Key Points
BOECs from VWD patients provide novel insight into the cellular mechanisms of the disease.
Abstract
Von Willebrand disease (VWD) is a heterogeneous bleeding disorder caused by decrease or dysfunction of von Willebrand factor (VWF). A wide range of mutations in the VWF gene have been characterized; however, their cellular consequences are still poorly understood. Here we have used a recently developed approach to study the molecular and cellular basis of VWD. We isolated blood outgrowth endothelial cells (BOECs) from peripheral blood of 4 type 1 VWD and 4 type 2 VWD patients and 9 healthy controls. We confirmed the endothelial lineage of BOECs, then measured VWF messenger RNA (mRNA) and protein levels (before and after stimulation) and VWF multimers. Decreased mRNA levels were predictive of plasma VWF levels in type 1 VWD, confirming a defect in VWF synthesis. However, BOECs from this group of patients also showed defects in processing, storage, and/or secretion of VWF. Levels of VWF mRNA and protein were normal in BOECs from 3 type 2 VWD patients, supporting the dysfunctional VWF model. However, 1 type 2M patient showed decreased VWF synthesis and storage, indicating a complex cellular defect. These results demonstrate for the first time that isolation of endothelial cells from VWD patients provides novel insight into cellular mechanisms of the disease.
Introduction
Von Willebrand disease (VWD) is the most common inherited bleeding disorder in humans, caused by mutations in the von Willebrand factor (VWF) gene. The disease is divided into 3 main types, based on the plasma phenotype: types 1 and 3 VWD are associated with partial or total quantitative deficiency of VWF, whereas type 2 VWD is associated with dysfunctional VWF. Type 1 VWD is the most common form of the disorder1 but can be difficult to diagnose due to heterogeneity of its clinical and laboratory phenotypes, which reflect heterogeneity in the genetic basis and pathogenic mechanisms of this disease.2 Moreover, the present classification system does not accurately predict response to desmopressin (DDAVP), which induces the release of VWF from endothelial cells (ECs) and is the principal therapeutic option for this subtype. Several large cohort studies have attempted to define the genetic basis of type 1 VWD, and numerous mutations have been reported. However, our understanding of the molecular and cellular basis of VWD is still limited.
VWF is a large plasma glycoprotein that mediates platelet adhesion to sites of endothelial damage and stabilizes circulating coagulation factor VIII (FVIII). It is produced mainly by ECs, where it is stored in Weibel-Palade bodies (WPBs) and from where it is released basally3 or upon stimulation.4 Endothelial VWF is also involved in the regulation of inflammation and angiogenesis,5 indicating that this multidomain protein exerts diverse roles in vascular biology. Until now, limited access to patients’ ECs has been a barrier to the understanding of the cellular pathophysiology of VWD. Studies on the cellular effects of VWF mutants have been performed using transfections of recombinant VWF in non-EC lines, which cannot entirely recapitulate the physiological endothelial milieu. For this reason, we isolated blood outgrowth endothelial cells (BOECs) from VWD patients to investigate the cellular mechanisms of VWD, an approach we have recently used to demonstrate the role of VWF in angiogenesis.5 Here we use BOECs to explore how molecular defects in VWF affect its synthesis, processing, storage, and/or secretion in patients with type 1 and type 2 VWD. We find that patients with similar plasma VWF phenotypes but different mutations in the VWF gene can have distinct cellular phenotypes, confirming the heterogeneity in this disease. These data demonstrate that BOECs offer a unique insight into the molecular basis of VWD.
Materials and methods
Unless otherwise specified, all reagents were purchased from Sigma-Aldrich (Gillingham, UK).
BOEC isolation
BOECs were isolated as described.5,6 The study was approved by the ethics committees of the Hammersmith, Queen Charlotte’s, and Royal Marsden hospitals; informed consent was obtained from all individuals in accordance with the Declaration of Helsinki.
Immunofluorescence (IF)
The endothelial identity of the BOECs was confirmed by IF analysis as described5,7 using a range of endothelial markers with antibodies to VWF (polyclonal: A0082; Dako, Ely, UK), vascular endothelial (VE)–cadherin (clone: 55-7H1; BD Biosciences, Oxford, UK), and Erg (clone: C20; Santa Cruz, Heidelberg, Germany). Secondary antibodies used were Alexa-fluor-488 (green) and Alexa-fluor 546 (red). Nuclei were visualized with TO-PRO-3 (blue). Coverslips were mounted with Fluoromount-G (eBioscience, Hatfield, UK) and imaged at room temperature with a Plan-Apochromat 63×, 1.4 N.A., Oil lens as described.5,7
Cell treatments
For measurement of VWF release, BOECs from VWD patients or healthy controls were seeded onto collagen-coated 6-well plates at a density of 10 000 cells per cm2. Forty-eight hours postseeding, the media were replaced with endothelial cell growth medium 2 (Lonza, Basel, Switzerland) without serum; confluent monolayers were stimulated with 100 ng/mL phorbol 12-myristate 13-acetate (PMA), or dimethylsulfoxide control, for 75 minutes at 37°C. Conditioned media and cell lysates were then collected. For IF, cells were seeded onto collagen-coated coverslips at 53 000 cells per cm2 and treated ± PMA, as described previously, 24 hours postseeding. Nine healthy donors were used to establish normal mean and range of VWF protein levels in BOEC lysates or supernatants; each VWD sample obtained from single isolations was tested in triplicate.
Enzyme-linked immunosorbent assay (ELISA) and immunoblotting
VWF concentrations were measured by ELISA.8 Immunoblotting of whole-cell lysates for propeptide and mature VWF were performed as described,8,9 using antibodies to VWF (Dako).
Real-time reverse transcription–polymerase chain reaction (RT-PCR)
RNA was prepared from confluent BOECs as previously described.5 Quantitative RT-PCR was carried out using the following primers to VWF (5′-GCAGTGGAGAACAGTGGTG-3′, 5′-GTGGCAGCGGGCAAAC-3′) and glyceraldehyde-3-phosphate dehydrogenase (5′-CAAGGTCATCCATGACAACTTTG-3′, 5′-GGGCCATCCACAGTCTTCTG-3′). Four healthy donors were used to establish normal mean and range; each VWD sample obtained from single isolations was tested in triplicate.
VWF gene sequencing
Forty-seven primer sets were used to amplify the 52 exons and flanking intronic regions of the VWF gene by standard techniques (primer sequences available on request). Sequencing was performed using BigDye terminator chemistry (v3.1) on a Prism_3130xl genetic analyzer (Applied Biosystems, Warrington, UK). All primers were designed based on the sequence of VWF NG_009072.1, to ensure specific amplification of VWF and not the VWF pseudogene, and were checked for single nucleotide polymorphisms using SNPCheck (https://ngrl.manchester.ac.uk/SNPCheckV3/snpcheck.htm). Sequence files were aligned to the reference sequence VWF NG_009072.1 using DNA variant analysis software (Mutation Surveyor v 3.2.4; SoftGenetics, State College, PA). All detected variants were referenced against the ISTH-SSC VWF Online Database (http://www.vwf.group.shef.ac.uk/).
Electron microscopy (EM)
EM was performed as described10 and viewed with a transmission electron microscope (Technai 12 G2 Spirit; FEI, Eindhoven, The Netherlands). Images were acquired using a Morada camera and iTEM software (Olympus SIS, Munster, Germany).
VWF multimer analysis
Multimer analysis of VWF from BOEC lysates or patients’ plasma was performed as previously described and processed with Photoshop software (Adobe Systems Europe Ltd, Maidenhead, UK) to permit optimal visualization of the multimeric banding pattern.11
Statistical analysis
All numerical data are expressed as means ± standard error of the mean (SEM). We analyzed data sets for significance with the Student t test, Wilcoxon signed rank, or Pearson correlation analysis. P values < .05 were considered statistically significant.
Results
VWD patients and healthy controls
Cells were isolated from 9 healthy controls (age, 45.2 ± 3.3, mean ± SEM; 6:3 male/female) and 4 type 1 VWD and 4 type 2 VWD patients. The patients’ laboratory characteristics are displayed in Table 1. The nucleotide changes in the VWF gene or the predicted amino acid changes are shown in Table 1 and supplemental Figure 1 (see the Blood Web site). The sequence variations in the patients’ VWF gene were all present in a heterozygous state; patient VWD-10 was a compound heterozygote for 2 changes.
Table 1.
