virus assembly. Similarly, it is also unclear if the failure of UL37 mutant proteins to rescue KΔUL37 may be attributed to aberrant conformational structures that inhibit UL37 from performing essential function(s) during infection. Several truncation mutants that are capable of binding UL36 and/or self-association in transfection-immunoprecipitation assays also did not rescue the UL37 deletion virus. These results may also suggest that UL37 performs a function, in addition to self-association and UL36 binding, that maps to residues 568–1123 and is necessary for productive infection. The specific essential function(s) that UL37 plays during virus assembly remains to be determined.
UL37.568-1123HA was the only truncation mutant analyzed that was able to partially complement KΔUL37. To determine if this complementation might possibly be attributable to the ability of UL37.568-1123HA to interact with UL36, it was necessary to demonstrate the interaction of UL37.568-1123HA with full-length virus-expressed UL36. Coimmunoprecipitation from transfected-infected cytoplamsic lysates lacking capsid structures verified that UL37.568-1123HA is capable of interacting with virus-expressed full-length UL36 (Fig. 10), in addition to binding UL36.512-767GFP in transfected cells (Fig. 1). Based on these results, it seems likely that UL37.568-1123HA also interacts with virus-expressed UL36 in KΔUL37-infected cells. The interaction of UL37.568-1123HA and virus-expressed UL36 may or may not contribute to the partial complementation of KΔUL37 seen in figure 9.
It is also intriguing if self-association of UL37 plays an essential role in infection. It is clear that UL37 and UL37 truncation mutants self-associate in transfected cells (Figs. 5, 6 and 7), but the occurrence of the interaction is more difficult to interpret in infected cells. UL37HA and UL37GFP readily coimmunoprecipitate from cotransfected cells subsequently infected with HSV-1 K23Z, KΔUL37 or KΔUL36, a virus that fails to express most of the UL36 open reading frame (Desai, 2000) (data not shown). However, in this experimental scenario, expression of plasmid-encoded UL37 is presumably altered in kinetics and increased in amount as compared to virus-expressed UL37. Attempts were also made to coimmunoprecipitate UL37.568-1123HA from K23Z-infected cells. In these analyses UL37.568-1123HA showed minimal levels of binding to virus-expressed UL37 (data not shown). However, virus-expressed UL37 and UL37HA differ in only 9 amino acids in size and migrate at the same location on an SDS-PAGE gel. Therefore, the low level of binding of UL37.568-1123HA cannot be directly compared with a level of coimmunoprecipitation of transiently expressed full-length UL37HA in the virus-infected cells and thus, lacks clear interpretation. In spite of several attempts, it remains to be determined if self-association of virus-expressed UL37 occurs within the virus-infected cell and/or plays an essential role in virus infection.
MATERIALS AND METHODS
Cells and Viruses
Vero cells (ATCC CCL-81) were grown in Dulbecco’s modified Eagles Medium (DMEM) supplemented with 5% fetal bovine serum (FBS), 2.25% sodium bicarbonate, 25mM HEPES buffer, glutamine (300 µg/ml), penicillin (100 µg/ml) and streptomycin (131 µg/ml). Infected cells were grown in DMEM with a reduced FBS concentration of 2%. HSV-1 KOS strain was used as the wild type strain (SMITH, 1964). A UL37 deletion virus, KΔUL37, and Vero cell line that stably expresses UL37 (BD45), were kind gifts from Prashant Desai (Desai et al., 2001). BD45 cells were grown in DMEM with 5% FBS. K23Z HSV-1 was also a kind gift from Prashant Desai (Desai et al., 1993).
Antibodies
Rabbit anti-HA antisera used for Western blotting and immunoprecipitation was purchased from Sigma. Goat anti-HA antibody used for Western blotting was purchased from Santa Cruz. Rabbit polyclonal anti-GFP antibody used for Western blotting was kindly provided by John Wills, Penn State College of Medicine. Goat polyclonal anti-GFP antibody used for Western blotting and immunoprecipitation was purchased from Rockland. Rabbit TrueBlot HRP-conjugated anti-rabbit antibody was purchased from eBioscience. Rabbit polyclonal anti-UL36 antibody was described previously (McNabb & Courtney, 1992).
Cloning
The UL37 gene was amplified from the HSV-1 KOS genome by PCR amplification using Platinum Pfx polymerase (Invitrogen). A forward primer containing a BglII site 50 bp upstream of the start codon of UL37 and a reverse primer containing an EcoRI site in place of the stop codon of UL37 were used for amplification. The DNA product was digested with EcoRI and BglII and ligated into the vector pEGFP-n2 (Clontech) also digested with the same restriction enzymes. Two separate clones produced from separate PCR reactions were sequenced and compared. Amino acid sequences of the UL37 codons were identical and one plasmid was selected and designated UL37GFP.
The UL37HA plasmid was generated by excising the UL37 fragment from the UL37GFP plasmid, described above, using EcoRI and BglII. This fragment was ligated into a hemagglutinin (HA) containing plasmid, UL46HA, (Murphy et al., 2008) that was digested with the same restriction enzymes. The parental vector of the UL46HA plasmid was the pEGFP-n2 GFP vector that was previously modified to replace the GFP sequence with the HA sequence (Loomis et al., 2006). The newly constructed plasmid, UL37HA, was sequenced to ensure that UL37 was in frame with HA.
UL37 C-terminal GFP and HA truncation mutants were constructed by using the forward primer used to make the UL37GFP plasmid and a reverse primer that contains an EcoRI site immediately downstream of the truncation site. The PCR products were digested with BglII and EcoRI and ligated into UL37GFP or UL46HA plasmids digested with the same restriction enzymes. The resulting inserts were sequenced to ensure that they contained the specified UL37 truncation sequences and were in frame with GFP or HA.
To construct the UL37HA N-terminal truncation mutants, the UL37HA plasmid was modified to contain a HindIII restriction enzyme site immediately upstream of the start codon of UL37. The QuickChange II XL Site-Directed Mutagenesis kit (Stratagene) was used according to the manufacturer’s instructions to insert the HindIII site into the UL37HA plasmid to generate the UL37HA-HindIII plasmid. UL37HA-HindIII plasmid contains 50 bp of genomic sequence upstream of the UL37 start site, followed by a HindIII restriction site immediately upstream of the first UL37 amino acid. UL37HA N-terminal truncation mutants were amplified with forward primers containing a HindIII site and start codon immediately upstream of the first codon of the specified truncation mutant. The reverse primer used was the same reverse primer utilized to create the UL37HA plasmid and contains an EcoRI restriction site. The above mentioned primers were used to amplify the mutant UL37 inserts, which were then digested with HindIII and EcoRI and ligated into UL37HA-HindIII plasmid previously digested with the same enzymes. The resulting N-terminal UL37HA truncation mutants contained 50 bp of genomic upstream sequence, a start codon, the truncated amino acid sequence and the HA tag. The UL37HA truncation mutant plasmids were sequenced to verify the correct mutant UL37 coding regions. The UL37GFP N-terminal truncation mutants were constructed by excising the UL37 mutant fragments from the respective UL37HA plasmids using EcoRI and BglII and then ligating the sequences into UL37GFP plasmid previously digested with the same enzymes.
Plasmids containing internal sequences of UL37 were made by amplifying the UL37 sequence with forward primers containing a HindIII site and start codon immediately before the first UL37 codon of the mutant protein. Reverse primers contained an EcoRI site that was immediately downstream of the last UL37 codon of the mutant protein. The inserts were amplified by PCR, digested with HindIII and EcoRI and ligated into UL37.568-1123HA plasmid previously digested with the same enzymes. The resulting plasmids contain 50 bp of HSV-1 sequence upstream of the UL37 start codon, followed by a methionine start codon, the internal UL37 sequence and the HA tag.
UL37Δ568-809.HA and UL37Δ568-809. GFP were generated using the QuickChange II XL Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer’s instructions. PCR primers complementary to nucleotides encoding amino acids 563–567 and 810–814 were used to generate a PCR product from UL37HA and UL37GFP templates, looping out codons 568–809 of UL37. Clones were screened by restriction enzyme digestion and sequenced to verify the removal of codons 568–809.
The UL36.512-767 gene fragment was amplified from HSV-1 KOS genomic DNA by PCR amplification using Platinum Pfx polymerase (Invitrogen). A forward primer containing a BglII site and an ATG start sequence immediately before codon 512 and a reverse primer containing a MfeI site directly after codon 767 was used to amplify codons 512–767 of UL36. The amplified DNA was digested with BglII and MfeI and ligated into the vector pEGFP-n2 (Clontech) digested with BglII and EcoRI. The resulting plasmid, UL36.512-767GFP, was sequenced to verify the correct amino acid sequence and that the gene fragment was in frame with the downstream GFP.
