Abstract
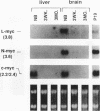
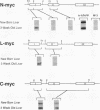
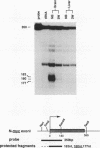
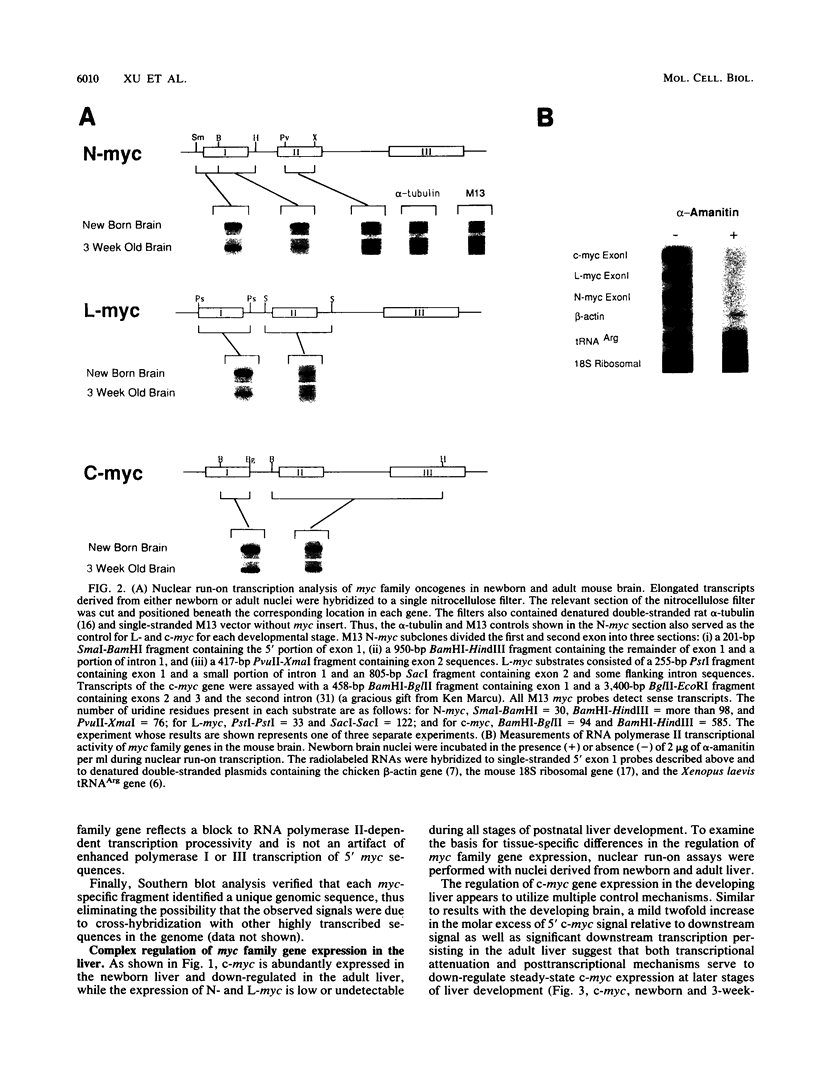
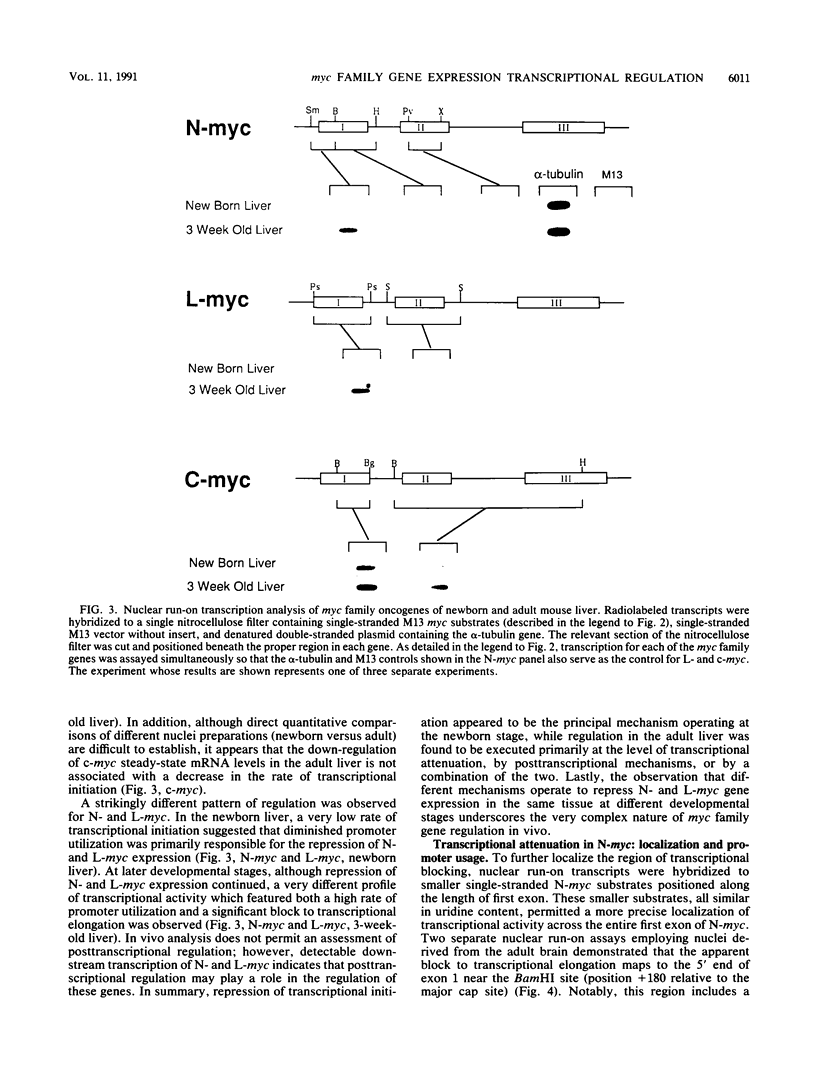
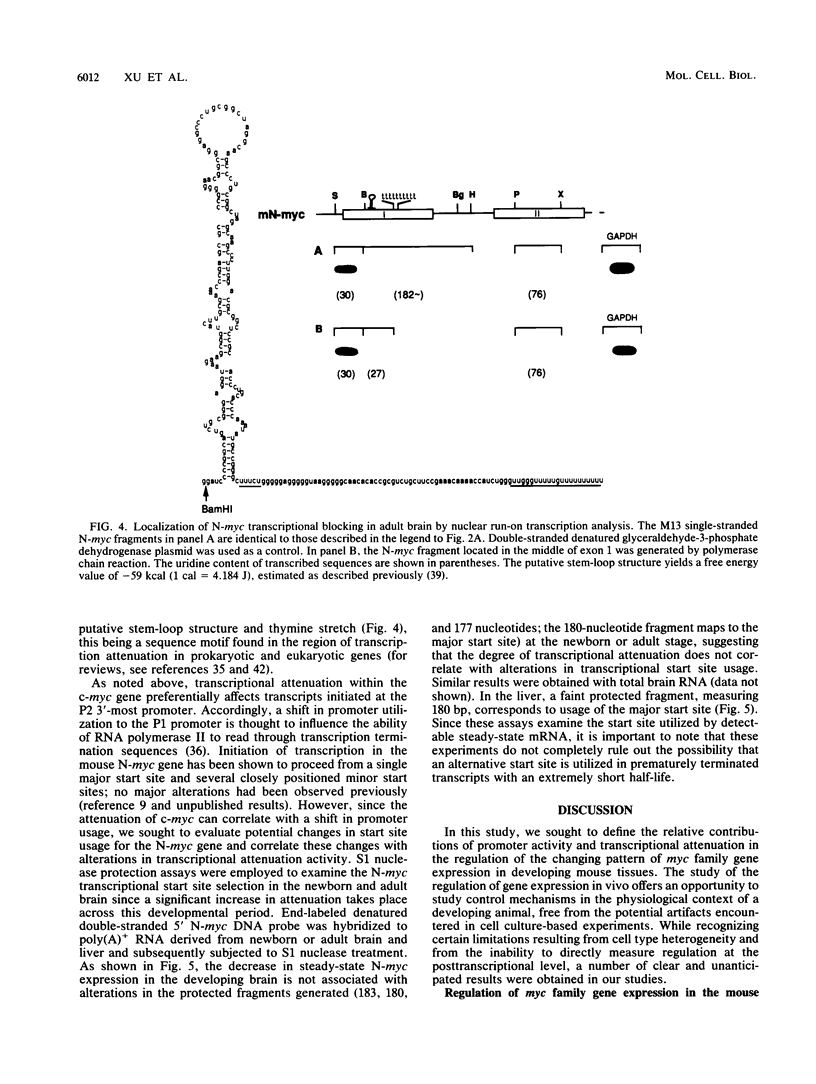
myc family genes (c-, N-, and L-myc) have been shown to be differentially expressed with respect to tissue type and developmental stage. To define and compare the regulatory mechanisms governing their differential developmental expression, we examined the transcriptional regulation of each myc family member during murine postnatal brain and liver development. Nuclear run-on transcription assays demonstrated that both the rate of transcriptional initiation and the degree of transcriptional blocking contribute in a complex manner to the regulation of all three genes. During postnatal brain development, the relative contribution of each transcriptional control mechanism to the regulation of myc family gene expression was found to be different for each gene. For instance, while modulation of transcriptional attenuation did not appear to contribute to the down-regulation of L-myc expression, attenuation was found to be the dominant mechanism by which steady-state N-myc mRNA levels were down-regulated. Different transcriptional strategies were found to be employed in newborn versus adult developing liver for repression of N- and L-myc expression. Undetectable steady-state N- and L-myc mRNA levels in newborn liver were associated with a very low rate of transcriptional initiation, whereas the lack of N- and L-myc expression at the adult stage was accompanied by a high rate of initiation and a striking degree of transcriptional attenuation. Transcriptional attenuation in the N-myc gene was found to map to a region encoding a potential stem-loop structure followed by a thymine tract within the first exon and was not dependent on the use of a specific transcriptional start site.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Babiss L. E., Friedman J. M. Regulation of N-myc gene expression: use of an adenovirus vector to demonstrate posttranscriptional control. Mol Cell Biol. 1990 Dec;10(12):6700–6708. doi: 10.1128/mcb.10.12.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Chung J., Sussman D. J., Zeller R., Leder P. The c-myc gene encodes superimposed RNA polymerase II and III promoters. Cell. 1987 Dec 24;51(6):1001–1008. doi: 10.1016/0092-8674(87)90586-1. [DOI] [PubMed] [Google Scholar]
- Clayton D. F., Darnell J. E., Jr Changes in liver-specific compared to common gene transcription during primary culture of mouse hepatocytes. Mol Cell Biol. 1983 Sep;3(9):1552–1561. doi: 10.1128/mcb.3.9.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Lopata M. A., MacDonald R. J., Cowan N. J., Rutter W. J., Kirschner M. W. Number and evolutionary conservation of alpha- and beta-tubulin and cytoplasmic beta- and gamma-actin genes using specific cloned cDNA probes. Cell. 1980 May;20(1):95–105. doi: 10.1016/0092-8674(80)90238-x. [DOI] [PubMed] [Google Scholar]
- Cleveland J. L., Huleihel M., Bressler P., Siebenlist U., Akiyama L., Eisenman R. N., Rapp U. R. Negative regulation of c-myc transcription involves myc family proteins. Oncogene Res. 1988;3(4):357–375. [PubMed] [Google Scholar]
- DePinho R. A., Legouy E., Feldman L. B., Kohl N. E., Yancopoulos G. D., Alt F. W. Structure and expression of the murine N-myc gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1827–1831. doi: 10.1073/pnas.83.6.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePinho R. A., Schreiber-Agus N., Alt F. W. myc family oncogenes in the development of normal and neoplastic cells. Adv Cancer Res. 1991;57:1–46. doi: 10.1016/s0065-230x(08)60994-x. [DOI] [PubMed] [Google Scholar]
- Derman E., Krauter K., Walling L., Weinberger C., Ray M., Darnell J. E., Jr Transcriptional control in the production of liver-specific mRNAs. Cell. 1981 Mar;23(3):731–739. doi: 10.1016/0092-8674(81)90436-0. [DOI] [PubMed] [Google Scholar]
- Eick D., Berger R., Polack A., Bornkamm G. W. Transcription of c-myc in human mononuclear cells is regulated by an elongation block. Oncogene. 1987;2(1):61–65. [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Expression of normal and translocated c-myc alleles in Burkitt's lymphoma cells: evidence for different regulation. EMBO J. 1989 Jul;8(7):1965–1972. doi: 10.1002/j.1460-2075.1989.tb03602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrlander P. D., Piechaczyk M., Marcu K. B. Chromatin structure of the murine c-myc locus: implications for the regulation of normal and chromosomally translocated genes. EMBO J. 1985 Dec 1;4(12):3195–3202. doi: 10.1002/j.1460-2075.1985.tb04065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzburg I., Behar L., Givol D., Littauer U. Z. The nucleotide sequence of rat alpha-tubulin: 3'-end characteristics, and evolutionary conservation. Nucleic Acids Res. 1981 Jun 25;9(12):2691–2697. doi: 10.1093/nar/9.12.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Soellner C., Scholz I. Characterization of a cloned ribosomal fragment from mouse which contains the 18S coding region and adjacent spacer sequences. Nucleic Acids Res. 1979 Apr;6(4):1351–1369. doi: 10.1093/nar/6.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson S., Sundaresan S., Jin P., Francke U., Asker C., Sümegi J., Klein G., Sejersen T. Chromosome localization and expression pattern of Lmyc and Bmyc in murine embryonal carcinoma cells. Oncogene. 1988 Dec;3(6):679–685. [PubMed] [Google Scholar]
- Kakkis E., Prehn J., Calame K. An active chromatin structure acquired by translocated c-myc genes. Mol Cell Biol. 1986 Apr;6(4):1357–1361. doi: 10.1128/mcb.6.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. Intrinsic sites of transcription termination and pausing in the c-myc gene. Mol Cell Biol. 1988 Oct;8(10):4389–4394. doi: 10.1128/mcb.8.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal G. W., Armstrong B. C., Battey J. F. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol Cell Biol. 1990 Aug;10(8):4180–4191. doi: 10.1128/mcb.10.8.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal G., Birrer M., Way J., Nau M., Sausville E., Thompson C., Minna J., Battey J. Multiple mechanisms for transcriptional regulation of the myc gene family in small-cell lung cancer. Mol Cell Biol. 1988 Aug;8(8):3373–3381. doi: 10.1128/mcb.8.8.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legouy E., DePinho R., Zimmerman K., Collum R., Yancopoulos G., Mitsock L., Kriz R., Alt F. W. Structure and expression of the murine L-myc gene. EMBO J. 1987 Nov;6(11):3359–3366. doi: 10.1002/j.1460-2075.1987.tb02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell T. J., Weinberg F., Morris P. W., Roeder R. G., Rutter W. J. Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science. 1970 Oct 23;170(3956):447–449. doi: 10.1126/science.170.3956.447. [DOI] [PubMed] [Google Scholar]
- Loose D. S., Shaw P. A., Krauter K. S., Robinson C., Englard S., Hanson R. W., Gluecksohn-Waelsch S. Trans regulation of the phosphoenolpyruvate carboxykinase (GTP) gene, identified by deletions in chromosome 7 of the mouse. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5184–5188. doi: 10.1073/pnas.83.14.5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A., Smith R. K., Tesfaye A., Achacoso P., Dildrop R., Rosenberg N., Alt F. W. Mechanism of endogenous myc gene down-regulation in E mu-N-myc tumors. Mol Cell Biol. 1991 Jan;11(1):440–444. doi: 10.1128/mcb.11.1.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango S. E., Schuler G. D., Steele M. E., Cole M. D. Germ line c-myc is not down-regulated by loss or exclusion of activating factors in myc-induced macrophage tumors. Mol Cell Biol. 1989 Aug;9(8):3482–3490. doi: 10.1128/mcb.9.8.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu K. B. Regulation of expression of the c-myc proto-oncogene. Bioessays. 1987 Jan;6(1):28–32. doi: 10.1002/bies.950060108. [DOI] [PubMed] [Google Scholar]
- McBurney M. W., Jones-Villeneuve E. M., Edwards M. K., Anderson P. J. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature. 1982 Sep 9;299(5879):165–167. doi: 10.1038/299165a0. [DOI] [PubMed] [Google Scholar]
- Nepveu A., Levine R. A., Campisi J., Greenberg M. E., Ziff E. B., Marcu K. B. Alternative modes of c-myc regulation in growth factor-stimulated and differentiating cells. Oncogene. 1987;1(3):243–250. [PubMed] [Google Scholar]
- Nisen P. D., Zimmerman K. A., Cotter S. V., Gilbert F., Alt F. W. Enhanced expression of the N-myc gene in Wilms' tumors. Cancer Res. 1986 Dec;46(12 Pt 1):6217–6222. [PubMed] [Google Scholar]
- Penn L. J., Brooks M. W., Laufer E. M., Land H. Negative autoregulation of c-myc transcription. EMBO J. 1990 Apr;9(4):1113–1121. doi: 10.1002/j.1460-2075.1990.tb08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Bressler P., Kelly K. Two distinct mechanisms of transcriptional control operate on c-myc during differentiation of HL60 cells. Mol Cell Biol. 1988 Feb;8(2):867–874. doi: 10.1128/mcb.8.2.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990 Jun;5(6):777–785. [PubMed] [Google Scholar]
- Spencer C. A., LeStrange R. C., Novak U., Hayward W. S., Groudine M. The block to transcription elongation is promoter dependent in normal and Burkitt's lymphoma c-myc alleles. Genes Dev. 1990 Jan;4(1):75–88. doi: 10.1101/gad.4.1.75. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Watt R., Marcu K. B. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983 Jun 2;303(5916):401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- Thiele C. J., Deutsch L. A., Israel M. A. The expression of multiple proto-oncogenes is differentially regulated during retinoic acid induced maturation of human neuroblastoma cell lines. Oncogene. 1988 Sep;3(3):281–288. [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tso J. Y., Sun X. H., Kao T. H., Reece K. S., Wu R. Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: genomic complexity and molecular evolution of the gene. Nucleic Acids Res. 1985 Apr 11;13(7):2485–2502. doi: 10.1093/nar/13.7.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., Bishop J. M. DNA sequences that mediate attenuation of transcription from the mouse protooncogene myc. Proc Natl Acad Sci U S A. 1989 Jan;86(2):505–509. doi: 10.1073/pnas.86.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. Transcription attenuation. J Biol Chem. 1988 Jan 15;263(2):609–612. [PubMed] [Google Scholar]