Abstract
Dehydrin is a plant disordered protein whose functions are not yet totally understood. Here it is reported that a KS-type dehydrin can reduce the formation of reactive oxygen species (ROS) from Cu. AtHIRD11, which is the Arabidopsis KS-type dehydrin, inhibited generation of hydrogen peroxide and hydroxyl radicals in the Cu–ascorbate system. The radical-reducing activity of AtHIRD11 was stronger than those of radical-silencing peptides such as glutathione and serum albumin. The addition of Cu2+ reduced the disordered state, decreased the trypsin susceptibility, and promoted the self-association of AtHIRD11. Domain analyses indicated that the five domains containing histidine showed ROS-reducing activities. Histidine/alanine substitutions indicated that histidine is a crucial residue for reducing ROS generation. Using the 27 peptides which are related to the KnS-type dehydrins of 14 plant species, it was found that the strengths of ROS-reducing activities can be determined by two factors, namely the histidine contents and the length of the peptides. The degree of ROS-reducing activities of a dehydrin can be predicted using these indices.
Key words: Circular dichroism, dehydrin, disordered protein, heavy metal, histidine, reactive oxygen species.
Introduction
Dehydrins (group 2 late embryogenesis abundant proteins) are plant proteins that are responsive to abiotic stresses such as drought, extreme temperature, and high salinity. Various plant species accumulate dehydrins during embryogenesis and stress responses. Studies on the conserved domains, expression, localization, conformational characteristics, and functions of dehydrins have been summarized (see reviews by Close, 1996; Svensson et al., 2002; Rorat, 2006; Tunnacliffe and Wise, 2007; Battaglia et al., 2008; Hundertmark and Hincha, 2008; Hara, 2010; Eriksson and Harryson, 2011). Dehydrins possess conserved K-segments (EKKGIMDKIKEKLPG or similar sequences), which are proposed to form an amphipathic helix (Close, 1996; Svensson et al., 2002; Rorat, 2006; Tunnacliffe and Wise, 2007; Battaglia et al., 2008; Hundertmark and Hincha, 2008; Hara, 2010; Eriksson and Harryson, 2011). Other unique domains, namely a Y-segment (a typical sequence; DEYGNP) and an S-segment (LHRSGSSSSSSSEDD or related sequences), frequently appear in dehydrin sequences. Using the three segments, dehydrins are conveniently classified by the following shorthand: SKn, YnSKn, YnKn, KnS, etc. Because dehydrins are composed of charged and polar amino acids, they are believed to have highly flexible structures (Close, 1996; Svensson et al., 2002; Rorat, 2006; Tunnacliffe and Wise, 2007; Battaglia et al., 2008; Hundertmark and Hincha, 2008; Hara, 2010; Eriksson and Harryson, 2011). Dehydrins show stability to boiling, abnormal mobility in electrophoresis, and high proteolytic sensitivity. Circular dichroism (CD), Fourier transform infrared spectroscopy, and nuclear magnetic resonance indicated that dehydrins are intrinsically disordered proteins (Tompa, 2009).
Dehydrins are ubiquitously found in various subcellular compartments, including the cytoplasm, nucleus, plasma membrane, tonoplast, plastid, mitochondrion, endoplasmic reticulum, and plasmodesmata (Close, 1996; Svensson et al., 2002; Rorat, 2006; Tunnacliffe and Wise, 2007; Battaglia et al., 2008; Hundertmark and Hincha, 2008; Hara, 2010; Eriksson and Harryson, 2011). Dehydrins have been detected mainly in and/or near the vasculature (Godoy et al., 1994; Danyluk et al., 1998; Bravo et al., 1999; Nylander et al., 2001, Hara et al., 2011). Studies using transgenics and mutants have reported the relationship between dehydrin expression and stress tolerance in plants. The overexpression of dehydrin genes in plants enhanced their tolerance to stresses, such as low temperature (Hara et al., 2003; Houde et al., 2004; Puhakainen et al., 2004; Yin et al., 2006; Xing et al., 2011; Ochoa-Alfaro et al., 2012), osmotic stress (Cheng et al., 2002; Figueras et al., 2004; Brini et al., 2007; Wang et al., 2011), and high salinity (Shekhawat et al., 2011). Reduction of dehydrin contents lowered seed longevity in Arabidopsis (Hundertmark et al., 2011). Functional studies at the molecular level have attempted to elucidate how dehydrins participate in promoting stress tolerance. Dehydrins showed cryoprotection (Close, 1996; Svensson et al., 2002; Rorat, 2006; Tunnacliffe and Wise, 2007; Battaglia et al., 2008; Hundertmark and Hincha, 2008; Hara, 2010; Eriksson and Harryson, 2011), antifreeze activity (Wisniewski et al., 1999), phospholipid binding (Kovacs et al., 2008; Koag et al., 2009; Eriksson et al., 2011), nucleic acid binding (Hara et al., 2009; Lin et al., 2012), and calcium binding (Heyen et al., 2002; Alsheikh et al., 2005). However, how these in vitro functions are associated with enhancing the stress tolerance in plants remains unknown.
One of the typical phenomena observed in transgenic plants expressing dehydrins is the reduction of lipid peroxidation under stress conditions (Hara et al., 2003; Shekhawat et al., 2011; Xing et al., 2011). It has been shown that the lipid peroxidation results from reactive oxygen species (ROS) generated in stressed plants (Shen et al., 1997; Iturbe-Ormaetxe et al., 1998). Generally, transition metals are thought to be the origins of ROS generation in vivo (Iturbe-Ormaetxe et al., 1998). Many dehydrins can bind heavy metals with their histidine-rich sequences (Svensson et al., 2000; Krüger et al., 2002; Hara et al., 2005; Rahman et al., 2011). Although it is postulated that dehydrins may control the ROS generation from transition heavy metals such as Cu, the ROS-controlling functions of dehydrins have not been demonstrated yet.
In order to investigate whether dehydrins can control the generation of ROS from heavy metals, the KS-type dehydrins were the focus of this study for the following reasons. (i) The KS-type dehydrins have been well characterized to bind heavy metals. The Ricinus communis KS-type dehydrin ITP was identified as a metal transporter that moves through the phloem of young plants (Krüger et al., 2002). The Arabidopsis KS-type dehydrin AtHIRD11 (At1g54410), which accumulated in the cambial zone of the vasculature, also bound metals (Hara et al., 2011). (ii) The KS-type dehydrins are the shortest subfamily in terms of length (~100 amino acids) and therefore this type has a simple domain constitution, suggesting that, if the KS-type dehydrins can control ROS generation, the domains that are related to the control can be identified with comparative ease. In this study, it was found that the KS-type dehydrins can reduce ROS generation from Cu. It is also proposed that the histidine contents and the length of the peptides are fundamental factors that influence the strength of ROS reduction by the KS-type dehydrins.