Coimmunoprecipitation
Vero cells grown to approximately 90–95% confluency in 60 mm or 100 mm plates were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Approximately 20–24 h post-transfection cells were washed twice with phosphate-buffered saline (PBS) and harvested by scraping. Cells were pelleted by centrifugation at 1000 × g for 10 min at 4°C and resuspended in 1% NP40 lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 2mM MgCl, 1% NP40, 0.1% Sigma protease inhibitor cocktail). Cells were then incubated on ice for 30 min and nuclei were pelleted by centrifugation at 1000 × g for 10 min at 4°C. The cytoplasmic fraction was clarified further by centrifugation at 14,000 rpm for 5 min at 4°C in a microfuge. The cytoplasmic lysate was precleared for approximately 1 h with protein G-agarose beads (Roche) that were washed three times in lysis buffer. Protein G-agarose beads were pelleted at 14,000 rpm for 5 min at 4°C in a microfuge. Immunoprecipitation antibody was added to the precleared samples and rocked at 4°C for a minimum of 1 h. Protein G-agarose beads were added to the samples and rocked overnight at 4°C. Immune complexes were washed three times in lysis buffer. 2× sample buffer (3.6% SDS, 18% βME, 114 mM Tris pH 6.8, 0.05% bromophenol blue and 18% glycerol) was added to the beads and the immunoprecipitated material was separated by SDS-PAGE, transferred to nitrocellulose membranes and analyzed by Western blotting using the antibodies mentioned above, ECL reagents, and autoradiography using Kodak BioMax XAR film. For sequential probing of the same nitrocellulose membrane, blots were submerged in stripping buffer (60 mM Tris-HCl pH 8.0, 2% SDS and 0.75% βME) at 56°C for approximately 35 minutes to remove bound antibodies. Although results and interpretations are consistent between repetitions of experiments, quantification of immunoprecipitated material was attempted but unsuccessful. Comparison of relative levels of binding requires densitometric quantification of four separate Western blots for each repetition of an experiment. In some of these experiments the density of the protein bands was not within linear range for all four separate Western blots of an experiment. Therefore quantification and normalization could not be accurately performed.
Transfection-Infection Assay
Vero cells grown in 100 mm plates were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Approximately 24 h post-transfection, cells were infected with HSV-1 K23Z (Desai et al., 1993) with an MOI of 10. Approximately 24 h after infection, cells were harvested by scraping, washed twice with phosphate-buffered saline (PBS) and immunoprecipitation, SDS-PAGE and Western blotting were performed as described above.
Trans-Complementation Assay
Vero cells grown in 60 mm plates to approximately 90–95% confluency were transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Approximately 24 h after transfection cells were infected with KΔUL37 HSV-1 at an MOI of 1. Infected cells were grown in 3.5 ml of DMEM containing 2% FBS. At 48 h after infection, media and cells were collected. Cells were lysed by three cycles of freeze thaw and cell debris was pelleted by centrifugation at 750 × g for 10 min at 4°C. Cellular debris was analyzed by SDS-PAGE and Western blotting to verify expression of the transfected plasmids. Media supernatant was stored at −80°C before analysis by plaque assay. The amount of infectious virions in the media supernatant was determined by plaque titration assays on BD45 cells. Titrations were performed in duplicate. Each plasmid was analyzed for trans-complementation a minimum of three times and results of the independent titrations were averaged and presented as percent PFU/ml as compared to full length UL37HA.
Acknowledgments
We thank John Wills, David Hughes and Carol Dickerson for helpful discussions. We also appreciate the generous gifts of HSV-1 K23Z, KΔUL37 and BD45 cells from Prashant Desai (Johns Hopkins University). We thank the Penn State College of Medicine Macromolecular Core Facility and Molecular Genetics Core Facility for oligonucleotide synthesis and plasmid sequencing, respectively. This work was supported by National Institutes of Health (NIH) grants CA42460 and AI083834.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albright AG, Jenkins FJ. The herpes simplex virus UL37 protein is phosphorylated in infected cells. J.Virol. 1993;67(8):4842–4847. doi: 10.1128/jvi.67.8.4842-4847.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinone SE, Smith GA. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J.Virol. 2010;84(3):1504–1512. doi: 10.1128/JVI.02029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtel JT, Shenk T. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J.Virol. 2002;76(3):1043–1050. doi: 10.1128/JVI.76.3.1043-1050.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll W, Ohno H, Songyang Z, Rapoport I, Cantley LC, Bonifacino JS, Kirchhausen T. Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 1996;15(21):5789–5795. [PMC free article] [PubMed] [Google Scholar]
- Bucks MA, O'Regan KJ, Murphy MA, Wills JW, Courtney RJ. Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology. 2007;361(2):316–324. doi: 10.1016/j.virol.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller KE, Lee JI, Ueda A, Smith GA. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J.Virol. 2007;81(21):11790–11797. doi: 10.1128/JVI.01113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P, DeLuca NA, Glorioso JC, Person S. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J.Virol. 1993;67(3):1357–1364. doi: 10.1128/jvi.67.3.1357-1364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P, Sexton GL, Huang E, Person S. Localization of herpes simplex virus type 1 UL37 in the Golgi complex requires UL36 but not capsid structures. J.Virol. 2008;82(22):11354–11361. doi: 10.1128/JVI.00956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P, Sexton GL, McCaffery JM, Person S. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J.Virol. 2001;75(21):10259–10271. doi: 10.1128/JVI.75.21.10259-10271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai PJ. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J.Virol. 2000;74(24):11608–11618. doi: 10.1128/jvi.74.24.11608-11618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossum E, Friedel CC, Rajagopala SV, Titz B, Baiker A, Schmidt T, Kraus T, Stellberger T, Rutenberg C, Suthram S, Bandyopadhyay S, Rose D, von Brunn A, Uhlmann M, Zeretzke C, Dong YA, Boulet H, Koegl M, Bailer SM, Koszinowski U, Ideker T, Uetz P, Zimmer R, Haas J. Evolutionarily conserved herpesviral protein interaction networks. PLoS.Pathog. 2009;5(9):e1000570. doi: 10.1371/journal.ppat.1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs W, Granzow H, Klupp BG, Kopp M, Mettenleiter TC. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J.Virol. 2002;76(13):6729–6742. doi: 10.1128/JVI.76.13.6729-6742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs W, Klupp BG, Granzow H, Mettenleiter TC. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J.Virol. 2004;78(21):11879–11889. doi: 10.1128/JVI.78.21.11879-11889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W, Roizman B. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J.Virol. 1972;10(5):1044–1052. doi: 10.1128/jvi.10.5.1044-1052.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzow H, Klupp BG, Mettenleiter TC. The pseudorabies virus US3 protein is a component of primary and of mature virions. J.Virol. 2004;78(3):1314–1323. doi: 10.1128/JVI.78.3.1314-1323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302(5649):1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- Heine JW, Honess RW, Cassai E, Roizman B. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J.Virol. 1974;14(3):640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess RW, Roizman B. Proteins specified by herpes simplex virus. XI. Identification and relative molar rates of synthesis of structural and nonstructural herpes virus polypeptides in the infected cell. J.Virol. 1973;12(6):1347–1365. doi: 10.1128/jvi.12.6.1347-1365.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovasevic V, Liang L, Roizman B. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J.Virol. 2008;82(7):3311–3319. doi: 10.1128/JVI.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol.Cell. 2005;19(4):547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Kelly BJ, Fraefel C, Cunningham AL, Diefenbach RJ. Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res. 2009;145(2):173–186. doi: 10.1016/j.virusres.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Adaptors for clathrin-mediated traffic. Annu.Rev.Cell Dev.Biol. 1999;15:705–732. doi: 10.1146/annurev.cellbio.15.1.705. 705–732. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Bonifacino JS, Riezman H. Linking cargo to vesicle formation: receptor tail interactions with coat proteins. Curr.Opin.Cell Biol. 1997;9(4):488–495. doi: 10.1016/s0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- Klupp BG, Fuchs W, Granzow H, Nixdorf R, Mettenleiter TC. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J.Virol. 2002;76(6):3065–3071. doi: 10.1128/JVI.76.6.3065-3071.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klupp BG, Granzow H, Mundt E, Mettenleiter TC. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J.Virol. 2001;75(19):8927–8936. doi: 10.1128/JVI.75.19.8927-8936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko DH, Cunningham AL, Diefenbach RJ. The major determinant for addition of tegument protein pUL48 (VP16) to capsids in herpes simplex virus type 1 is the presence of the major tegument protein pUL36 (VP1/2) J.Virol. 2010;84(3):1397–1405. doi: 10.1128/JVI.01721-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Vittone V, Diefenbach E, Cunningham AL, Diefenbach RJ. Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology. 2008;378(2):347–354. doi: 10.1016/j.virol.2008.05.035. [DOI] [PubMed] [Google Scholar]
- Leege T, Granzow H, Fuchs W, Klupp BG, Mettenleiter TC. Phenotypic similarities and differences between UL37-deleted pseudorabies virus and herpes simplex virus type 1. J.Gen.Virol. 2009;90(Pt 7):1560–1568. doi: 10.1099/vir.0.010322-0. [DOI] [PubMed] [Google Scholar]
- Leuzinger H, Ziegler U, Schraner EM, Fraefel C, Glauser DL, Heid I, Ackermann M, Mueller M, Wild P. Herpes simplex virus 1 envelopment follows two diverse pathways. J.Virol. 2005;79(20):13047–13059. doi: 10.1128/JVI.79.20.13047-13059.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Fitzgerald K, Kurt-Jones E, Finberg R, Knipe DM. Herpesvirus tegument protein activates NF-kappaB signaling through the TRAF6 adaptor protein. Proc.Natl.Acad.Sci.U.S.A. 2008;105(32):11335–11339. doi: 10.1073/pnas.0801617105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis JS, Courtney RJ, Wills JW. Packaging determinants in the UL11 tegument protein of herpes simplex virus type 1. J.Virol. 2006;80(21):10534–10541. doi: 10.1128/JVI.01172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Haverlock S, Coller KE, Antinone SE, Pincetic A, Smith GA. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc.Natl.Acad.Sci.U.S.A. 2005;102(16):5832–5837. doi: 10.1073/pnas.0500803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Lee JI, Haverlock-Moyns S, Schober JM, Smith GA. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J.Virol. 2006;80(1):201–209. doi: 10.1128/JVI.80.1.201-209.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J. The abundance of the herpes simplex virus type 1 UL37 tegument protein in virus particles is closely controlled. J.Gen.Virol. 1997:78189–78194. doi: 10.1099/0022-1317-78-1-189. [DOI] [PubMed] [Google Scholar]
- McLauchlan J, Liefkens K, Stow ND. The herpes simplex virus type 1 UL37 gene product is a component of virus particles. J.Gen.Virol. 1994;75(Pt 8):2047–2052. doi: 10.1099/0022-1317-75-8-2047. [DOI] [PubMed] [Google Scholar]
- McNabb DS, Courtney RJ. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology. 1992;190(1):221–232. doi: 10.1016/0042-6822(92)91208-c. [DOI] [PubMed] [Google Scholar]
- Meckes DG, Jr, Wills JW. Dynamic Interactions of the UL16 Tegument Protein with the Capsid of Herpes Simplex Virus. J.Virol. 2007;81(23):13028–13036. doi: 10.1128/JVI.01306-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG, Jr, Wills JW. Structural Rearrangement within an Enveloped Virus upon Binding to the Host Cell. J.Virol. 2008;82(21):10429–10435. doi: 10.1128/JVI.01223-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: an update. Virus Res. 2009;143(2):222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- Mijatov B, Cunningham AL, Diefenbach RJ. Residues F593 and E596 of SV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology. 2007;368(1):26–31. doi: 10.1016/j.virol.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Murphy MA, Bucks MA, O'Regan KJ, Courtney RJ. The HSV-1 tegument protein pUL46 associates with cellular membranes and viral capsids. Virology. 2008;376(2):279–289. doi: 10.1016/j.virol.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Naldinho-Souto R, Browne H, Minson T. Herpes simplex virus tegument protein VP16 is a component of primary enveloped virions. J.Virol. 2006;80(5):2582–2584. doi: 10.1128/JVI.80.5.2582-2584.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Brown JC. Time-dependent transformation of the herpesvirus tegument. J.Virol. 2009;83(16):8082–8089. doi: 10.1128/JVI.00777-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Brown JC. Structure and capsid association of the herpesvirus large tegument protein UL36. J.Virol. 2010;84(18):9408–9414. doi: 10.1128/JVI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padula ME, Sydnor ML, Wilson DW. Isolation and preliminary characterization of herpes simplex virus 1 primary enveloped virions from the perinuclear space. J.Virol. 2009;83(10):4757–4765. doi: 10.1128/JVI.01927-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasdeloup D, Beilstein F, Roberts AP, McElwee M, McNab D, Rixon FJ. Inner tegument protein pUL37 of herpes simplex virus type 1 is involved in directing capsids to the trans-Golgi network for envelopment. J.Gen.Virol. 2010;91(Pt 9):2145–2151. doi: 10.1099/vir.0.022053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J.Virol. 2009;83(13):6610–6623. doi: 10.1128/JVI.02655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke K, Kieneke D, Wolfstein A, Michael K, Steffen W, Scholz T, Karger A, Sodeik B. Plus-and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS.Pathog. 2010;6(7):e1000991. doi: 10.1371/journal.ppat.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read GS, Patterson M. Packaging of the virion host shutoff (Vhs) protein of herpes simplex virus: two forms of the Vhs polypeptide are associated with intranuclear B and C capsids, but only one is associated with enveloped virions. J.Virol. 2007;81(3):1148–1161. doi: 10.1128/JVI.01812-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AE, Wills EG, Roller RJ, Ryckman BJ, Baines JD. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J.Virol. 2002;76(17):8939–8952. doi: 10.1128/JVI.76.17.8939-8952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AP, Abaitua F, O'Hare P, McNab D, Rixon FJ, Pasdeloup D. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J.Virol. 2009;83(1):105–116. doi: 10.1128/JVI.01032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B, Knipe DM, Whitley RJ. Herpes Simplex Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia, Pa: Lippincott, Williams and Wilkins; 2007. pp. 2501–2601. [Google Scholar]
- Rozen R, Sathish N, Li Y, Yuan Y. Virion-wide protein interactions of Kaposi's sarcoma-associated herpesvirus. J.Virol. 2008;82(10):4742–4750. doi: 10.1128/JVI.02745-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C, Korbel GA, Kattenhorn LM, Ploegh HL. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J.Virol. 2005;79(24):15582–15585. doi: 10.1128/JVI.79.24.15582-15585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz JB, Albright AG, Kinchington PR, Jenkins FJ. The UL37 protein of herpes simplex virus type 1 is associated with the tegument of purified virions. Virology. 1995;206(2):1055–1065. doi: 10.1006/viro.1995.1028. [DOI] [PubMed] [Google Scholar]
- Shelton LS, Pensiero MN, Jenkins FJ. Identification and characterization of the herpes simplex virus type 1 protein encoded by the UL37 open reading frame. J.Virol. 1990;64(12):6101–6109. doi: 10.1128/jvi.64.12.6101-6109.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KO. Relationship between the envelope and the infectivity of herpes simplex virus. Proc.Soc.Exp.Biol.Med. 1964:115814–115816. doi: 10.3181/00379727-115-29045. [DOI] [PubMed] [Google Scholar]
- Spear PG, Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J.Virol. 1972;9(1):143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-Filled HSV-1 capsids. Mol.Cell. 2007;26(4):479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Dong YA, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E, Haas J. Herpesviral protein networks and their interaction with the human proteome. Science. 2006;311(5758):239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- Vittone V, Diefenbach E, Triffett D, Douglas MW, Cunningham AL, Diefenbach RJ. Determination of interactions between tegument proteins of herpes simplex virus type 1. J.Virol. 2005;79(15):9566–9571. doi: 10.1128/JVI.79.15.9566-9571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Molloy SS, Thomas L, Liu G, Xiang Y, Rybak SL, Thomas G. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell. 1998;94(2):205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- Watanabe D, Ushijima Y, Goshima F, Takakuwa H, Tomita Y, Nishiyama Y. Identification of nuclear export signal in UL37 protein of herpes simplex virus type 2. Biochem.Biophys.Res.Commun. 2000;276(3):1248–1254. doi: 10.1006/bbrc.2000.3600. [DOI] [PubMed] [Google Scholar]
- Wild P, Engels M, Senn C, Tobler K, Ziegler U, Schraner EM, Loepfe E, Ackermann M, Mueller M, Walther P. Impairment of nuclear pores in bovine herpesvirus 1-infected MDBK cells. J.Virol. 2005;79(2):1071–1083. doi: 10.1128/JVI.79.2.1071-1083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfstein A, Nagel CH, Radtke K, Dohner K, Allan VJ, Sodeik B. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic. 2006;7(2):227–237. doi: 10.1111/j.1600-0854.2005.00379.x. [DOI] [PubMed] [Google Scholar]
- Xia D, Srinivas S, Sato H, Pesnicak L, Straus SE, Cohen JI. Varicella-zoster virus open reading frame 21, which is expressed during latency, is essential for virus replication but dispensable for establishment of latency. J.Virol. 2003;77(2):1211–1218. doi: 10.1128/JVI.77.2.1211-1218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J.Virol. 1999;73(4):3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]