Abstract
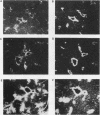
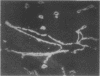
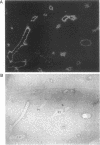
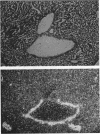
We previously demonstrated that glutamine synthetase (GS) and ornithine aminotransferase (OAT) mRNAs are expressed in the mouse liver acinus preferentially in pericentral hepatocytes, that is, those immediately surrounding terminal central veins (A.L. Bennett, K.E. Paulson, R.E. Miller, and J.E. Darnell, Jr., J. Cell Biol. 105:1073-1085, 1987, and F.C. Kuo, W.L. Hwu, D. Valle, and J.E. Darnell, Jr., Proc. Natl. Acad. Sci. USA, in press). We now show that hepatocytes surrounding large collecting hepatic veins but not portal veins also express these two mRNAs. The pericentral hepatocytes are the most distal hepatocytes with respect to acinar blood flow, whereas this is not necessarily the case for hepatocytes next to the large collecting hepatic veins. This result implies that it is contact with some hepatic venous element which signals positional expression. In an effort to induce conditions that change relationships between hepatocytes and blood vessels, regenerating liver was studied. After surgical removal of two-thirds or more of the liver, there was no noticeable change in GS or OAT expression in the remaining liver tissue during regeneration. However, treatment with carbon tetrachloride (CCl4), which specifically kills pericentral hepatocytes, completely removed GS- and OAT-containing cells and promptly halted hepatic transcription of GS. Repair of CCl4 damage is associated with invasion of inflammatory and scavenging cells, which remove dead hepatocytes to allow regrowth. Only when hepatocytes resumed contact with pericentral veins were the pretreatment levels of OAT and GS mRNA and high levels of GS transcription restored.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belayew A., Tilghman S. M. Genetic analysis of alpha-fetoprotein synthesis in mice. Mol Cell Biol. 1982 Nov;2(11):1427–1435. doi: 10.1128/mcb.2.11.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett A. L., Paulson K. E., Miller R. E., Darnell J. E., Jr Acquisition of antigens characteristic of adult pericentral hepatocytes by differentiating fetal hepatoblasts in vitro. J Cell Biol. 1987 Sep;105(3):1073–1085. doi: 10.1083/jcb.105.3.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger H. J., Gebhardt R., Mayer C., Mecke D. Different capacities for amino acid transport in periportal and perivenous hepatocytes isolated by digitonin/collagenase perfusion. Hepatology. 1989 Jan;9(1):22–28. doi: 10.1002/hep.1840090105. [DOI] [PubMed] [Google Scholar]
- Chang T. W., Goldberg A. L. The metabolic fates of amino acids and the formation of glutamine in skeletal muscle. J Biol Chem. 1978 May 25;253(10):3685–3693. [PubMed] [Google Scholar]
- Clayton D. F., Harrelson A. L., Darnell J. E., Jr Dependence of liver-specific transcription on tissue organization. Mol Cell Biol. 1985 Oct;5(10):2623–2632. doi: 10.1128/mcb.5.10.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. M., Chung E. Y., Darnell J. E., Jr Gene expression during liver regeneration. J Mol Biol. 1984 Oct 15;179(1):37–53. doi: 10.1016/0022-2836(84)90305-x. [DOI] [PubMed] [Google Scholar]
- GRISHAM J. W. A morphologic study of deoxyribonucleic acid synthesis and cell proliferation in regenerating rat liver; autoradiography with thymidine-H3. Cancer Res. 1962 Aug;22:842–849. [PubMed] [Google Scholar]
- Gaasbeek Janzen J. W., Gebhardt R., ten Voorde G. H., Lamers W. H., Charles R., Moorman A. F. Heterogeneous distribution of glutamine synthetase during rat liver development. J Histochem Cytochem. 1987 Jan;35(1):49–54. doi: 10.1177/35.1.2878950. [DOI] [PubMed] [Google Scholar]