Materials and methods
Preparation of recombinant AtHIRD11
A recombinant AtHIRD11 protein was produced by a method reported previously (Hara et al., 2011). In brief, the open reading frame (ORF) of AtHIRD11 was inserted into the pET-30 Escherichia coli expression system (Novagen, WI, USA). This expression system synthesizes a recombinant protein which has His-tag and S-tag sequences at the N-terminus. The E. coli strain BL21 having the expression construct was pre-cultured at 37 °C. Protein expression, which was induced by the addition of isopropyl-β-d-thiogalactopyranoside (1mM), proceeded for an additional 3h at 28 °C. Bacterial cells (800ml of culture) were lysed by BugBuster reagent (Novagen). The lysate clarified by centrifugation was heated at 90 °C for 20min, and then centrifuged again. The supernatant containing the tagged AtHIRD11 was digested with Factor Xa (Novagen) to remove the tags (His-tags and S-tags). The tag-less AtHIRD11 protein was purified subsequently by a HiTrap Chelating HP column (1ml, GE Healthcare, Tokyo, Japan) immobilizing Ni2+ and then an anion-exchange column (10ml, DEAE-Toyopearl 650M, Tosoh, Tokyo, Japan). The sample was desalted using a NAP-25 column (GE Healthcare) and freeze-dried. The dried AtHIRD11 was weighed and stored as a water solution (10mg ml–1) at –80 °C until use. The protein was identified as AtHIRD11 using matrix-assisted laser desorption/ionization-time of flight-mass spectrometry (MALDI-TOF-MS).
Peptide synthesis
In this study, 32 peptides other than AtHIRD11 were chemically synthesized. Their sequences appear in Table 1. Each synthetic peptide has a tryptophan residue at the N-terminus which allows detection by UV (280nm) to monitor the synthetic peptide quantitatively. The peptides, which were prepared by an automated solid phase peptide synthesizer (Tetras, Advanced ChemTech, KY, USA), were purified using C18 reversed-phase column chromatography (LC-20AB, Shimadzu, Kyoto, Japan) to 98% homogeneity with a linear gradient of acetonitrile (from 20% to 40%) in 0.1% trifluoroacetic acid solution over 20min. The purified peptides were identified by MS (LCMS-2020, Shimadzu).
Table 1.
Histidine contents and radical-reducing activities of KS-type dehydrin-related peptides. The 50% inhibitory dose (ID50) values were determined by the ROS generation reaction using the Cu–ascorbate system. Ascorbate (300 µM) and CuCl2 (4.6 µM) were used. The results are from four replicates.
| Peptide names | Sequences | Species | AANs | No. of histidines | Histidine contents (%) | ID50 (µM) | ID50×AAN (µM) | Used in Fig. 6 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Average | SD | Average | SD | |||||||
| AtHIRD11 and domains | ||||||||||
| AtHIRD11 | D1+D2+D3+D4+D5+D6+D7 | Arabidopsis thaliana | 98 | 13 | 13.3 | 0.58 | 0.18 | 56.85 | 17.66 | ✓ |
| D1 | WMAGLINKIGDAL HIGGGNKEG | Arabidopsis thaliana | 22 | 1 | 4.5 | 9.59 | 0.67 | 211.0 | 14.7 | ✓ |
| D2 | WEHKKEEEHKK HVDEHKSGE | Arabidopsis thaliana | 20 | 4 | 20.0 | 1.50 | 0.02 | 30.03 | 0.50 | ✓ |
| D3 | WHKEGIVDKIKDKIHG | Arabidopsis thaliana | 16 | 2 | 12.5 | 3.32 | 0.11 | 53.08 | 1.78 | ✓ |
| D4 | WGEGKSHDGEGKSHDG | Arabidopsis thaliana | 16 | 2 | 12.5 | 3.78 | 0.14 | 60.52 | 2.24 | ✓ |
| D5 | WEKKKKKDKKEKK | Arabidopsis thaliana | 13 | 0 | 0.0 | 195 | 3 | 2535 | 39 | |
| D6 | WHHDDGHH | Arabidopsis thaliana | 8 | 4 | 50.0 | 1.88 | 0.05 | 15.07 | 0.42 | ✓ |
| D7 | WSSSSDSDSD | Arabidopsis thaliana | 10 | 0 | 0.0 | 75.1 | 4.2 | 751.0 | 42.0 | |
| D5+D6+D7 | WEKKKKKDKKEKKHHD DGHHSSSSDSDSD | Arabidopsis thaliana | 29 | 4 | 13.8 | 1.47 | 0.05 | 42.74 | 1.54 | ✓ |
| Modified domains | ||||||||||
| D2H/A | WEAKKEEEAKKAVDEAKSGE | 20 | 0 | 0.0 | 147 | 14 | 2940 | 270 | ||
| D3H/A | WAKEGIVDKIKDKIAG | 16 | 0 | 0.0 | 223 | 64 | 3568 | 1016 | ||
| D4H/A | WGEGKSADGEGKSADG | 16 | 0 | 0.0 | 144 | 32 | 2304 | 509 | ||
| D6H/A | WAADDGAA | 8 | 0 | 0.0 | 196 | 4 | 1568 | 28 | ||
| D6 modified | ||||||||||
| D6 D/A | WHHAAGHH | 8 | 4 | 50.0 | 3.20 | 0.03 | 25.59 | 0.28 | ✓ | |
| D6 D/N | WHHNNGHH | 8 | 4 | 50.0 | 1.85 | 0.10 | 14.78 | 0.82 | ✓ | |
| D6 D/H1 | WHHHDGHH | 8 | 5 | 62.5 | 3.13 | 0.10 | 25.01 | 0.80 | ✓ | |
| D6 D/H2 | WHHHHGHH | 8 | 6 | 75.0 | 2.32 | 0.19 | 18.55 | 1.49 | ✓ | |
| D6 D/H3 | WHHHHHHH | 8 | 7 | 87.5 | 3.14 | 0.41 | 25.14 | 3.26 | ✓ | |
| D6×2 | WHHDDGHHDDGHH | 13 | 6 | 46.2 | 1.35 | 0.13 | 17.52 | 1.63 | ✓ | |
| D6×3 | WHHDDGHHDDGHHDDGHH | 18 | 8 | 44.4 | 0.95 | 0.04 | 17.04 | 0.67 | ✓ | |
| D6 other plants | ||||||||||
| OsD6 | WHGEEGHHHDGH | Oryza sativa | 12 | 5 | 41.7 | 1.69 | 0.20 | 20.25 | 2.37 | ✓ |
| RrD6 | WHEHGHEHGHD | Retama raetam | 11 | 5 | 45.5 | 1.75 | 0.20 | 19.28 | 2.15 | ✓ |
| GmD6 | WHGHDHHGH | Glycine max | 9 | 5 | 55.6 | 2.11 | 0.17 | 19.01 | 1.50 | ✓ |
| SbD6 | WHGEGHDHDGH | Sorghum bicolor | 11 | 4 | 36.4 | 1.85 | 0.25 | 20.35 | 2.78 | ✓ |
| MsD6 | WHGEGHEHGH | Medicago sativa | 10 | 4 | 40.0 | 2.18 | 0.19 | 21.82 | 1.92 | ✓ |
| CpD6 | WHDEHGHDGH | Carica papaya | 10 | 4 | 40.0 | 2.52 | 0.06 | 25.22 | 0.64 | ✓ |
| BdD6 | WHGEGHKKEDGH | Brachypodium distachyon | 12 | 3 | 25.0 | 2.75 | 0.33 | 33.03 | 3.95 | ✓ |
| VvD6 | WHEDGHDHGG | Vaccinium vitis | 10 | 3 | 30.0 | 3.01 | 0.50 | 30.09 | 5.01 | ✓ |
| CmD6 | WHGEGHKHG | Corylus mandshurica | 9 | 3 | 33.3 | 2.40 | 0.12 | 21.64 | 1.09 | ✓ |
| RcD6 | WHEHGH | Ricinus communis | 6 | 3 | 50.0 | 2.87 | 0.11 | 17.19 | 0.68 | ✓ |
| HvD6 | WDGEGHKDDDGH | Hordeum vulgare | 12 | 2 | 16.7 | 2.66 | 0.07 | 31.86 | 0.80 | ✓ |
| PmD6 | WHGEGHDGG | Plantago major | 9 | 2 | 22.2 | 3.18 | 0.18 | 28.62 | 1.65 | ✓ |
| CuD6 | WHEDGHE | Citrus unshiu | 7 | 2 | 28.6 | 4.19 | 0.44 | 29.32 | 3.07 | ✓ |
AAN, amino acid number.
Reduction of ROS generation
The inhibiting effects of AtHIRD11 and its related domains on ROS generation were measured by the Cu–ascorbate system, which was established for researching the ROS-reducing activities of peptides by detecting hydroxyl radicals (Guilloreau et al., 2007). This system consisted of 1/10 phosphate-buffered saline (PBS) pH 7.4 (13.7mM NaCl, 0.27mM KCl, 1mM Na2HPO4, and 0.176mM KH2PO4), desferrioxamine (1 µM), test samples [0–1.85 µM for AtHIRD11, 0–30 µM for peptides containing histidine, 0–300 µM for peptides without histidine, 0–5 µM for bovine serum albumin (BSA), 0–10 µM for EDTA, 0–100 µM for glutathione (GSH), 0–500 µM for histidine, and 0–2mM for glycine], CuCl2 (4.6 µM), coumarin-3-carboxylic acid (3-CCA, 10mM), which is a hydroxyl radical detector, and sodium ascorbate (300 µM) in a total volume of 200 µl. The ROS generation was started by the addition of sodium ascorbate, and then the increase in fluorescence (395nm excitation, 452nm emission) was monitored for 10min using a Varioskan Flash microplate reader (Thermo Scientific, Tokyo, Japan) in the kinetic mode. The initial velocity of the increase in fluorescence was measured. In a previous report, the addition of desferrioxamine was recommended because an increase in background fluorescence may occur due to the presence of trace metals in the water used in the experiment (Guilloreau et al., 2007). Although the present experimental conditions did not result in such a background increase, desferrioxamine was used for completeness.
Since hydrogen peroxide is also generated in the Cu–ascorbate system (Guilloreau et al., 2007), the hydrogen peroxide generation was quantified by the titanium sulphate method (Eisenberg, 1943) with modifications. The test mixture contained 1/10 PBS pH 7.4, desferrioxamine (1 µM), AtHIRD11 (0–1.85 µM), CuCl2 (4.6 µM), and sodium ascorbate (300 µM) in a total volume of 200 µl. After reacting for 3min, 50 µl of 3% titanium sulphate solution was added to the mixture, and then absorbance at 450nm was measured using the Varioskan Flash microplate reader. The blank in each case was the mixture containing neither CuCl2 nor sodium ascorbate. Preliminary experiments showed that a reaction period of 3min was appropriate for measuring the initial velocity of hydrogen peroxide generation. A calibration curve was produced with the authentic hydrogen peroxide solution.
CD analyses
AtHIRD11 was subjected to a spectropolarimeter (J-820, JASCO) in the presence of metals. AtHIRD11 (4.6 µM) and various concentrations (2.3, 23, and 230 µM) of metals, such as CaCl2, MgCl2, MnCl2, CoCl2, NiCl2, CuCl2, and ZnCl2, were combined in 1/10 PBS pH 7.4. The scan was performed from 195nm to 250nm. The scan speed, resolution, and cell width were 100nm min–1, 1nm, and 2mm, respectively. The obtained data were analysed by DICHROWEB (http://dichroweb.cryst.bbk.ac.uk/html/home.shtml), which is an online server for predicting secondary structures of proteins (Whitmore and Wallace, 2004).
Protease sensitivity of AtHIRD11
AtHIRD11 (4.6 µM), metals such as CaCl2, MgCl2, MnCl2, CoCl2, NiCl2, CuCl2, and ZnCl2 (2.3, 23, and 230 µM), and trypsin (0.05 µM) were combined in 1/10 PBS pH 7.4. A digestive reaction was started by the addition of trypsin. After the samples were incubated at room temperature for 10min, the reactions were terminated by heating at 95 °C. The AtHIRD11 in the samples was resolved by SDS–PAGE, and then the gel was stained with colloidal Coomassie blue (Bio-Safe, Bio-Rad, Tokyo, Japan). After the digital images were taken, the intensities of the AtHIRD11 bands were determined by NIH-Image software (http://rsbweb.nih.gov/nih-image/). The intensity of the non-digested AtHIRD11 with no metal was standardized (100%).
Self-association of AtHIRD11
The degree of association of AtHIRD11 was determined as follows. AtHIRD11 (4.6 µM) was mixed with metals such as CaCl2, MgCl2, MnCl2, CoCl2, NiCl2, CuCl2, and ZnCl2 (2.3, 23, and 230 µM) in 1/10 PBS pH 7.4 (total volume 40 µl) in siliconized plastic tubes. After incubating at room temperature for 5min, the samples were centrifuged at 10 000 g for 15min at 4 °C. The supernatants were totally transferred to the new tubes, and then the pellets were resuspended in 40 µl of 1/10 PBS by vortexing. AtHIRD11 was resolved by SDS–PAGE. The gel was stained with colloidal Coomassie blue (Bio-Safe). The intensities of the AtHIRD11 bands in the digital images were determined by NIH-Image software. The amount of AtHIRD11 in the pellet was expressed as a percentage. In each sample, the sum of the intensities of AtHIRD11 in the supernatant and the pellet was standardized (100%).
Data analysis
Data for P-values were analysed by Student’s t-test at a significance level of 0.05. To fit curves through points, the curve-fitting tools in Microsoft Excel 2007 were used.
Results
Reduction of ROS generation from Cu by AtHIRD11
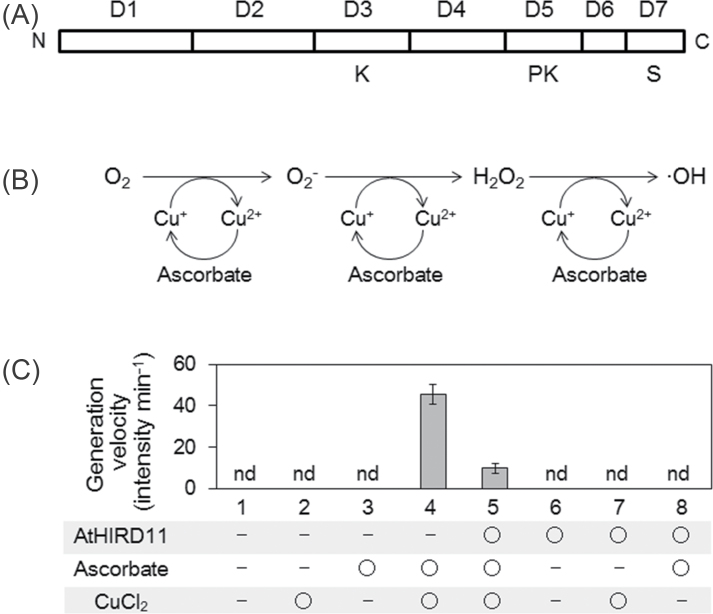
AtHIRD11 (At1g54410) is a KS-type dehydrin consisting of 98 amino acids (Hara et al., 2011). AtHIRD11 has a simple domain constitution including K-, PK-, and S-segments (Fig. 1A). Histidine residues frequently occur throughout the sequence of AtHIRD11. The histidine content of AtHIRD11 (13.3%) is the seventh highest in the ORFs of the Arabidopsis genome (Hara et al., 2010). Orthologues of AtHIRD11 are widely spread in higher plants such as Ricinus communis, Glycine max, Solanum sogarandinum, Oryza sativa, Medicago sativa, Vitis vinifera, etc. (Rorat et al., 2004; Hara et al., 2011).
Fig. 1.
Reduction of ROS generation from Cu by AtHIRD11. (A) Domain constitution of AtHIRD11. The amino acid sequence was divided into seven domains (D1–D7). D3, D5, and D7 are K-, PK-, and S-segments, respectively, which are found in many dehydrins. (B) A scheme of the Cu–ascorbate system used in this study. (C) Hydroxyl radical generation under different conditions of the ROS generation system. Eight combinations were tested. AtHIRD11 (0.93 µM), ascorbate (300 µM), and CuCl2 (4.6 µM) were used. Values and bars indicate means and SD of four measurements, respectively.
In order to test whether AtHIRD11 affects ROS generation, the recombinant AtHIRD11 protein that is produced by E. coli was prepared. A common ROS-generating reaction, namely the Cu–ascorbate system, which was established to investigate ROS-silencing activity (Guilloreau et al., 2007), was used. Ascorbate reduces Cu2+ to Cu+, and then Cu+ subsequently reduces oxygen to hydroxyl radicals via the formation of superoxide anions and hydrogen peroxide as intermediates under aerobic conditions (Fig. 1B). Through the radical generation, Cu+ is regenerated from Cu2+ by ascorbate. According to the theory, hydroxyl radicals were generated when ascorbate (300 µM) was combined with Cu2+ (4.6 µM) (Fig. 1C, condition 4). However, the addition of AtHIRD11 (0.93 µM) attenuated the radical generation of Cu2+ with ascorbate (Fig. 1C, condition 5). In this system, Cu2+, ascorbate, or AtHIRD11 did not generate hydroxyl radicals alone (Fig. 1C, conditions 2, 3, and 6). Neither the combination of AtHIRD11 and Cu nor that of AtHIRD11 and ascorbate generated hydroxyl radicals (Fig. 1C, conditions 7 and 8). These results show that hydroxyl radical formation, which occurred under the co-existence of ascorbate and Cu2+, was reduced by AtHIRD11.
The reduction of hydroxyl radical generation by AtHIRD11 was dose dependent, and the 50% inhibitory dose (ID50) was 0.58±0.18 µM (n=4) (Fig. 2A, left graph). AtHIRD11 also attenuated hydrogen peroxide generation in a dose-dependent manner at the ID50 of 0.52±0.16 µM (n=4) (Fig. 2A, right graph). The ID50 values for the reduction of hydroxyl radical generation were compared between AtHIRD11 and BSA, EDTA, GSH, histidine, or glycine (Fig. 2B). EDTA is a strong quencher of ROS generation from the metal–ascorbate system (Saran and Bors, 1991). The data indicate that AtHIRD11 showed the lowest ID50 value among the compounds tested (Fig. 2B).
Fig. 2.
Reducing activities of ROS generation by AtHIRD11 and other compounds. (A) Dose-dependent reductions of the ROS generation by AtHIRD11. Results regarding hydroxyl radicals (left graph) and hydrogen peroxide (right graph) are shown. Ascorbate (300 µM) and CuCl2 (4.6 µM) were used. (B) Reducing activities of the generation of hydroxyl radicals by different compounds. AH11, BSA, EDTA, GSH, His, and Gly represent AtHIRD11, bovine serum albumin, ethylene diamine tetra-acetic acid, glutathione, histidine, and glycine, respectively. Values and bars indicate means and SD of four measurements, respectively. Significant differences (P < 0.05) in comparison with the value of AtHIRD11 were determined by Student’s t-test (* in B).
Effect of Cu on the conformation of AtHIRD11
Since it has been reported that several dehydrins changed their conformations when binding to metals (Hara et al., 2009; Mu et al., 2011; Rahman et al., 2011), experiments were carried out to confirm whether AtHIRD11 also shows a conformational change induced by Cu2+. The CD analysis showed that AtHIRD11 was probably disordered, because a large negative peak at 200nm was observed (Fig. 3A, grey broken line). The addition of Cu2+ mitigated the degree of the negative peak at 200nm in a dose-dependent manner (Fig. 3B). This suggests that in the interaction between AtHIRD11 and Cu2+, more Cu2+ results in a greater decrease in disorder. Although such conformational changes of AtHIRD11 also occurred with Co2+, Ni2+, and Zn2+, no change occurred with Ca2+, Mg2+, and Mn2+ (Fig. 3B). Because AtHIRD11 bound Co2+, Ni2+, Cu2+, and Zn2+, whereas it did not bind Ca2+, Mg2+, and Mn2+ (Hara et al., 2011), this indicated that the decrease in disorder was promoted only by the metals which bound to AtHIRD11.
Fig. 3.
Conformational alterations of AtHIRD11 by metals. (A) Circular dichroism (CD) analyses using AtHIRD11 with different concentrations of Cu2+. AtHIRD11 alone (4.6 µM) is shown by a grey broken line. The [AtHIRD11]:[Cu2+] ratios are 1:0.5 (4.6 µM:2.3 µM, grey solid line), 1:5 (4.6 µM:23 µM, black broken line), and 1:50 (4.6 µM:230 µM, black solid line). Values are means of four measurements. (B) Effects of different metal cations on conformational changes of AtHIRD11. CD values at 200nm are compared. The white column showing the value without metal (NM) is standardized (100%). The [AtHIRD11]:[Cu2+] ratios are 1:0.5 (light grey columns), 1:5 (dark grey columns), and 1:50 (black columns). The concentrations of AtHIRD11 and Cu2+ were the same as in A. (C) Composition of secondary structures in AtHIRD11 as predicted by DICHROWEB (http://dichroweb.cryst.bbk.ac.uk/html/home.shtml). H1, H2, S1, S2, T, and U indicate regular helix, distorted helix, regular β-strand, distorted β-strand, turn, and unordered contents, respectively. The white columns represent the value without metal. The [AtHIRD11]:[Cu2+] ratios are 1:0.5 (light grey columns), 1:5 (dark grey columns), and 1:50 (black columns). The concentrations of AtHIRD11 and Cu2+ were the same as in A. In B and C, values and bars indicate means and SD of four measurements, respectively. *Significant differences (P < 0.05) in comparison with the value without metal were determined by Student’s t-test.
The DICHROWEB analysis also indicated that the disordered state of AtHIRD11 was reduced by supplying Cu2+ (Fig. 3C, U). On the other hand, the analysis showed a decrease in distorted helices (Fig. 3C, H2) and increases in regular and distorted β-strands (Fig. 3C; Supplementary Fig. S1, S2 available at JXB online) as the Cu2+ concentration increased, whereas the disordered state was predominant even when the highest concentration of Cu2+ was added to AtHIRD11.
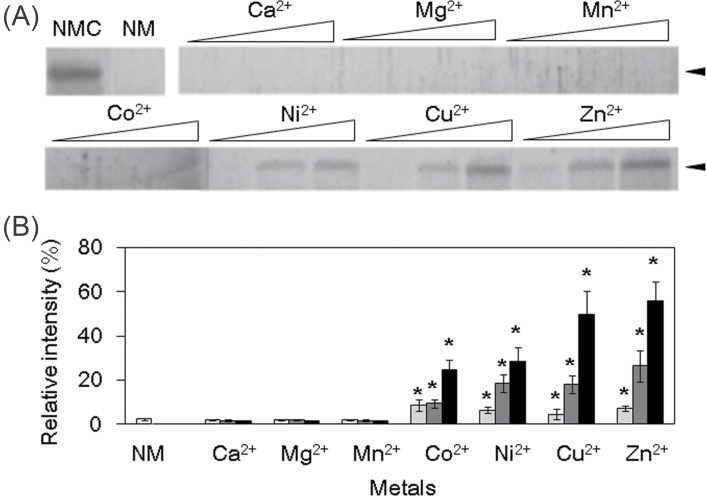
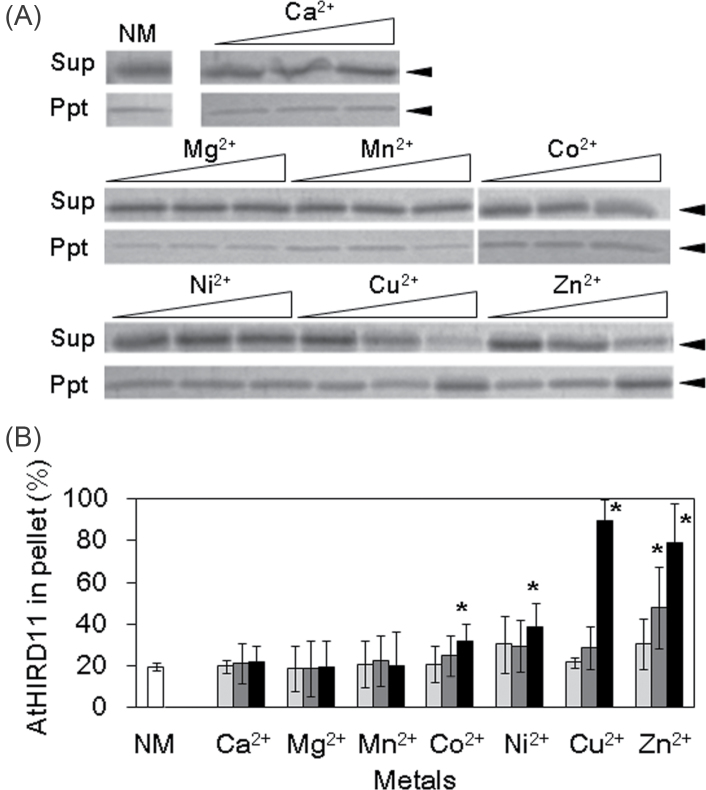
Metals including Cu2+ can induce not only conformational changes but also protease resistance in some disordered proteins such as prion (Lehmann, 2002). Therefore, it is assumed that AtHIRD11 may be converted to protease-resistant forms. Although AtHIRD11 is highly susceptible to trypsin, AtHIRD11 became resistant to protease by the addition of Co2+, Ni2+, Cu2+, and Zn2+ (Fig. 4A). On the other hand, Ca2+, Mg2+, and Mn2+, which cannot bind to AtHIRD11, did not enhance the protease resistance. If the molar ratios of Cu2+ to AtHIRD11 increased, the degree of trypsin susceptibility decreased (Fig. 4B). Similar results were obtained in the case of Co2+, Ni2+, and Zn2+. Moreover, the addition of Cu2+ increased the association species of AtHIRD11 in a dose-dependent manner (Fig. 5). Co2+, Ni2+, and Zn2+ promoted the association of AtHIRD11 as Cu2+ did. However, Ca2+, Mg2+, and Mn2+ did not.
Fig. 4.
Effects of metals on trypsin resistance of AtHIRD11. AtHIRD11 (4.6 µM) was treated with trypsin (0.05 µM) after metal ions were added. (A) AtHIRD11 treated with trypsin was resolved by SDS–PAGE. The gel was stained with colloidal Coomassie blue. Open triangles represent levels of metal concentrations. For each metal, the concentration increases from left to right in three steps. The [AtHIRD11]:[Cu2+] ratios are 1:0.5 (4.6 µM:2.3 µM, left), 1:5 (4.6 µM:23 µM, middle), and 1:50 (4.6 µM:230 µM, right). NMC indicates AtHIRD11 alone which was treated with neither metal nor trypsin. NM denotes AtHIRD11 treated with trypsin but without metal. Arrowheads show the size of AtHIRD11. (B) Relative intensities of the AtHIRD11 bands. The band intensity of the NMC condition is standardized (100%). The [AtHIRD11]:[metals] ratios are 1:0.5 (light grey columns), 1:5 (dark grey columns), and 1:50 (black columns). The concentrations of AtHIRD11 and Cu2+ were the same as in A. Values and bars indicate means and SD of four measurements, respectively. *Significant differences (P < 0.05) in comparison with the NM condition (white bar) were determined by Student’s t-test.
Fig. 5.
Effects of metals on association species formation of AtHIRD11. Different kinds of metals were added to the AtHIRD11 solutions (4.6 µM), and then the mixtures were centrifuged. The resultant supernatants (Sup) and pellets (Ppt) were resolved by SDS–PAGE. (A) The SDS–polyacrylamide gel was stained with colloidal Coomassie blue. Open triangles represent levels of metal concentrations. For each metal, the concentration increases from left to right in three steps. The [AtHIRD11]:[Cu2+] ratios are 1:0.5 (4.6 µM:2.3 µM, left), 1:5 (4.6 µM:23 µM, middle), and 1:50 (4.6 µM:230 µM, right). NM indicates AtHIRD11 alone (without metal). Arrowheads show the size of AtHIRD11. (B) Relative intensities of the AtHIRD11 bands in the pellet fractions. The sums of band intensities in supernatants and those in pellets are expressed as 100%. The [AtHIRD11]:[metals] ratios are 1:0.5 (light grey columns), 1:5 (dark grey columns), and 1:50 (black columns). The concentrations of AtHIRD11 and Cu2+ were the same as in A. Values and bars indicate means and SD of four measurements, respectively. *Significant differences (P < 0.05) in comparison with the NM condition (a white bar) were determined by Student’s t-test.
Domains contributing to the ROS silencing
In order to postulate the mechanisms regarding the reducing activity of the Cu-promoted ROS generation by AtHIRD11, an attempt was made to determine the functional domains which contribute to the activity. The AtHIRD11 amino acid sequence was divided into seven domains, D1–D7 (Fig. 1A). D1 and D2 are N-terminal sequences which do not contain any conserved segments found in dehydrins. D3, D5, and D7 are conserved K-, polylysine (PK)-, and S-segments, respectively. D4 and D6 are junction regions between the conserved segments (D4, between the K-segment and the PK-segment; D6, between the PK-segment and the S-segment). The seven domains (D1– D7) and the whole AtHIRD11 sequence were subjected to the Cu–ascorbate system (Table 1; AtHIRD11 and domains). It was indicated that five (D1–D4 and D6) out of the seven domains showed apparent ROS-reducing activities (ID50 of <10 µM), suggesting that the functional domains exist throughout the whole sequence of AtHIRD11. However, it was assumed that D6 may be one of the core sequences for expressing the ROS-reducing activity, because this domain showed strong activity despite having the shortest sequence.
Factors determining the ROS-silencing activity
Since the domains containing histidine apparently showed the ROS-reducing activity as described above, it was suggested that histidine may be a crucial residue for the activity. To confirm this, mutant domains were prepared in which histidine residues in the corresponding original domains were changed to alanine residues (Table 1; Modified domains). The mutant domains were D2H/A, D3H/A, D4H/A, and D6H/A, whose original domains were D2, D3, D4, and D6, respectively. As expected, the activities of all four mutant domains were remarkably lower than those of the corresponding original domains. This finding suggests that the presence of histidine in the domains of AtHIRD11 is important to express efficient ROS-reducing activities.
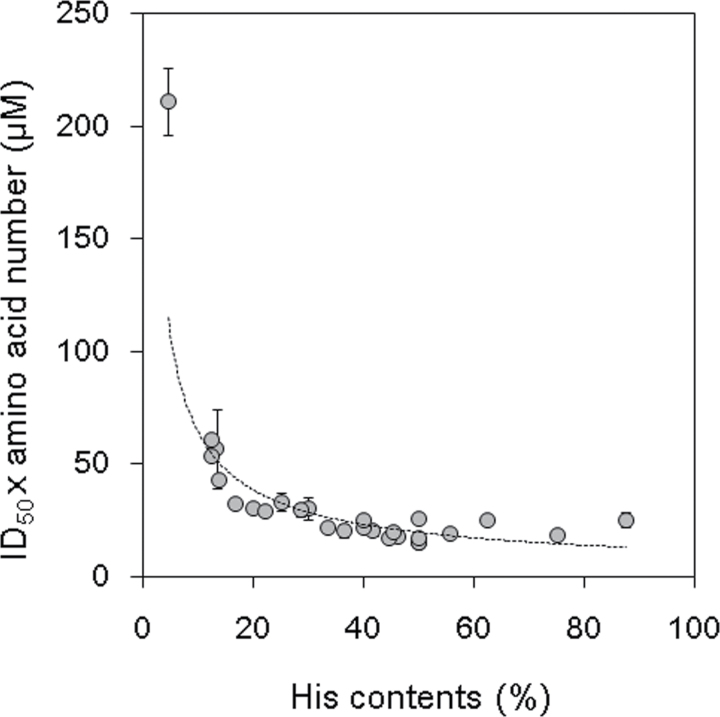
To search for the factors that determine the magnitude of the ROS-reducing activities, more data were gathered regarding the ROS-reducing activities of the KS-type dehydrin-related peptides which contain various numbers of histidine residues. In addition to the peptides tested above, 21 other peptides were prepared, namely a sequence of D5+D6+D7, seven peptides that were mutant D6 sequences of AtHIRD11, and D6 sequences found in the KS-type dehydrins of 13 plant species (Table 1). The sequences of the D6 domains and their adjacent sites of the KnS-type dehydrins used in this study are shown in Supplementary Fig. S1 at JXB online. The data regarding the sequences, amino acid numbers, the numbers of histidine residues, and the ID50 values of the 27 peptides that possess at least one histidine residue are represented in Table 1 with the symbol ‘✓’. Using these data, the combinations of the items of data that showed good correlations were searched. Finally, it was found that when the indices of the ID50×amino acid number (µM) were plotted against the histidine contents (%), it is likely that the dots fit the continuous curve for the most part (Fig. 6). Several approximation models were applied to fit curves through the dots. Comparison of the R2 values indicated that a power approximation showed the best fit, namely ID50×amino acid number (µM)=352×histidine content (%)–0.74 (R2=0.788) (Fig. 6, broken line). This curve indicated that the value of ID50×amino acid number greatly decreased as the histidine contents increased in the range from ~5% to 15%. In the histidine contents range extending from ~15% to 50%, however, the decreasing slope of the ID50×amino acid number curve was much smaller. When the histidine contents were >50%, the value of ID50×amino acid number was nearly constant regardless of the histidine contents. Taken together, Fig. 6 suggests that, if the peptide lengths are assumed to be uniform, the ID50 values of the peptides decrease as their histidine contents increase, whereas the decrease of the ID50 values reaches a plateau at more than ~50% of the histidine contents.
Fig. 6.
Relationships between histidine contents (%) and ID50×amino acid number values (µM) in the 27 KS-dehydrin-related peptides. The histidine contents (x-axis) were plotted against the ID50×amino acid number values (y-axis). Values in Table 1 were used to draw this graph. Values and bars indicate means and SD of four measurements, respectively. The regression line (y=352x–0.74, R2=0.788) is shown with a broken line.
Discussion
Although many studies have reported that dehydrin expression provided an enhancement of abiotic stress tolerances in plants, the stress enhancement mechanisms have not been fully elucidated. In plants, one of the common symptoms in abiotic stress responses is physiological damage caused by ROS (Shen et al., 1997; Iturbe-Ormaetxe et al., 1998). It is believed that reactive transition metals which are released from organelles and enzymes under abiotic stresses are the sources of ROS generation (Iturbe-Ormaetxe et al., 1998). Previous results have indicated that dehydrins bound metals (Svensson et al., 2000; Krüger et al., 2002; Hara et al., 2005; Rahman et al. 2011), suggesting that dehydrins may stabilize the transition metals by binding them (Hara et al., 2005; Sun and Lin, 2010). However, there has been no report which experimentally demonstrated this proposed stabilization. In this study, the focus was on elucidating the ROS-silencing activity of the KS-type dehydrins. A common ROS-generating reaction, namely the Cu–ascorbate system, was used. The ID50 values were 0.58 µM and 0.52 µM for the reductions in hydroxyl radical generation and hydrogen peroxide generation, respectively. Since the present Cu–ascorbate system was implemented with 4.6 µM Cu2+, these ID50 values were obtained when the ratio of [AtHIRD11] to [Cu2+] was ~1:8. Previous data showed that the maximum binding capacity (B max) of AtHIRD11 for Cu2+ was 8 (Hara et al., 2011). This suggests that AtHIRD11 can efficiently reduce ROS generation from Cu when the range of the Cu2+ concentration is within the binding capacity of AtHIRD11. The typical Cu content in plants is ~90 µmol kg–1 dry weight, whereas the value is changeable under different growth conditions (Palmer and Guerinot, 2009). On the other hand, the extractable amount of AtHIRD11 protein from the above-ground part of the Arabidopsis plant was found to be ~10 µmol kg–1 dry weight (Hara et al., 2011). This suggests that AtHIRD11 may effectively reduce the ROS generated from Cu in planta, because the ROS-reducing activity is maintained if 1mol of AtHIRD11 binds 9mol of Cu2+. It was reported that the Musa KS-type dehydrin MpDhn12 complemented the copper sensitivity of the yeast mutant delta sod1, which lacked Cu/Zn superoxide dismutase (Mu et al., 2011). This phenomenon might be caused by the ROS-silencing activity of MpDhn12 as for AtHIRD11.
It was shown here that the conformational changes of AtHIRD11 increased as the concentration of Cu2+ was elevated. DICHROWEB analyses suggest that the disordered state decreased while the β-strand content increased when AtHIRD11 was treated with Cu2+ (Fig. 3C). The disordered content, however, was still dominant even when Cu2+ was supplied at the highest concentration. As the disordered state decreased, AtHIRD11 that was treated with Cu2+ showed some association states, because the AtHIRD11 protein was precipitated by centrifugation (Fig. 5) and formed protease-resistant species (Fig. 4). However, it was unlikely that this association state was a typical aggregation for the following reasons. First, the visible turbidity was not found in the AtHIRD11 solution containing Cu2+. Secondly, α-helix aggregation, which is monitored on the basis of the decrease in CD at 222nm (Zhong and Johnson, 1992), was not detected (Supplementary Fig. S2 at JXB online). Thirdly, the result of the 1-anilino-8-naphthalene sulphonate test for indicating the structural transition from disorder to an orderly aggregated state (Tompa, 2009) was negative (Supplementary Fig. S3). Based on these combined findings, it was hypothesized that when AtHIRD11 interacts with Cu2+, AtHIRD11 may self-associate by maintaining a respectably disordered state.
The self-association was accelerated when the ratio of [AtHIRD11] to [Cu2+] reached 1:50 (Fig. 5). At this concentration ratio, AtHIRD11 no longer reduced ROS formation. As described above, the reduction of Cu-promoted ROS generation by AtHIRD11 was more effective when the ratio of [AtHIRD11] to [Cu2+] was larger. These results suggest that the magnitude of the ROS-silencing activities of KS-type dehydrins is negatively correlated with the degree of conformational changes in the proteins.
In order to investigate the ROS-reducing domains of AtHIRD11, the reducing activities of the seven domains of AtHIRD11 were determined. Five domains (D1, D2, D3, D4, and D6) which contain histidine showed ROS-reducing activities (Table 1; AtHIRD11 and domains). The mutant domains corresponding to D2, D3, D4, and D6, which contain no histidine, manifested much lower activities than the original domains (Table 1; Modified domains). This indicates that histidine is indispensable for the domains to express their efficient ROS-reducing activities. Since dehydrins can bind metals via their histidine residues (Hara et al., 2005; Sun and Lin, 2010), it is likely that the chelating action of Cu2+ by histidine provides the ROS-reducing activities of AtHIRD11. Moreover, it was found that the levels of the ROS-reducing activities of the peptides were reflected by the histidine contents and the numbers of amino acids; namely, the indices of ID50×amino acid number (µM) were highly related to the histidine contents (%) (Fig. 6). A comparison of the ROS-reducing activities of the peptides on the basis of constant amino acid numbers showed that the ID50 values decreased as the histidine contents increased. Intriguingly, however, the decrease in the ID50 values weakened when the histidine contents surpassed 20%, and then the decrease reached a plateau at a low level when the histidine contents surpassed 50%. This indicates that the effects of the histidine contents on the enhancement of the ROS-reducing activity levelled off at >20% of the contents. Since histidine is one of the most expensive amino acids to biosynthesize (Rees et al., 2009), the production of peptides possessing extremely high histidine contents is likely to be costly for plants. Considering the metabolic cost of histidine biosynthesis, a level of histidine contents of ~20% is likely to be sufficient for efficient ROS reduction. Indeed, the ORF which shows the highest histidine contents in the Arabidopsis genome was At5g53590, whose histidine content was 19.7% (Hara et al., 2010).
The sixth domain, D6, which showed high ROS-silencing activity, was located between the PK- and S-segments of AtHIRD11 (Fig. 1A). The D6-like sequences were found in all KS-type dehydrins that were checked in the open databases. Various D6 sequences which are shown in Supplementary Fig. S1 at JXB online are suggested to contain mainly histidine, and subsequently aspartate and glycine. Such histidine-rich sequences are found in the metal transporters of many organisms. For instance, Arabidopsis AtMTP1 belonging to the cation diffusion facilitator family has a histidine-rich loop which may function as a buffering pocket for Zn2+ (Kawachi et al., 2008). The present results suggest that the histidine-rich loop may have a role in protecting the transportation machinery from damage by ROS by binding transition metals.
The KS-type dehydrin is the smallest subfamily that consists of simple domain constitutions. This suggests that the KS-type dehydrin may be a prototype of other dehydrin subfamilies. Accordingly, the present results regarding the ROS reduction by KS-type dehydrins may be useful for finding dehydrins that show higher ROS-silencing activities. Arabidopsis possesses 10 copies of dehydrin genes (Hundertmark and Hincha, 2008). Among them, the histidine-rich dehydrin Lti30 (At3g50970), which is responsive to cold stress, shows the highest histidine content (13.5%). Although the histidine content of Lti30 is similar to that of AtHIRD11 (13.3%), the size of Lti30 (193 amino acids) was approximately double that of AtHIRD11 (98 amino acids). This suggests that Lti30 may show more potent ROS-silencing activity (i.e. a lower ID50 value) than AtHIRD11, if the correlation between the indices of ID50×amino acid number (µM) and the histidine contents (%) described in Fig. 6 is taken into consideration. Since it was recently reported that Lti30 could bind to membranes (Eriksson et al., 2011), Lti30 may protect membranes from the metal-promoting lipid peroxidation by binding the metals on the surfaces of the membranes. Alternatively, in the presence of metals, Lti30 may be released from the membranes by sequestering the metals from the membranes, because histidine residues that are associated with the membrane binding may be shielded by the metals.
In conclusion, it is proposed that histidine-rich peptides which inhibit the generation of ROS from metals exist in the plant kingdom. Some kinds of dehydrins including the KS-types may be such histidine-rich ROS-silencing peptides. Moreover, a common method of predicting the levels of the ROS-reducing activities of such peptides was found by using indices of the histidine contents and the amino acid numbers. These findings may be useful in elucidating the functions of the histidine-rich proteins and domains.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Amino acid sequences of D5s, D6s, and D7s in different KnS-type dehydrins.
Figure S2. Effect of Cu2+ on CD at 222nm of AtHIRD11.
Figure S3. Effect of Cu2+ on the formation of ordered aggregation in AtHIRD11.
Acknowledgements
We thank Professor Naoto Oku (University of Shizuoka) for helpful discussions. This study was supported in part by a Grant-in-Aid (no. 23380192) for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
References
- Alsheikh MK, Svensson JT, Randall SK. 2005. Phosphorylation regulated ion-binding is a property shared by the acidic subclass dehydrins. Plant, Cell and Environment 28, 1114–1122 [Google Scholar]
- Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA. 2008. The enigmatic LEA proteins and other hydrophilins. Plant Physiology 148, 6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo LA, Close TJ, Corcuera LJ, Guy CL. 1999. Characterization of an 80-kDa dehydrin-like protein in barley responsive to cold acclimation. Physiologia Plantarum 106, 177–183 [Google Scholar]
- Brini F, Hanin M, Lumbreras V, Amara I, Khoudi H, Hassairi A, Pagès M, Masmoudi K. 2007. Overexpression of wheat dehydrin DHN-5 enhances tolerance to salt and osmotic stress in Arabidopsis thaliana. Plant Cell Reports 26, 2017–2026 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Targolli J, Huang X, Wu R. 2002. Wheat LEA genes, PMA80 and PMA1959 enhance dehydration tolerance of transgenic rice (Oryza sativa L.). Molecular Breeding 10, 71–82 [Google Scholar]
- Close TJ. 1996. Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiologia Plantarum 97, 795–803 [Google Scholar]
- Danyluk J, Perron A, Houde M, Limin A, Fowler B, Benhamou N, Sarhan F. 1998. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. The Plant Cell 10, 623–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg G. 1943. Colorimetric determination of hydrogen peroxide. Industrial and Engineering Chemistry, Analytical Edition 15, 327–328 [Google Scholar]
- Eriksson SK, Harryson P. 2011. Dehydrins: molecular biology, structure and function. In: Lüttge U, Beck E, Bartels D, eds. Plant desiccation tolerance. Berlin: Springer, 289–305 [Google Scholar]
- Eriksson SK, Kutzer M, Procek J, Gröbner G, Harryson P. 2011. Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. The Plant Cell 23, 2391–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueras M, Pujal J, Saleh A, Save R, Pagès M, Goday A. 2004. Maize Rabl7 overexpression in Arabidopsis plants promotes osmotic stress tolerance. Annals of Applied Biology 144, 251–257 [Google Scholar]
- Godoy JA, Lunar R, Torres-Schumann S, Moreno J, Rodrigo RM, Pintor-Toro JA. 1994. Expression, tissue distribution and subcellular localization of dehydrin TAS14 in salt-stressed tomato plants. Plant Molecular Biology 26, 1921–1934 [DOI] [PubMed] [Google Scholar]
- Guilloreau L, Combalbert S, Sournia-Saquet A, Mazarguil H, Faller P. 2007. Redox chemistry of copper-amyloid-beta: the generation of hydroxyl radical in the presence of ascorbate is linked to redox-potentials and aggregation state. Chembiochem 23, 1317–1325 [DOI] [PubMed] [Google Scholar]
- Hara M. 2010. The multifunctionality of dehydrins: an overview. Plant Signaling and Behavior 5, 503–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M, Fujinaga M, Kuboi T. 2005. Metal binding by citrus dehydrin with histidine-rich domains. Journal of Experimental Botany 56, 2695–2703 [DOI] [PubMed] [Google Scholar]
- Hara M, Kashima D, Horiike T, Kuboi T. 2010. Metal-binding characteristics of the protein which shows the highest histidine content in the Arabidopsis genome. Plant Biotechnology 27, 475–480 [Google Scholar]
- Hara M, Shinoda Y, Kubo M, Kashima D, Takahashi I, Kato T, Horiike T, Kuboi T. 2011. Biochemical characterization of the Arabidopsis KS-type dehydrin protein, whose gene expression is constitutively abundant rather than stress dependent. Acta Physiologiae Plantarum 33, 2103–2116 [Google Scholar]
- Hara M, Shinoda Y, Tanaka Y, Kuboi T. 2009. DNA binding of citrus dehydrin promoted by zinc ion. Plant, Cell and Environment 32, 532–541 [DOI] [PubMed] [Google Scholar]
- Hara M, Terashima S, Fukaya T, Kuboi T. 2003. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 217, 290–298 [DOI] [PubMed] [Google Scholar]
- Heyen BJ, Alsheikh MK, Smith EA, Torvik CF, Seals DF, Randall SK. 2002. The calcium-binding activity of a vacuole-associated, dehydrin-like protein is regulated by phosphorylation. Plant Physiology 130, 675–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde M, Dallaire S, N’Dong D, Sarhan F. 2004. Overexpression of the acidic dehydrin WCOR410 improves freezing tolerance in transgenic strawberry leaves. Plant Biotechnology Journal 2, 381–387 [DOI] [PubMed] [Google Scholar]
- Hundertmark M, Buitink J, Leprince O, Hincha DK. 2011. The reduction of seed-specific dehydrins reduces seed longevity in Arabidopsis thaliana. Seed Science Research 21, 165–173 [Google Scholar]
- Hundertmark M, Hincha DK. 2008. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9, 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I, Escuredo PR, Arrese-Igor C, Becana M. 1998. Oxidative damage in pea plants exposed to water deficit or paraquat. Plant Physiology 116, 173–181 [Google Scholar]
- Kawachi M, Kobae Y, Mimura T, Maeshima M. 2008. Deletion of a histidine-rich loop of AtMTP1, a vacuolar Zn2+/H+ antiporter of Arabidopsis thaliana, stimulates the transport activity. Journal of Biological Chemistry 283, 8374–8383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koag MC, Wilkens S, Fenton RD, Resnik J, Vo E, Close TJ. 2009. The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiology 150, 1503–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs D, Kalmar E, Torok Z, Tompa P. 2008. Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiology 147, 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger C, Berkowitz O, Stephan UW, Hell R. 2002. A metal-binding member of the late embryogenesis abundant protein family transports iron in the phloem of Ricinus communis L. Journal of Biological Chemistry 277, 25062–25069 [DOI] [PubMed] [Google Scholar]
- Lehmann S. 2002. Metal ions and prion diseases. Current Opinion in Chemical Biology 6, 187–192 [DOI] [PubMed] [Google Scholar]
- Lin CH, Peng PH, Ko CY, Markhart AH, Lin TY. 2012. Characterization of a novel Y2K-type dehydrin VrDhn1 from Vigna radiate. Plant and Cell Physiology 53, 930–942 [DOI] [PubMed] [Google Scholar]
- Mu P, Feng D, Su J, Zhang Y, Dai J, Jin H, Liu B, He Y, Qi K, Wang H, Wang J. 2011. Cu2+ triggers reversible aggregation of a disordered His-rich dehydrin MpDhn12 from Musa paradisiaca. Journal of Biochemistry 150, 491–499 [DOI] [PubMed] [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV. 2001. Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Molecular Biology 45, 263–279 [DOI] [PubMed] [Google Scholar]
- Ochoa-Alfaro AE, Rodríguez-Kessler M, Pérez-Morales MB, Delgado-Sánchez P, Cuevas-Velazquez CL, Gómez-Anduro G, Jiménez-Bremont JF. 2012. Functional characterization of an acidic SK3 dehydrin isolated from an Opuntiastreptacantha cDNA library. Planta 235, 565–578 [DOI] [PubMed] [Google Scholar]
- Palmer CM, Guerinot ML. 2009. Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nature Chemical Biology 5, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhakainen T, Hess MW, Mäkelä P, Svensson J, Heino P, Palva ET. 2004. Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Molecular Biology 54, 743–753 [DOI] [PubMed] [Google Scholar]
- Rahman LN, Smith GS, Bamm VV, Voyer-Grant JA, Moffatt BA, Dutcher JR, Harauz G. 2011. Phosphorylation of Thellungiella salsuginea dehydrins TsDHN-1 and TsDHN-2 facilitates cation-induced conformational changes and actin assembly. Biochemistry 50, 9587–9604 [DOI] [PubMed] [Google Scholar]
- Rees JD, Ingle RA, Smith JA. 2009. Relative contributions of nine genes in the pathway of histidine biosynthesis to the control of free histidine concentrations in Arabidopsis thaliana. Plant Biotechnology Journal 7, 499–511 [DOI] [PubMed] [Google Scholar]
- Rorat T. 2006. Plant dehydrins: tissue location, structure and function. Cellular and Molecular Biology Letters 11, 536–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorat T, Grygorowicz WJ, Irzykowski W, Rey P. 2004. Expression of KS-type dehydrins is primarily regulated by factors related to organ type and leaf developmental stage during vegetative growth. Planta 218, 878–885 [DOI] [PubMed] [Google Scholar]
- Saran M, Bors W. 1991. Direct and indirect measurements of oxygen radicals. Klinische Wochenschrift 69, 957–964 [DOI] [PubMed] [Google Scholar]
- Shekhawat UK, Srinivas L, Ganapathi TR. 2011. MusaDHN-1, a novel multiple stress-inducible SK(3)-type dehydrin gene, contributes affirmatively to drought- and salt-stress tolerance in banana. Planta 234, 915–932 [DOI] [PubMed] [Google Scholar]
- Shen B, Jensen RG, Bohnert HJ. 1997. Mannitol protects against oxidation by hydroxyl radicals. Plant Physiology 115, 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Lin HH. 2010. Role of plant dehydrins in antioxidation mechanisms. Biologia 65, 755–759 [Google Scholar]
- Svensson J, Ismail AM, Palva ET, Close TJ. 2002. Dehydrins. In: Storey KB, Storey JM, eds. Sensing, signaling and cell adaptation. Amsterdam: Elsevier, 155–171 [Google Scholar]
- Svensson J, Palva ET, Welin B. 2000. Purification of recombinant Arabidopsis thaliana dehydrins by metal ion affinity chromatography. