Abstract
A novel dopamine-plus-HSA (human serum albumin) approach was developed to functionalize iron oxide nanoparticles (IONPs), yielding nanoconjugates that are highly efficient in labeling various types of cell lines, which was demonstrated by in vivo MR imaging on xenograft and focal cerebral ischemia models.
There has been fast progress in cell transplantation research, which is believed to have a revolutionary impact on the current therapeutics.1–3 With an on-going worldwide research interest, a new cycle of breakthroughs in this realm is expected. In the past two decades, many iron oxide nanoparticle (IONP) formulas have been developed and were utilized as contrast agents for magnetic resonance imaging (MRI). The application of MRI in conjunction with IONP based cell labeling techniques has provided an excellent solution4,5 to the non-invasive tracking of implanted cells in the host organism. One major challenge for this technique, however, is to induce a sufficient amount of particles into the cells to compensate the dilution effect caused by cell division, while not affecting the normal cellular functions.4 Several approaches have been raised and investigated, with the most utilized one being the employment of commercial Feridex in combination with transfection agents.6,7 However, lacking a universal formula, tedious trial and error tests have to be made with individual cell lines to achieve optimal transfection.4
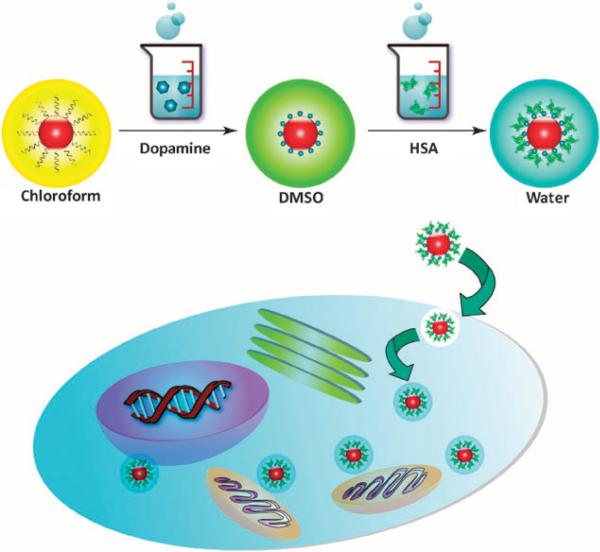
In view of these challenges, herein we report a novel IONP based formula that can safely and effectively label versatile cell types, without the application of any excipients. In brief, 15 nm IONPs made by pyrolysis were surface modified with dopamine8,9 and were subsequently adsorbed with one layer of human serum albumin (HSA) (Fig. 1). The yielded nanostructures were stable in PBS and many other buffer solutions, with a hydrodynamic size of around 30 nm (Fig. S1, Supporting Information †), which is close to the assumption of 15 nm core plus 5–6 nm dopamine/HSA coating. Such HSA-IONPs were evaluated to have a r2 relaxivity of 314.5 mM–1 s–1, 2.5 times higher than that of Feridex (123.6 mM–1 s–1, Fig. S2, Supporting Information †). Such enhancement in relaxivity was observed previously by other groups and was attributed to a better crystallinity of particles made from pyrolysis.9 Cellular toxicity assay was performed with representative cell lines, and within the studied concentration range (from 5 to 150 μg Fe/ml), no adverse effect was found on cell growth of all types (Fig. S2, Supporting Information †).
Fig. 1.
Schematic illustration of the particle formation and cell labeling.
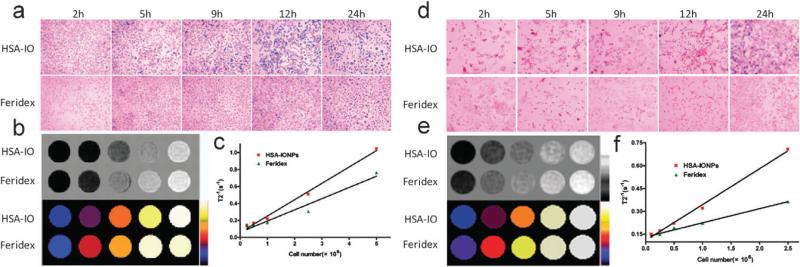
To evaluate the cell labeling efficiency, HSA-IONPs were incubated with various cell lines at a concentration of 20 μg Fe/ml for 24 h without any excipient. At selective time intervals (2, 5, 9, 12, and 24 h), the incubation was stopped and Prussian blue staining was performed to depict the particle internalization status (Fig. 2 and Fig. S3–S6, Supporting Information †). Aside from macrophages, which showed decent uptake at early time points and reached a plateau at 12 h, all the other cells displayed a steady and slow particle internalization profile. For all the studied cell lines, over 95% of the cells were found labeled with HSA-IONPs after 24 h incubation. On the contrary, incubation of Feridex under the same conditions resulted in suboptimal or even marginal particle uptake.
Fig. 2.
(a) Prussian blue staining of the HSA-IONPs/Feridex labeled macrophages. (b) Upper panel: the MRI phantoms of the iron-laden macrophages; bottom panel: colour map of the phantom results. (c) T2–1 vs. macrophage number curve. The linear correlation suggests good MRI sensitivity of the iron-laden cells. (d) Prussian blue staining of the HSA-IONP/Feridex labeled neural stem cells (NSCs). (e) The black-white and colour maps of the MRI phantom results of the iron-laden NSCs. (f) T2–1 vs. cell number curve of the iron-laden NSCs.
The particle internalization of Feridex is mainly mediated by phagocytosis.10 For nonphagocytic cells, Feridex need to be pre-complexed with positively charged transfection agents, otherwise, the cell labeling is insufficient, as observed in our studies. Although detailed mechanism of HSA-IONP uptake is unclear, it is obviously not species-specific, and is likely also through endocytosis/phagocytosis, as indicated by the TEM analysis results (Fig. S7, Supporting Information †), which found populations of particles within endosomes/lysosoms. The overall zeta potential of HSA-IONPs is slightly negative (–9.46 ± 1.86 mv in PBS, Fig. S8, Supporting Information †), but it is possible that the HSA sheath was incomplete, and the intermediate dopamine coating was partially exposed which facilitated the cellular uptake. Another possibility is that the uptake was mediated by HSA coating. It has been reported previously that HSA can interact with glycoprotein (gp60) receptor (albondin) and/or SPARC (secreted protein acid and rich in cysteine) that are expressed on various types of cell surface and lead to facile transportation.11,12
Phantom studies were performed to evaluate the visibility of the iron-laden cells under MRI. After 24 h incubation, the macrophages or neural stems cells (NSCs) were redispersed in agarose gel at various cell concentrations (0.1, 0.25, 0.5, 1, 2.5, 5×106 cell in 300 μL agarose gel) and were subjected to MR imaging. Perfect liner relationships were found between T2–1 and cell numbers (R2 = 0.993 and 0.995, respectively, Fig. 2c and f), suggesting good MRI sensitivity of the labeled cells. Owing to a higher magnetic moment and a better internalization rate, the HSA-IONP group outperformed the Feridex group with higher curve slopes in both cases. To further evaluate the labeled cell visibility in vivo, 5 × 104 NSCs labeled with either HSA-IONPs or Feridex, were grafted into the right (HSA-IONPs, R) and left (Feridex, L) strata of mice, and T2-weighted MRI scans were performed 6 h later (Fig. S9, Supporting Information †). It was found that the cell implanted regions were visualized as hypointensities on T2-weighted images, with the signal drop percentages being 44.7% (R) vs. 24.2% (L) compared to the normal tissues.
Next, we used MRI to study the in vivo dynamics of HSA-IONP labeled macrophages (Raw 264.7) on two models on mice. The first one is a xenograft U87MG model. The design of the experiment is based on the knowledge that macrophages play an important role in tumor cell proliferation and angiogenesis, and are heavily recruited to the tumor site during tumor progression.13 About 5 × 106 HSA-IONP labeled macrophages were intravaneously (i.v.) injected, and starting from the 6 h time point, hypointensities were found at the tumor site, with a signal reduction of 49.9% (Fig. 3). The contrast was gradually faded out over time, and the signal loss was restored to 23.2% at 1 d and 17.8% at 2 d, and finally became undetectable after one week. Such contrast loss could be a result of the macrophage division and particle degradation. Also, it may be contributed by a dilution effect of the newly generated tumor mass as well as the endogenous macrophages that were recruited to the tumor area.14 Positive Prussian blue staining on the tumor sections confirmed the remains of IONPs at the tumor sites after 7 d (Fig. S10, Supporting Information †). Further F4/80 and Prussian blue double staining showed good co-localization, suggesting that the particles stayed within the macrophages after day 7. On the other hand, iron-laden cells were found frequently in the leading edge of CD31-positive vessel lumen, indicating that the macrophages might be actively involved in the tumor neovascularization.13
Fig. 3.
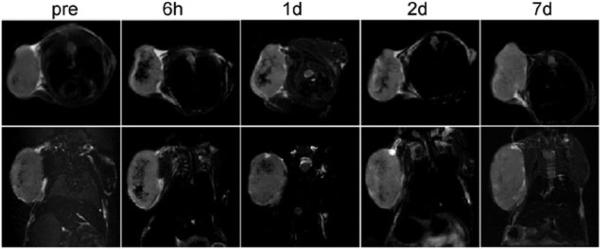
Using MRI to track the i.v. injected iron-laden macrophages on xenograft U87MG model. Top row: the axial view. Bottom row: the coronal view.
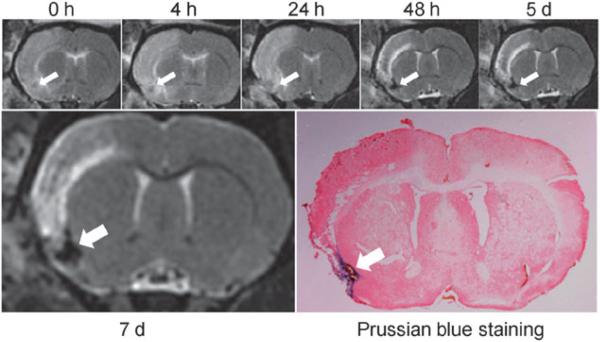
The dynamics of such HSA-IONP labeled macrophages were also investigated on a focal cerebral ischemia model. It was previously reported that ischemic brain injury may induce an inflammatory process that contributes to the delayed progression of the injury,15,16 and macrophage activation is part of the process.17,18 The stroke model was induced following the previously published protocols.19 One day after the stroke onset, 5 × 106 HSA-IONP labeled macrophages were i.v. injected, and MR scans were performed to longitudinally track the in vivo cell fate. Shown in Fig. 4, the ischemic lesion appeared as hyperintensities on T2-weighted images; on the contrary, the HSA-IONP labeled macrophages were displayed as hypointensities, which started accumulating at the stroke region from the 48 h time point, inducing a dramatic signal loss (87.3%). The contrast effect lasted over the remaining observation period, with the signal reductions being 88.5% and 91.7% on the 5th and 7th day. It was previously reported that the macrophage activation occurs 1–5 days post the stroke onset20,21 and may persist for a long period of time,15,22 which are in good accord with our observations.
Fig. 4.
Longitudinal MRI studies and Prussian blue staining on a rat focal cerebral ischemia model injected with iron-laden macrophages.
In summary, we have developed a dopamine-plus-HSA approach to modify IONPs. This approach allows a facile preparation of nanoconstructions that are non-toxic and are efficient in labeling various cell types in a nonspecific manner. Compared with conventional Feridex nanoparticles, these HSA-IONPs show better T2 contrast. More importantly, unlike Feridex, which need to complex with toxic transfection agents to label nonphagocytic cells, no excipients are required in HSA-IONP cell labeling. Further evaluation of this novel iron oxide nanoparticle formula for stem cell labeling and tracking in vivo is currently in progress.
Supplementary Material
Acknowledgments
This research was supported by Intramural Research Programs of the National Institute of Biomedical Imaging and Bioengineering (NIBIB).
Footnotes
Electronic supplementary information (ESI) available: The procedure of nanoparticle preparation and cell labeling. The experimental details of in vitro and in vivo characterizations. See DOI: 10.1039/b917195a
Notes and references
- 1.Bordignon C, Carlo-Stella C, Colombo MP, De Vincentiis A, Lanata L, Lemoli RM, Locatelli F, Olivieri A, Rondelli D, Zanon P, Tura S. Haematologica. 1999;84:1110–1149. [PubMed] [Google Scholar]
- 2.Kapp M, Rasche L, Einsele H, Grigoleit GU. Curr. Opin. Hematol. 2009;16:437–443. doi: 10.1097/MOH.0b013e32832f57d4. [DOI] [PubMed] [Google Scholar]
- 3.Amos TA, Gordon MY. Cell Transplant. 1995;4:547–569. doi: 10.1177/096368979500400605. [DOI] [PubMed] [Google Scholar]
- 4.Modo M, Hoehn M, Bulte JW. Mol. Imaging. 2005;4:143–164. doi: 10.1162/15353500200505145. [DOI] [PubMed] [Google Scholar]
- 5.Bulte JW, Kraitchman DL. NMR Biomed. 2004;17:484–499. doi: 10.1002/nbm.924. [DOI] [PubMed] [Google Scholar]
- 6.Daldrup-Link HE, Rudelius M, Oostendorp RA, Settles M, Piontek G, Metz S, Rosenbrock H, Keller U, Heinzmann U, Rummeny EJ, Schlegel J, Link TM. Radiology. 2003;228:760–767. doi: 10.1148/radiol.2283020322. [DOI] [PubMed] [Google Scholar]
- 7.van den Bos EJ, Wagner A, Mahrholdt H, Thompson RB, Morimoto Y, Sutton BS, Judd RM, Taylor DA. Cell Transplant. 2003;12:743–756. doi: 10.3727/000000003108747352. [DOI] [PubMed] [Google Scholar]
- 8.Xie J, Chen K, Lee HY, Xu C, Hsu AR, Peng S, Chen X, Sun S. J. Am. Chem. Soc. 2008;130:7542–7543. doi: 10.1021/ja802003h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie J, Xu C, Kohler N, Hou Y, Sun S. Adv. Mater. 2007;19:3648–3652. [Google Scholar]
- 10.Raynal I, Prigent P, Peyramaure S, Najid A, Rebuzzi C, Corot C. Invest. Radiol. 2004;39:56–63. doi: 10.1097/01.rli.0000101027.57021.28. [DOI] [PubMed] [Google Scholar]
- 11.Desai N, Trieu V, Yao Z, Louie L, Ci S, Yang A, Tao C, De T, Beals B, Dykes D, Noker P, Yao R, Labao E, Hawkins M, Soon-Shiong P. Clin. Cancer Res. 2006;12:1317–1324. doi: 10.1158/1078-0432.CCR-05-1634. [DOI] [PubMed] [Google Scholar]
- 12.Porter PL, Sage EH, Lane TF, Funk SE, Gown AM. J. Histochem. Cytochem. 1995;43:791–800. doi: 10.1177/43.8.7622842. [DOI] [PubMed] [Google Scholar]
- 13.Condeelis J, Pollard JW. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Murdoch C, Giannoudis A, Lewis CE. Blood. 2004;104:2224–2234. doi: 10.1182/blood-2004-03-1109. [DOI] [PubMed] [Google Scholar]
- 15.Weber R, Wegener S, Ramos-Cabrer P, Wiedermann D, Hoehn M. Magn. Reson. Med. 2005;54:59–66. doi: 10.1002/mrm.20532. [DOI] [PubMed] [Google Scholar]
- 16.Stoll G, Jander S, Schroeter M. Prog. Neurobiol. 1998;56:149–171. doi: 10.1016/s0301-0082(98)00034-3. [DOI] [PubMed] [Google Scholar]
- 17.Henning EC, Ruetzler CA, Gaudinski MR, Hu TC, Latour LL, Hallenbeck JM, Warach S. J. Cereb. Blood Flow Metab. 2009;29:1229–1239. doi: 10.1038/jcbfm.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kochanek PM, Hallenbeck JM. Stroke. 1992;23:1367–1379. doi: 10.1161/01.str.23.9.1367. [DOI] [PubMed] [Google Scholar]
- 19.Zhao H, Shimohata T, Wang JQ, Sun G, Schaal DW, Sapolsky RM, Steinberg GK. J. Neurosci. 2005;25:9794–9806. doi: 10.1523/JNEUROSCI.3163-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato H, Kogure K, Liu XH, Araki T, Itoyama Y. Brain Res. 1996;734:203–212. [PubMed] [Google Scholar]
- 21.Clark RK, Lee EV, Fish CJ, White RF, Price WJ, Jonak ZL, Feuerstein GZ, Barone FC. Brain Res. Bull. 1993;31:565–572. doi: 10.1016/0361-9230(93)90124-t. [DOI] [PubMed] [Google Scholar]
- 22.Schroeter M, Franke C, Stoll G, Hoehn M. Acta Neuropathol. 2001;101:114–122. doi: 10.1007/s004010000262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.