VWD patient phenotypes
| Patient | VWD diagnosis | Age | Sex | VWF:Ag IU/dL | VWF:RCo IU/dL | FVIII:C IU/dL | VWF:Ag post-DDAVP IU/dL | VWF:RCo post-DDAVP IU/dL | FVIII:C post-DDAVP IU/dL | VWF gene | Blood group |
|---|---|---|---|---|---|---|---|---|---|---|---|
| VWD-10 | 1 | 58 | M | 7 | 8 | 18 | 7 | 10 | 18 | c.2438dupG and c.2281+4A>G (intronic) | B |
| VWD-8 | 1 | 61 | F | 15 | 14 | 68 | 28 | 47 | 283 | D75-G178 del | O |
| VWD-1 | 1 | 41 | F | 13 | 12 | 23 | 39 | N.A. | 94 | I1343V, V1360A, F1369I, S1378F, R1379C | O |
| VWD-12 | 1 | 43 | F | 22 | 15 | 59 | 90 | N.A. | 300 | Y1584C | O |
| VWD-13 | 2A | 28 | M | 38 | 14 | 50 | 142 | 81 | 245 | R1597W | O |
| VWD-2 | 2M | 49 | F | 26 | 9 | 48 | 84 | 36 | 150 | I1416T | A |
| VWD-3 | 2M | 27 | M | 26 | 6 | 42 | 101 | 48 | 83 | I1416T | N.A. |
| VWD-17 | 2M | 73 | M | 14 | <5 | 40 | N.A. | N.A. | N.A. | R1315C | B |
The parameters defining the clinical phenotypes of the patients studied are displayed. Patients’ plasma VWF:Ag and VWF:RCo at baseline and pre- and post-DDAVP treatment are shown. Sequence changes in the VWF gene of patients are indicated with the predicted amino acid change or, if intronic, the nucleotide change. Normal range for VWF:Ag and VWF:RCo = 50 to 150 IU/dL and for FVIII:C = 45 to 150 IU/dL.
N.A., not available.
Most of the patients had been treated previously with DDAVP. Type 1 VWD patients showed variable responses to DDAVP and in 1 case no response at all. DDAVP response data from 1 type 2M patient (VWD-17) were unavailable (Table 1).
BOEC isolation and culture
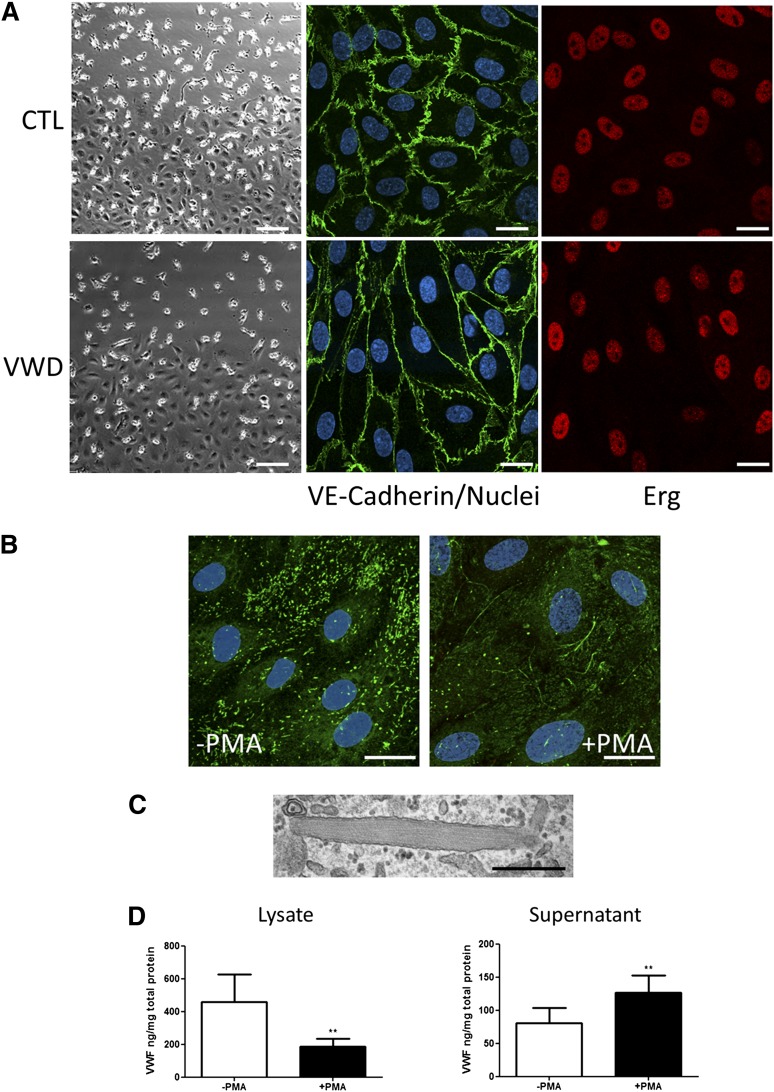
BOECs isolated and expanded in culture formed confluent monolayers with characteristic endothelial cobblestone morphology (Figure 1A). No morphologic difference in BOECs from healthy donors and VWD patients was observed (Figure 1 A-C). The endothelial nature of BOECs was confirmed by confocal IF microscopy, using EC markers including VE-cadherin, the transcription factor Erg, and intercellular adhesion molecule 2 (Figure 1A and Starke et al5). BOECs were negative for the common leukocyte antigen CD45 (Starke et al5 and data not shown).
Figure 1.
BOEC characterization and activation with PMA. (A) Representative images at the edge of a confluent colony of BOECs from a healthy control (top far left) and VWD patient (bottom far left). Scale bars represent 100 µm. Colonies of confluent BOECs were stained for VE-cadherin and Erg. Nuclei were stained with TO-PRO3 (scale bars represent 20 µm). (B) Representative image of healthy control BOECs stimulated with PMA or control to induce WPB exocytosis. BOECs were stained for IF with antibody to VWF and TO-PRO3 to identify the nuclei. Strings of VWF can be seen associated with the cell surface in the PMA-treated samples (scale bars represent 20 µm). (C) Representative EM image of a single WPB from a healthy control BOEC (scale bar represents 500 nm). (D) BOEC VWF was quantified by ELISA in total cellular lysate (left) or in cell culture supernatant (right) in the presence or absence of PMA stimulation (mean ± SEM from 9 healthy controls). **P < .01.
VWF expression, storage, and release in BOECs from healthy donors
VWF protein levels were measured in BOEC cell lysate and culture supernatant from 9 healthy donors, in the presence or absence of PMA, a potent secretagogue for WPB release (Figure 1D). The amount of intracellular VWF in BOECs varied significantly between donors (mean, 458 ng/mg; range, 59-1390). Basal VWF release was also variable (mean, 81 ng/mg; range, 7-214). As expected, PMA treatment caused a significant increase in the levels of VWF in the cell culture media (mean, 127 ng/mg; range, 27-229) and a significant decrease in VWF levels in the cellular lysate (mean, 186 ng/mg; range, 32-400) (Figure 1D). Repeated isolations from the same donor showed similar VWF levels, ± PMA stimulation (supplemental Figure 2), indicating good reproducibility within subject. Multimer gel analysis of BOECs from healthy donors showed the typical ladder of low–molecular weight (MW) and high-MW VWF, with a predominance of the lower-MW species, similar to reports from other cellular sources (Figure 2E).12,13 Confocal IF microscopy (Figure 1B) demonstrated that in healthy BOECs the majority of VWF staining was localized in WPBs, which appear as cigar-shaped rod-like structures, similar to those normally observed in other ECs in culture4 and tissues14,15 (see Figure 1B). EM confirmed the presence of rod-shaped WPBs with intraorganelle striations seen in longitudinal sections, reflecting the presence of VWF tubules (Figure 1C). Upon PMA stimulation, intracellular VWF staining was decreased, with a loss of WPB staining and the appearance of VWF strings at the cell surface (Figure 1B), as expected.16
Figure 2.