- Gaasbeek Janzen J. W., Lamers W. H., Moorman A. F., de Graaf A., Los J. A., Charles R. Immunohistochemical localization of carbamoyl-phosphate synthetase (ammonia) in adult rat liver; evidence for a heterogeneous distribution. J Histochem Cytochem. 1984 Jun;32(6):557–564. doi: 10.1177/32.6.6373912. [DOI] [PubMed] [Google Scholar]
- Gebhardt R. Altered acinar distribution of glutamine synthetase and different growth response of cultured enzyme-positive and -negative hepatocytes after partial hepatectomy. Cancer Res. 1990 Jul 15;50(14):4407–4410. [PubMed] [Google Scholar]
- Gebhardt R., Burger H. J., Heini H., Schreiber K. L., Mecke D. Alterations of hepatic enzyme levels and of the acinar distribution of glutamine synthetase in response to experimental liver injury in the rat. Hepatology. 1988 Jul-Aug;8(4):822–830. doi: 10.1002/hep.1840080421. [DOI] [PubMed] [Google Scholar]
- Gebhardt R., Cruise J., Houck K. A., Luetteke N. C., Novotny A., Thaler F., Michalopoulos G. K. Differential effect of growth factors on growth stimulation and phenotypic stability of glutamine-synthetase-positive and -negative hepatocytes in primary culture. Differentiation. 1986;33(1):45–55. doi: 10.1111/j.1432-0436.1986.tb00409.x. [DOI] [PubMed] [Google Scholar]
- Gebhardt R. Different proliferative activity in vitro of periportal and perivenous hepatocytes. Scand J Gastroenterol Suppl. 1988;151:8–18. doi: 10.3109/00365528809095909. [DOI] [PubMed] [Google Scholar]
- Goodenough D. A., Revel J. P. A fine structural analysis of intercellular junctions in the mouse liver. J Cell Biol. 1970 May;45(2):272–290. doi: 10.1083/jcb.45.2.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta Y., Kato T. Ornithine aminotransferase distribution in ocular tissues and retinas of cat and mouse. Invest Ophthalmol Vis Sci. 1989 Jun;30(6):1173–1177. [PubMed] [Google Scholar]
- Häussinger D. Glutamine metabolism in the liver: overview and current concepts. Metabolism. 1989 Aug;38(8 Suppl 1):14–17. doi: 10.1016/0026-0495(89)90133-9. [DOI] [PubMed] [Google Scholar]
- Iannaccone P. M. Fractal geometry in mosaic organs: a new interpretation of mosaic pattern. FASEB J. 1990 Mar;4(5):1508–1512. doi: 10.1096/fasebj.4.5.2307328. [DOI] [PubMed] [Google Scholar]
- Jungermann K., Katz N. Functional hepatocellular heterogeneity. Hepatology. 1982 May-Jun;2(3):385–395. doi: 10.1002/hep.1840020316. [DOI] [PubMed] [Google Scholar]
- Kafatos F. C., Jones C. W., Efstratiadis A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 1979 Nov 24;7(6):1541–1552. doi: 10.1093/nar/7.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. F., Darnell J. E., Jr Mouse glutamine synthetase is encoded by a single gene that can be expressed in a localized fashion. J Mol Biol. 1989 Jul 5;208(1):45–56. doi: 10.1016/0022-2836(89)90086-7. [DOI] [PubMed] [Google Scholar]
- Kuo C. F., Paulson K. E., Darnell J. E., Jr Positional and developmental regulation of glutamine synthetase expression in mouse liver. Mol Cell Biol. 1988 Nov;8(11):4966–4971. doi: 10.1128/mcb.8.11.4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechnagel R. O., Glende E. A., Jr Carbon tetrachloride hepatotoxicity: an example of lethal cleavage. CRC Crit Rev Toxicol. 1973 Nov;2(3):263–297. doi: 10.3109/10408447309082019. [DOI] [PubMed] [Google Scholar]
- Schöls L., Mecke D., Gebhardt R. Reestablishment of the heterogeneous distribution of hepatic glutamine synthetase during regeneration after CCl4-intoxication. Histochemistry. 1990;94(1):49–54. doi: 10.1007/BF00266789. [DOI] [PubMed] [Google Scholar]
- Windmueller H. G., Spaeth A. E. Respiratory fuels and nitrogen metabolism in vivo in small intestine of fed rats. Quantitative importance of glutamine, glutamate, and aspartate. J Biol Chem. 1980 Jan 10;255(1):107–112. [PubMed] [Google Scholar]