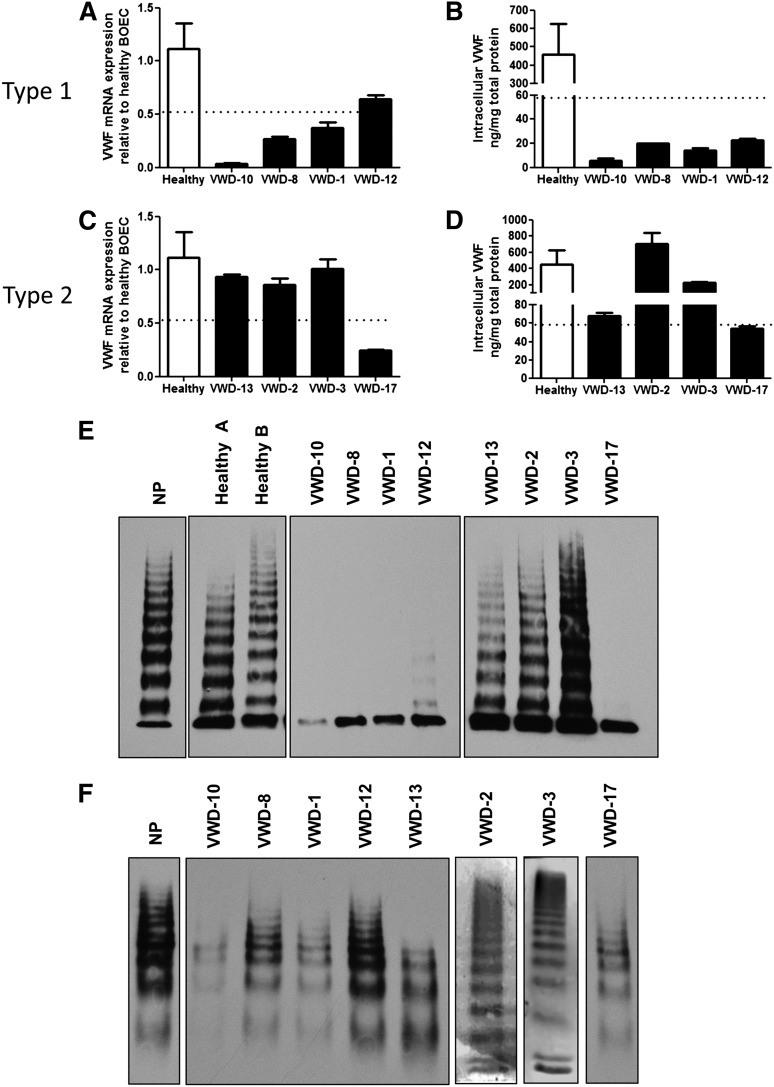
Expression of VWF in plasma and BOECs from type 1 and 2 VWD patients. (A,C) Real-time RT-PCR with primers specific for VWF was performed on RNA isolated from BOECs from 4 healthy controls and from type 1 (A) or type 2 (C) VWD patients. The VWF expression was normalized to glyceraldehyde-3-phosphate dehydrogenase, and all samples were normalized to 1 healthy control. The mean expression of controls was 1.12 (range, 0.52-1.70). (B,D) VWF protein was measured in total cellular lysates from healthy control BOECs (mean, 458 ng/mg total protein; range, 59-1390) and type 1 (B) or 2 (D) VWD BOECs with a VWF ELISA. In (A-D), the dotted line indicates the lowest value in healthy donors for mRNA and protein, as the lower limit of the normal range (mean ± SEM from 4 to 9 healthy controls, see “Materials and methods”). (E,F) VWF multimer analysis of VWF from BOEC lysates (E) or plasma (F).
BOEC phenotypes in type 1 VWD patients
Having established the cellular phenotype of VWF in BOECs from healthy donors, we next examined cells from type 1 VWD patients. BOECs were successfully isolated from all patients with VWD studied, with no apparent correlation between the isolation success and levels of VWF (data not shown).
Analysis of VWF messenger RNA (mRNA) levels by real-time RT-PCR from BOECs demonstrated a reduction of VWF expression in 3 of 4 type 1 patients compared with controls (n = 4) with considerable differences between patients (Figure 2A). Intracellular VWF protein was decreased in all type 1 VWD patients (Figure 2B) compared with controls (n = 9). Significant linear correlation was found between mRNA levels and plasma VWF:Antigen (Ag) levels (R = 0.9976, P = .0024; supplemental Figure 3A), suggesting that, in these type 1 VWD patients, plasma levels of VWF are linked to VWF endothelial gene transcription. Intracellular VWF protein levels showed a trend toward positive correlation with VWF:Ag levels (supplemental Figure 3B) as well as mRNA levels (supplemental Figure 3C). Detailed cellular analysis of each patient revealed complex and heterogeneous phenotypes, described below.
Patient VWD-10.
Plasma analysis demonstrated very low levels of VWF:Ag (7 IU/dL) and VWF:Ristocetin Cofactor assay (RCo) (8 IU/dL) (Table 1) with a very faint ladder of VWF multimers (Figure 2F). Because plasma VWF was detectable, although very low, and all multimeric forms were visible, this patient was classified as severe type 1 VWD. BOECs from this patient showed a particularly low expression of VWF mRNA (Figure 2A) and of intracellular VWF protein, nearly 10-fold below the normal range (Figure 2B), with undetectable release of VWF, either basal or upon agonist stimulation (Figure 3Ai). Multimer analysis of BOEC cell lysate revealed a weak band corresponding to the VWF dimer17 and absence of higher multimers (Figure 2E). Confocal IF microscopy demonstrated almost total absence of VWF-specific WPB staining (Figure 3Aii). Thus, the defect in this patient seems to be due to severely compromised endothelial synthesis of VWF. Sequencing of the VWF gene demonstrated that this patient is compound heterozygous for 2 different mutations: c.2438dupG, predicting loss of exon 17; and c.2281+4A>G, predicting incorporation of intron 17 sequence (Table 1; supplemental Figure 1).18
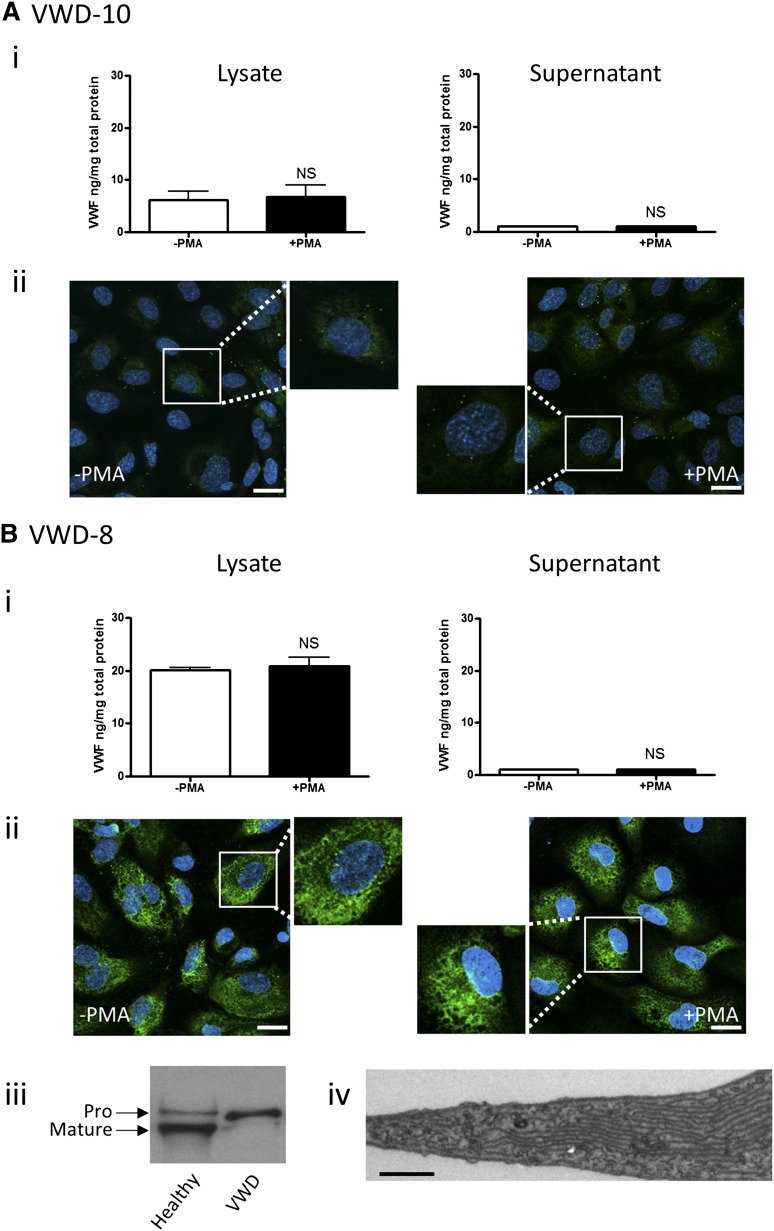
Figure 3.
BOEC characterization: type 1 VWD. Data from patient VWD-10 (A) and patient VWD-8 (B). (Ai,Bi) VWF was measured by ELISA in the total cell lysate (left) and cell culture supernatant (right) from BOECs isolated from the indicated patient, in the absence and presence of PMA stimulation (mean ± SEM from 3 replicates). (Aii,Bii) VWF expression from control or PMA-stimulated BOECs was visualized by confocal IF microscopy (scale bars represent 20 µm). Boxes show 2× magnified image. (Biii) Mature and propeptide-containing VWF was detected by western blotting from a healthy control or VWD patient VWD-8. Equal amounts of VWF were loaded per lane. (Biv) Representative image from EM analysis of BOECs from VWD patient VWD-8 shows extensive ER (scale bar represents 200 nm).
Patient VWD-8.
Slightly higher plasma VWF:Ag levels (15 IU/dL) and VWF:RCo (14 IU/dL) are characteristic of this patient (Table 1). Analysis of patient plasma showed a full range of VWF multimers (Figure 2F). BOECs from this patient showed significantly reduced VWF mRNA levels (Figure 2A) and intracellular VWF protein levels (Figure 2B), with undetectable basal or stimulated release of VWF (Figure 3Bi). Multimer analysis of BOEC cell lysate revealed a band corresponding to the VWF dimer and absence of higher multimers (Figure 2E). Confocal analysis of BOECs confirmed the presence of some intracellular VWF; however, the cells lacked characteristic WPB staining (Figure 3Bii), and no VWF strings were visible upon cellular stimulation with PMA. The meshlike pattern of intracellular staining of VWF suggested endoplasmic reticulum (ER) retention. Normal maturation of VWF requires cleavage of the propeptide by furin during its passage through the trans-Golgi, followed by the assembly of VWF into multimers.19 An ER retention phenotype would be compatible with the defect in multimerization and would result in low propeptide cleavage. We investigated the levels of immature VWF, containing the propeptide sequence, compared with mature, fully processed VWF. BOECs from patient VWD-8 showed an increased level of immature, propeptide-containing VWF (∼350 kDa), compared with control BOECs, and absence of mature VWF (∼250 kDa) (Figure 3Biii). EM confirmed the absence of WPB and showed an expansion of the ER (Figure 3Biv), previously shown to correlate with accumulated unprocessed VWF.20 Thus, in this type 1 VWD patient, a defect in VWF exit from the ER is combined with reduced mRNA synthesis. This patient was found to be heterozygous for the common exon 4-5 deletion in the VWF gene, which has been described in type 1 and type 3 VWD.21
Patient VWD-1.
VWF:Ag and VWF:RCo plasma levels are similarly reduced in this patient (13 and 12 IU/dL, respectively; see Table 1). Plasma analysis revealed a weak full range of VWF multimers (Figure 2F). In BOECs from this patient, VWF mRNA and protein levels were reduced compared with controls (Figure 2A-B), and no significant VWF release was observed upon stimulation with PMA (Figure 4Ai). Multimer analysis of BOEC cell lysate revealed that only dimeric VWF was detectable in the cell lysates, similar to the previous type 1 VWD BOECs (Figure 2E), suggesting a partial defect in multimerization. As in patient VWD-8, IF analysis of BOECs revealed some intracellular staining for VWF, mostly diffuse in an ER-like mesh, which was unchanged following PMA stimulation (Figure 4Aii). Closer inspection by EM analysis revealed some rare elongated WPBs (Figure 4Aiii). In this patient, the ratio between propeptide-containing and mature VWF appears increased compared with controls (Figure 4Aiv), suggesting abnormal VWF processing through the ER, although not as severe as in patient VWD-8. These data suggest that, as in the previous case, the defect in patient VWD-1 is due to reduced mRNA synthesis combined with abnormal processing. Sequencing of the VWF gene demonstrated that this patient is heterozygous for a gene conversion event between the normal VWF gene and the VWF pseudogene, predicting 5 amino acid substitutions in exon 28 of the VWF gene (I1343V, V1360A, F1369I, S1378F, and R1379C; Table 1 and supplemental Figure 1). All of these substitutions had been associated previously with VWD, although not as part of a single conversion event.
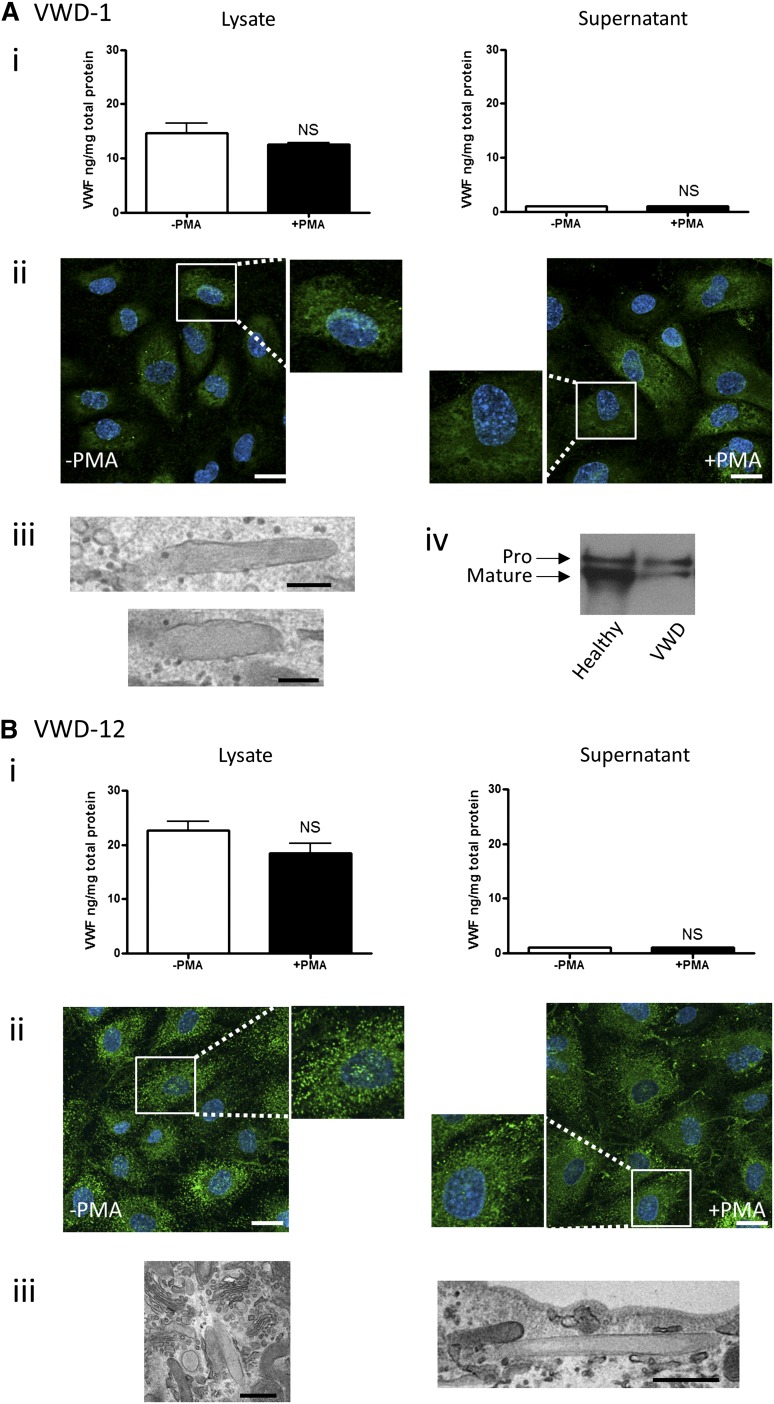
Figure 4.
BOEC characterization: type 1 VWD. Data from patient VWD-1 (A) and patient VWD-12 (B). (Ai,Bi) VWF was measured by ELISA in the total cell lysate (left) and cell culture supernatant (right) from BOECs isolated from the indicated patient in the absence and presence of PMA stimulation (mean ± SEM from 3 replicates). (Aii,Bii) VWF expression from control or PMA-stimulated BOECs was visualized by confocal IF microscopy (scale bars represent 20 µm). Boxes show 2× magnified image. EM analysis of BOECs show elongated (Aiii,Biii) or rounded WPBs (Biii) (scale bars represent 200 nm in Aiii and 500 nm in Biii). (Aiv) Mature and propeptide-containing VWF was detected by western blotting from a healthy control and VWD-1. Equal amounts of VWF were loaded per lane.
Patient VWD-12.
This type 1 VWD patient presents with higher VWF:Ag and VWF:RCo plasma levels (22 IU/dL and 15 IU/dL, respectively) compared with the other 3 studied. Plasma analysis revealed a full range of VWF multimers (Figure 2F). BOEC VWF mRNA levels were low but within the normal range, whereas BOEC VWF protein levels were below the limit of the normal range (Figure 2A-B). VWF release following PMA stimulation was below the limit of detection (Figure 4Bi). BOEC VWF multimer analysis showed a strong band representing the VWF dimer, as in other type 1 patients, as well as weak staining for intermediate MW forms (Figure 2E). IF analysis showed VWF staining, in punctate structures and in ER-like juxtanuclear material. Upon stimulation, a few VWF strings appeared at the cell surface, and the punctuate staining was slightly reduced (Figure 4Bii); however, intracellular VWF levels were not significantly decreased following PMA stimulation (Figure 4Bi, left panel). EM analysis showed the presence of a few cigar-shaped WPBs as well as abnormal, rounded WPBs (Figure 4Biii), possibly reflecting a defect of VWF assembly into tubules. Therefore, in this patient, the defect appears to be due to abnormal processing and/or storage combined with a small reduction in synthesis. Sequencing of the VWF gene showed that this patient is heterozygous for a common mutation causing an amino acid change in the A2 domain of VWF (Y1584C) (Table 1; supplemental Figure 1).
BOEC phenotypes in type 2 VWD patients
We next focused on type 2 VWD patients, where the defect is predominantly functional, although a quantitative VWF deficiency is also often seen.22 Of these, 1 was a type 2A (group 2) VWD (VWD-13), and 3 were type 2M VWD, of which 2 were members of the same family (VWD-2 and VWD-3) and the third (VWD-17) carried a different mutation (see Table 1). VWF mRNA levels in BOECs from these patients were normal in the first 3 but reduced in patient VWD-17 (Figure 2C) compared with 4 controls. Similarly, VWF intracellular levels were normal, despite considerable variability, in the first 3 patients and just below the normal range in patient VWD-17 (Figure 2D). There was no correlation between BOEC VWF mRNA or protein and plasma VWF:Ag (supplemental Figure 3D-E), suggesting that in these patients the levels of plasma VWF are not directly correlated to endothelial synthesis of VWF. In addition, mRNA did not correlate with VWF protein levels in BOECs (supplemental Figure 3F). Detailed cellular analysis of each patient is described below.
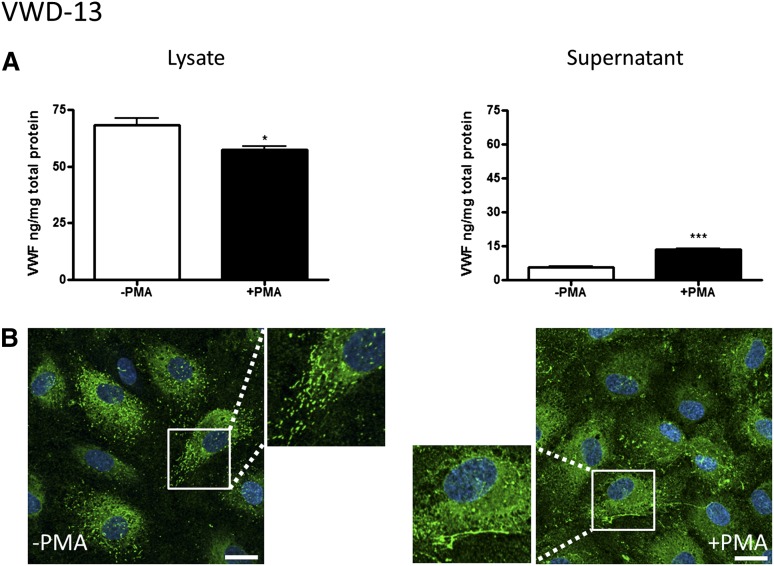
Patient VWD-13.
Patient VWD-13 has been diagnosed with type 2A VWD and carries a well-described mutation in the A2 domain of the VWF gene (R1597W) that causes enhanced cleavage by the protease ADAMTS13 (a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13).23,24 As expected, plasma VWF from this patient shows loss of high-MW multimers (Figure 2F). In this patient’s BOECs, levels of VWF protein were at the low end of the normal range (68 ng/mg total protein, Figure 2D). PMA induced a low but significant increase in VWF release (Figure 5A), as shown by the decrease in VWF intracellular levels and increase in VWF supernatant levels. Analysis of BOEC lysates showed a normal pattern of VWF multimers (Figure 2E), in agreement with the model that the loss of high-MW multimers in this patient’s plasma is due to enhanced cleavage by plasma ADAMTS13. Intracellular staining for VWF (Figure 5B) demonstrated some cigar-shaped staining, and upon cellular activation, VWF strings were visible on the cell surface. Therefore, the cellular data in patient VWD-13 show normal endothelial synthesis, processing, and storage of VWF and support the current notion of defective extracellular processing of VWF.
Figure 5.
BOEC characterization: type 2A VWD. (A-B) Data from patient VWD-13. (A) VWF was measured by ELISA in the total cell lysate (left) and cell culture supernatant (right) from BOECs isolated from a type 2A VWD patient in the absence and presence of PMA stimulation (mean ± SEM from 3 replicates). (B) VWF expression from control or PMA-stimulated BOECs was visualized by confocal IF microscopy (scale bars represent 20 µm). Boxes show 2× magnified image. *P < .05; ***P < .001.
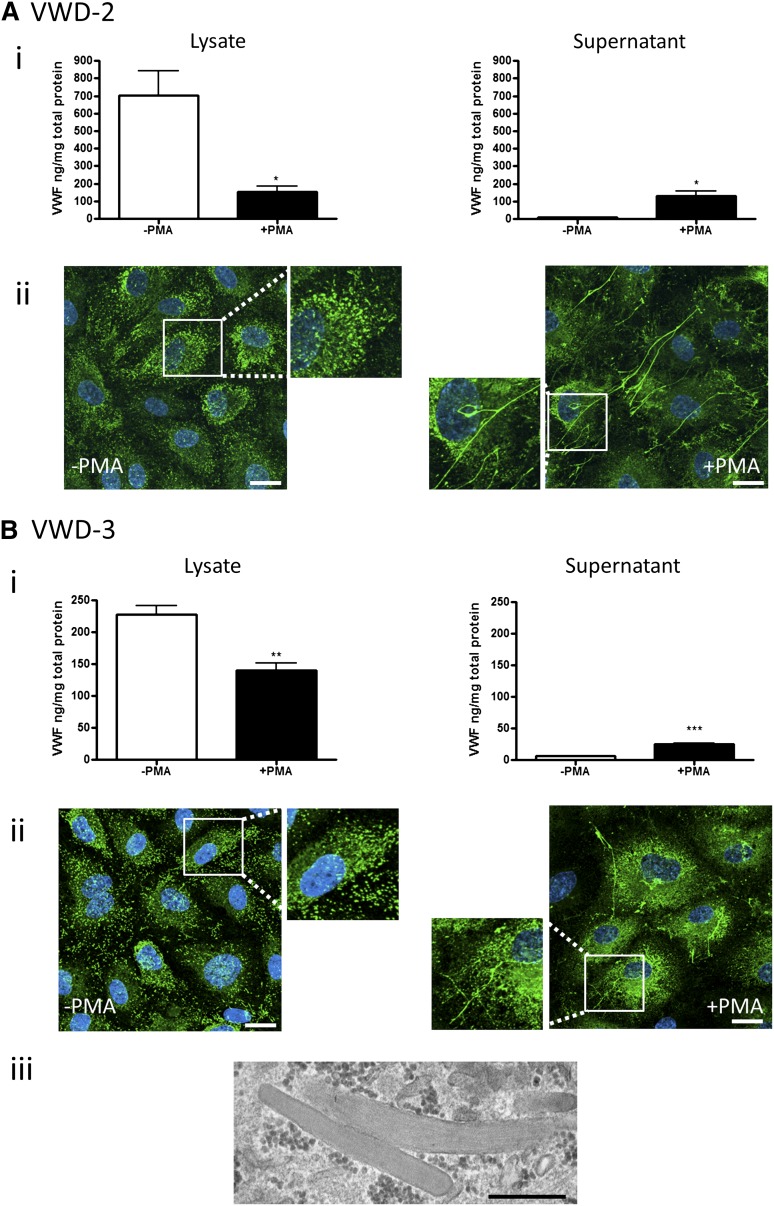
Patients VWD-2 and VWD-3.
Patients VWD-2 and VWD-3 are from the same family with type 2M VWD. The mutation in this family (I1416T) resides in the A1 domain of VWF and has been shown to reduce flow-based platelet adhesion.25 These patients show a very similar plasma phenotype, with normal multimeric composition (Figure 2F) and defective binding to glycoprotein Ib (see VWF:RCo, Table 1). In these patients, levels of BOEC VWF mRNA and protein were within the normal range (Figure 2C-D), and agonist stimulation induced significant increase in the release of VWF, with decrease in the cell lysate in both patients (Figure 6Ai,Bi). VWF multimers from BOECs appear normal, in line with the pattern seen in plasma (Figure 2E). Normal intracellular staining of VWF with cigar-shaped WPBs was observed by light microscopy and by EM (Figure 6Aii,Bii-iii); many VWF strings were released upon agonist stimulation (Figure 6Aii,Bii). Therefore, the cellular data suggest that this VWF mutation does not affect the ability of the cells to synthesize, store, and release cellular VWF upon stimulation and are consistent with a functional defect in VWF. As in the previous case, the reduction in plasma VWF levels does not appear to be linked to a defect in endothelial synthesis and storage.
Figure 6.
BOEC characterization: type 2M VWD family. Data from patient VWD-2 (A) and patient VWD-3 (B). (Ai,Bi) VWF was measured by ELISA in the total cell lysate (left) and cell culture supernatant (right) from BOECs isolated from the indicated patient in the absence and presence of PMA stimulation (mean ± SEM from 3 replicates). (Aii,Bii) VWF expression in control or PMA-stimulated BOECs was visualized by confocal IF microscopy (scale bars represent 20 µm). Boxes show 2× magnified image. (Biii) Representative image from EM analysis (scale bar represents 500 nm) of BOECs from patient VWD-3 illustrates the presence of morphologically normal WPBs. *P < .05; ** P < .01; ***P < .001.
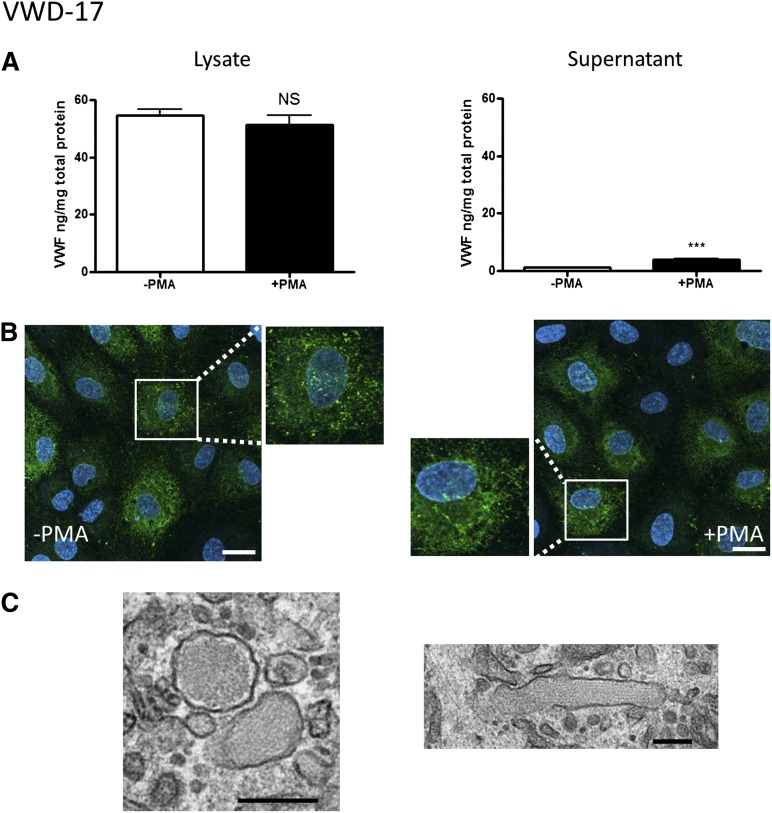
Patient VWD-17.
This patient showed low VWF:Ag and very low VWF:RCo plasma levels, with normal plasma VWF multimers, consistent with type 2M VWD (Table 1; Figure 2F). A mutation was identified in the A1 domain of the VWF gene (R1315C). At variance with the other type 2 patients described previously, VWF mRNA and, to a lesser extent, protein levels in BOECs from this patient were reduced (Figure 2C-D). A very small but significant increase in VWF release was detected upon agonist stimulation (Figure 7A). Surprisingly, repeated multimeric analysis of VWF from BOEC lysates (Figure 2E) showed only a strong single dimer band, and no higher multimeric forms, similar to the pattern observed in 3 type 1 VWD patients. Staining of BOECs for VWF demonstrated a combination of diffuse, ER-like, and punctate rather than cigar-shaped staining, with rare VWF strings on the cell surface upon cellular stimulation (Figure 7B). EM analysis confirmed the presence of abnormal, rounded WPBs (Figure 7C), alongside rare elongated WPBs, consistent with a defect in storage and release of VWF. Therefore, in this patient, despite the diagnosis of type 2M based on the plasma parameters, cellular defects in VWF synthesis and possibly processing and storage are also present.
Figure 7.
BOEC characterization: type 2M VWD. (A-C) Data from patient VWD-17. (A) VWF was measured by ELISA in the total cell lysate (left) and cell culture supernatant (right) from BOECs in the absence or presence of PMA stimulation (mean ± SEM from 3 replicates). (B) VWF expression from control and PMA-stimulated BOECs was visualized by confocal IF microscopy (scale bars represent 20 µm). Boxes show 2× magnified image. (C) EM analysis of BOECs (scale bars represent 200 nm) shows the presence of rounded WPBs (left), plus a rare elongated WPB (right). ***P < .001.
Discussion
Over the past 25 years, substantial progress has been made in the diagnosis and characterization of VWD, thanks mainly to advances in the genetic and molecular analysis of this common disease.2 However, understanding of the cellular defects of VWD has lagged behind because of the complexity of accessing the relevant cells from patients and the difficulties in establishing reliable in vitro models. Here we demonstrate that the study of ECs, the main source of plasma VWF,26 from patients with VWD, provides novel, important information on the pathogenesis of VWD and opens a new phase in the understanding of this complex disease.
BOECs (also called late outgrowth endothelial progenitor cells or endothelial colony-forming cells) are bone marrow–derived stem cells that have the ability to differentiate into mature ECs27 and have been implicated in postnatal vasculogenesis and endothelial repair at sites of endothelial damage.28 We have previously isolated BOECs from VWD patients5 and patients with chronic obstructive pulmonary disease29; another group had also previously reported isolation of BOECs from a patient with VWD.30 These studies indicate that despite the well-known heterogeneity of ECs from different vascular beds, BOECs represent a valid model to investigate the phenotype of ECs in patients.
Here we have used this approach to characterize the cellular defects of 4 patients with type 1 VWD and 4 with type 2 VWD, all with mutations in the VWF gene. The main results of this study are as follows: (1) Analysis of mRNA from BOECs shows that decreased VWF transcript levels are more common in patients with type 1 VWD compared with type 2 VWD. (2) In these type 1 VWD patients, levels of endothelial gene transcription are predictive of the levels of plasma VWF. (3) A combination of cellular defects may be present in patients with type 1 VWD. (4) Similar cellular phenotypes but variable intracellular VWF protein levels are found in members of the same type 2M family. (5) In some type 2M VWD patients, cellular defects in synthesis and/or storage are also present.
Access to patients’ BOECs allows the study of intrinsic cellular properties of the VWF protein, including trafficking, storage, and exocytosis, which cannot be observed using the standard plasma assays investigating VWF function. For example, lack or dysfunction of WPBs, as observed in BOECs from several type 1 VWD patients, may result in defective storage of other WPB components, such as regulators of inflammation and angiogenesis, which can themselves contribute to the disease.
Another unique feature of these studies is that they allow measurement of VWF mRNA levels in ECs, which has not been possible before, except for very rare cases where human umbilical vein ECs have been isolated from neonates with VWD.31,32 Whether the decrease in steady-state mRNA levels observed in type 1 VWD patients is the result of decreased gene transcription or rather a defect in mRNA stability needs to be determined. Mechanisms such as nonsense-mediated RNA decay, which is important for the removal of mutated mRNA, might be involved.33,34 In any case, the positive correlation between mRNA levels and intracellular and plasma VWF levels confirms the long-standing hypothesis that type 1 VWD is associated with a defective synthesis of VWF. Interestingly, recombinant interleukin 11 has been shown to increase mRNA expression for VWF in mild VWD and could be tested in patients’ BOECs prior to treatment.35 However, the type 1 VWD patients included in this study also showed defects in endothelial processing, storage, and/or secretion of VWF, suggesting a more complex cellular phenotype not simply linked to a decrease in VWF synthesis. Whether this applies to the majority of type 1 VWD cases will be determined by larger studies.
In all the patients selected for this study, we identified mutations in the VWF gene, a mix of common and rare mutations that give rise to a variety of clinical phenotypes. For several of these mutations (D75-G178del, Y1584C, R1597W, and R1315C), recombinant VWF protein has previously been expressed in nonendothelial models. Clearly, the study of endogenous mutant VWF in BOECs avoids potential artifacts that may be seen when expressing recombinant VWF in cell lines; it also avoids issues related to transfection efficiency or physiologically relevant expression levels and allows the study of mutants in the heterozygous state. Comparing the data on BOECs from VWD patients with the cellular data obtained with recombinant VWF with the same mutation provides a good indication of the relevance of such studies. From the examples discussed subsequently, there seems to be a broad agreement between the patients’ BOEC data and the data from recombinant models.
VWF multimer analysis on BOEC and plasma samples presented some intriguing results. In all the BOEC samples, VWF showed an abundance of lower-MW species compared with plasma VWF. In 3 out of 4 type 1 patients, only dimeric VWF was detectable. Several studies have shown that VWF from cell lysates is predominantly of the low-MW species, rather than the full range of multimers seen in cell culture supernatant or plasma.13,20,36 This has been suggested to be due to the rapid transport of multimerized VWF to the extracellular compartment after exit from the ER.13 Moreover, lower levels of VWF present in type 1 VWD may decrease the ability to detect higher-MW multimers. These reasons may explain the discrepancy between VWF multimers in BOEC lysates and plasma in these type 1 VWD patients.
Another possible reason for the presence of VWF dimers in BOECs may be a defect in ER processing of VWF, which appears to be common in the type 1 patients selected for this study, and most evident in VWD-8. In this patient, the mutation is a deletion of VWF exons 4 and 5, corresponding to the D1 region of the VWF propeptide,21,37 which is involved in multimerization. BOECs from this patient show defective processing of pre-pro VWF, which appears to be retained in the ER and fails to be properly packaged into WPBs. Thus, these data demonstrate for the first time an endothelial defect in VWF ER exit, linked to type 1 VWD, and are in line with the limited response to DDAVP treatment in this patient. Expression studies performed in HEK-293T cells had previously revealed that this mutation caused decreased secretion and defective multimerization of VWF, in line with our data from BOECs. No ER phenotype was reported in that study; however, dilation of the ER with associated impaired WPB packaging and cellular secretion has been seen in HEK-293T cells using other recombinant VWF mutants.12,20
Similar agreement between BOEC and recombinant model studies was found for the Y1584C mutation, identified in patient VWD-12 and previously reported in several type 1 VWD patients.37-41 Expression studies in different cell types consistently reported reduced secretion of this mutant VWF, but variable data on the levels of intracellular mutant VWF. This is in line with BOEC data from patient VWD-12, which showed reduced expression of VWF mRNA and protein, and decreased agonist-stimulated release of VWF, with the presence of malformed, possibly immature WPBs.
The results from the cellular studies in type 2 VWD patients also provide new insights into the disease. Three out of 4 of these patients showed normal levels of VWF mRNA, concordant with the model that type 2 VWD is mainly a defect of VWF function. As ever in VWD, the phenotypes can be complex. For example, in the 2 members of the same type 2M family, carrying a known mutation in the A1 domain (I1416T), the intracellular levels of VWF protein were quite different. Nevertheless, BOECs from both patients released normal levels of VWF (both with and without secretagogue activation). The reason for this variability is unclear and suggests that some cellular phenotypes may be influenced by other factors besides the VWF mutation. Plasma VWF levels are influenced by a wide variety of factors, including hormonal state, growth factors, inflammation, and coexisting diseases,2 which clearly cannot be captured by studies on patients’ ECs and may explain a discrepancy between normal endothelial synthesis and decreased plasma levels of VWF.
BOEC studies may also be useful in cases of difficult classification. Patient VWD-17 had an R1315C mutation in exon 28 of the VWF gene. Patients carrying this genetic defect have been classified as either type 1,40 2M,42 or 2A,43,44 reflecting the wide variety of plasma VWF phenotypes. Interestingly, reports of DDAVP response in these patients are also mixed, ranging from no response through partial to full response; in our case, the clinical data were not available. In patient VWD-17, the plasma phenotype was in line with a diagnosis of type 2M VWD (a ratio of VWF:Ag to VWF:RCo >0.7). Interestingly, multimer analysis of VWF in BOEC lysates from this patient showed only dimeric VWF, similar to the pattern observed when this mutant was expressed in COS-7 cells.44 Our study indicates that this mutation causes a defect in VWF processing; this, combined with the reduced levels of BOEC VWF, may be responsible for the decrease in intracellular higher-MW multimers. The data in this patient indicate a complex type 2 VWD phenotype with multiple cellular defects and suggest how BOEC studies can provide a useful additional tool for the categorization of patients with unclear phenotypes.
In conclusion, we have demonstrated a novel approach to the study of VWD, which has revealed a further level of complexity in the variety of endothelial cellular phenotypes that can be associated with VWF gene mutations. Future larger studies will provide significant new insight into the molecular and cellular basis of this disease and provide the foundation for improved management of this disease. Moreover, BOECs may be a useful tool to study the effect of novel treatments in VWD, prior to their use in the patient, based on the patient’s specific defect, leading the way to a personalized treatment of VWD.
Supplementary Material
Acknowledgments
The authors thank Prof Dorian Haskard, Dr Graeme Birdsey, Prof Justin Mason, and colleagues (Imperial College London) for their constructive criticism and support of this study; Prof Peter Barnes (Imperial College London) for support and encouragement; Dr Elspeth Payne and Dr Clare Wykes for help in establishing BOEC cultures; Dr Agata Nowak and Dr Susan Shapiro for help with VWF multimer gel analysis; and Dr Sarah Mangles for providing a healthy BOEC sample.
This work was supported by an intermediate fellowship from the British Heart Foundation (BHF) (FS/10/047/28393) (R.D.S.) and a project grant from the UK Medical Research Council (MRC) (G0600868). D.F.C., C.E.F.D., and K.J.H-L. were supported by UK MRC funding. T.A.J.M. was supported by a BHF fellowship (FS/06/064/21490). This work received support from the National Institute for Health Research Biomedical Research Centre Funding Scheme.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
There is an Inside Blood commentary on this article in this issue.
Authorship
Contribution: R.D.S. performed research, contributed to design of the study, carried out data interpretation, and wrote the paper; A.M.R. designed the study, carried out data interpretation, and wrote the paper; K.E.P. contributed to areas of research, data interpretation, and manuscript writing; K.J.H.-L. and C.E.F.D. performed EM analysis; J.A.C. performed genetic analysis of patients and contributed to data interpretation and manuscript writing; T.A.J.M. contributed to specific areas of research; C.M.M. recruited patients and contributed to manuscript writing; D.F.C. supervised EM analysis and contributed to data interpretation and manuscript writing; and M.A.L. recruited patients and contributed to design of the study and manuscript writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anna M. Randi, Vascular Sciences, National Heart and Lung Institute, Faculty of Medicine, Hammersmith Campus, Imperial College London, Du Cane Rd, London W12 0NN, United Kingdom; e-mail: a.randi@imperial.ac.uk.
References
- 1.Nichols WC, Ginsburg D. von Willebrand disease. Medicine (Baltimore) 1997;76(1):1–20. doi: 10.1097/00005792-199701000-00001. [DOI] [PubMed] [Google Scholar]
- 2.James PD, Lillicrap D. von Willebrand disease: clinical and laboratory lessons learned from the large von Willebrand disease studies. Am J Hematol. 2012;87(suppl 1):S4–S11. doi: 10.1002/ajh.23142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giblin JP, Hewlett LJ, Hannah MJ. Basal secretion of von Willebrand factor from human endothelial cells. Blood. 2008;112(4):957–964. doi: 10.1182/blood-2007-12-130740. [DOI] [PubMed] [Google Scholar]
- 4.Metcalf DJ, Nightingale TD, Zenner HL, et al. Formation and function of Weibel-Palade bodies. J Cell Sci. 2008;121(pt 1):19–27. doi: 10.1242/jcs.03494. [DOI] [PubMed] [Google Scholar]
- 5.Starke RD, Ferraro F, Paschalaki KE, et al. Endothelial von Willebrand factor regulates angiogenesis. Blood. 2011;117(3):1071–1080. doi: 10.1182/blood-2010-01-264507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ingram DA, Mead LE, Tanaka H, et al. Identification of a novel hierarchy of endothelial progenitor cells using human peripheral and umbilical cord blood. Blood. 2004;104(9):2752–2760. doi: 10.1182/blood-2004-04-1396. [DOI] [PubMed] [Google Scholar]
- 7.Birdsey GM, Dryden NH, Amsellem V, et al. Transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood. 2008;111(7):3498–3506. doi: 10.1182/blood-2007-08-105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Donnell J, Boulton FE, Manning RA, et al. Amount of H antigen expressed on circulating von Willebrand factor is modified by ABO blood group genotype and is a major determinant of plasma von Willebrand factor antigen levels. Arterioscler Thromb Vasc Biol. 2002;22(2):335–341. doi: 10.1161/hq0202.103997. [DOI] [PubMed] [Google Scholar]
- 9.Rosén H, Calafat J, Holmberg L, et al. Sorting of Von Willebrand factor to lysosome-related granules of haematopoietic cells. Biochem Biophys Res Commun. 2004;315(3):671–678. doi: 10.1016/j.bbrc.2004.01.106. [DOI] [PubMed] [Google Scholar]
- 10.Michaux G, Abbitt KB, Collinson LM, et al. The physiological function of von Willebrand’s factor depends on its tubular storage in endothelial Weibel-Palade bodies. Dev Cell. 2006;10(2):223–232. doi: 10.1016/j.devcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Baillod P, Affolter B, Kurt GH, et al. Multimeric analysis of von Willebrand factor by vertical sodium dodecyl sulphate agarose gel electrophoresis, vacuum blotting technology and sensitive visualization by alkaline phosphatase anti-alkaline phosphatase complex. Thromb Res. 1992;66(6):745–755. doi: 10.1016/0049-3848(92)90050-k. [DOI] [PubMed] [Google Scholar]
- 12.Wang JW, Valentijn KM, de Boer HC, et al. Intracellular storage and regulated secretion of von Willebrand factor in quantitative von Willebrand disease. J Biol Chem. 2011;286(27):24180–24188. doi: 10.1074/jbc.M110.215194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyons SE, Bruck ME, Bowie EJ, et al. Impaired intracellular transport produced by a subset of type IIA von Willebrand disease mutations. J Biol Chem. 1992;267(7):4424–4430. [PubMed] [Google Scholar]
- 14.Huber D, Cramer EM, Kaufmann JE, et al. Tissue-type plasminogen activator (t-PA) is stored in Weibel-Palade bodies in human endothelial cells both in vitro and in vivo. Blood. 2002;99(10):3637–3645. doi: 10.1182/blood.v99.10.3637. [DOI] [PubMed] [Google Scholar]
- 15.Kuo MC, Patschan D, Patschan S, et al. Ischemia-induced exocytosis of Weibel-Palade bodies mobilizes stem cells. J Am Soc Nephrol. 2008;19(12):2321–2330. doi: 10.1681/ASN.2007111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J, Roth R, Heuser JE, et al. Integrin alpha(v)beta(3) on human endothelial cells binds von Willebrand factor strings under fluid shear stress. Blood. 2009;113(7):1589–1597. doi: 10.1182/blood-2008-05-158584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner DD. Cell biology of von Willebrand factor. Annu Rev Cell Biol. 1990;6:217–246. doi: 10.1146/annurev.cb.06.110190.001245. [DOI] [PubMed] [Google Scholar]
- 18.Cutler JA, Hadonou M, Madan B, Laffan MA, Mitchell MJ. The identification of a repeat novel frameshift mutation in the VWF gene with an unpredicted effect [abstract]. J Thromb Haemost. 2009;7(suppl s2):PP-WE-635.
- 19.Sadler JE. von Willebrand factor assembly and secretion. J Thromb Haemost. 2009;7(suppl 1):24–27. doi: 10.1111/j.1538-7836.2009.03375.x. [DOI] [PubMed] [Google Scholar]
- 20.Michaux G, Hewlett LJ, Messenger SL, et al. Analysis of intracellular storage and regulated secretion of 3 von Willebrand disease-causing variants of von Willebrand factor. Blood. 2003;102(7):2452–2458. doi: 10.1182/blood-2003-02-0599. [DOI] [PubMed] [Google Scholar]
- 21.Sutherland MS, Cumming AM, Bowman M, et al. A novel deletion mutation is recurrent in von Willebrand disease types 1 and 3. Blood. 2009;114(5):1091–1098. doi: 10.1182/blood-2008-08-173278. [DOI] [PubMed] [Google Scholar]
- 22.Federici AB, Canciani MT, Forza I, et al. Ristocetin cofactor and collagen binding activities normalized to antigen levels for a rapid diagnosis of type 2 von Willebrand disease—single center comparison of four different assays. Thromb Haemost. 2000;84(6):1127–1128. [PubMed] [Google Scholar]
- 23.Hassenpflug WA, Budde U, Obser T, et al. Impact of mutations in the von Willebrand factor A2 domain on ADAMTS13-dependent proteolysis. Blood. 2006;107(6):2339–2345. doi: 10.1182/blood-2005-04-1758. [DOI] [PubMed] [Google Scholar]
- 24.Pruss CM, Golder M, Bryant A, et al. Use of a mouse model to elucidate the phenotypic effects of the von Willebrand factor cleavage mutants, Y1605A/M1606A and R1597W. J Thromb Haemost. 2012;10(5):940–950. doi: 10.1111/j.1538-7836.2012.04675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKinnon TA, Nowak AA, Cutler J, et al. Characterisation of von Willebrand factor A1 domain mutants I1416N and I1416T: correlation of clinical phenotype with flow-based platelet adhesion. J Thromb Haemost. 2012;10(7):1409–1416. doi: 10.1111/j.1538-7836.2012.04760.x. [DOI] [PubMed] [Google Scholar]
- 26.Bowie EJ, Solberg LA, Jr, Fass DN, et al. Transplantation of normal bone marrow into a pig with severe von Willebrand’s disease. J Clin Invest. 1986;78(1):26–30. doi: 10.1172/JCI112560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin Y, Weisdorf DJ, Solovey A, et al. Origins of circulating endothelial cells and endothelial outgrowth from blood. J Clin Invest. 2000;105(1):71–77. doi: 10.1172/JCI8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Critser PJ, Yoder MC. Endothelial colony-forming cell role in neoangiogenesis and tissue repair. Curr Opin Organ Transplant. 2010;15(1):68–72. doi: 10.1097/MOT.0b013e32833454b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paschalaki K, Starke RD, Mercado N, et al. Endothelial colony forming cells (ECFC) are senescent and dysfunctional in COPD due to reduced sirtuin-1 levels. Heart. 2011;97(20):e7. [Google Scholar]
- 30.Berber E, James PD, Hough C, et al. An assessment of the pathogenic significance of the R924Q von Willebrand factor substitution. J Thromb Haemost. 2009;7(10):1672–1679. doi: 10.1111/j.1538-7836.2009.03551.x. [DOI] [PubMed] [Google Scholar]
- 31.Ewenstein BM, Inbal A, Pober JS, et al. Molecular studies of von Willebrand disease: reduced von Willebrand factor biosynthesis, storage, and release in endothelial cells derived from patients with type I von Willebrand disease. Blood. 1990;75(7):1466–1472. [PubMed] [Google Scholar]
- 32.Federici AB, de Groot PG, Moia M, et al. Type I von Willebrand disease, subtype ‘platelet low’: decreased platelet adhesion can be explained by low synthesis of von Willebrand factor in endothelial cells. Br J Haematol. 1993;83(1):88–93. doi: 10.1111/j.1365-2141.1993.tb04636.x. [DOI] [PubMed] [Google Scholar]
- 33.Platè M, Duga S, Baronciani L, et al. Premature termination codon mutations in the von Willebrand factor gene are associated with allele-specific and position-dependent mRNA decay. Haematologica. 2010;95(1):172–174. doi: 10.3324/haematol.2009.012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castaman G, Platè M, Giacomelli SH, et al. Alterations of mRNA processing and stability as a pathogenic mechanism in von Willebrand factor quantitative deficiencies. J Thromb Haemost. 2010;8(12):2736–2742. doi: 10.1111/j.1538-7836.2010.04060.x. [DOI] [PubMed] [Google Scholar]
- 35.Ragni MV, Jankowitz RC, Chapman HL, et al. A phase II prospective open-label escalating dose trial of recombinant interleukin-11 in mild von Willebrand disease. Haemophilia. 2008;14(5):968–977. doi: 10.1111/j.1365-2516.2008.01827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayadas TN, Wagner DD. Vicinal cysteines in the prosequence play a role in von Willebrand factor multimer assembly. Proc Natl Acad Sci USA. 1992;89(8):3531–3535. doi: 10.1073/pnas.89.8.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodeve AC. The genetic basis of von Willebrand disease. Blood Rev. 2010;24(3):123–134. doi: 10.1016/j.blre.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 38.O’Brien LA, James PD, Othman M, et al. Association of Hemophilia Clinic Directors of Canada. Founder von Willebrand factor haplotype associated with type 1 von Willebrand disease. Blood. 2003;102(2):549–557. doi: 10.1182/blood-2002-12-3693. [DOI] [PubMed] [Google Scholar]
- 39.Pruss CM, Golder M, Bryant A, et al. Pathologic mechanisms of type 1 VWD mutations R1205H and Y1584C through in vitro and in vivo mouse models. Blood. 2011;117(16):4358–4366. doi: 10.1182/blood-2010-08-303727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.James PD, Notley C, Hegadorn C, et al. The mutational spectrum of type 1 von Willebrand disease: results from a Canadian cohort study. Blood. 2007;109(1):145–154. doi: 10.1182/blood-2006-05-021105.. [DOI] [PubMed] [Google Scholar]
- 41.Bowen DJ, Collins PW. An amino acid polymorphism in von Willebrand factor correlates with increased susceptibility to proteolysis by ADAMTS13. Blood. 2004;103(3):941–947. doi: 10.1182/blood-2003-05-1505. [DOI] [PubMed] [Google Scholar]
- 42.Casaña P, Martínez F, Espinós C, et al. Search for mutations in a segment of the exon 28 of the human von Willebrand factor gene: new mutations, R1315C and R1341W, associated with type 2M and 2B variants. Am J Hematol. 1998;59(1):57–63. doi: 10.1002/(sici)1096-8652(199809)59:1<57::aid-ajh11>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 43.Goodeve A, Eikenboom J, Castaman G, et al. Phenotype and genotype of a cohort of families historically diagnosed with type 1 von Willebrand disease in the European study, Molecular and Clinical Markers for the Diagnosis and Management of Type 1 von Willebrand Disease (MCMDM-1VWD). Blood. 2007;109(1):112–121. doi: 10.1182/blood-2006-05-020784. [DOI] [PubMed] [Google Scholar]
- 44.Ribba AN, Hilbert L, Lavergne JM, et al. The arginine-552-cysteine (R1315C) mutation within the A1 loop of von Willebrand factor induces an abnormal folding with a loss of function resulting in type 2A-like phenotype of von Willebrand disease: study of 10 patients and mutated recombinant von Willebrand factor. Blood. 2001;97(4):952–959. doi: 10.1182/blood.v97.4.952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.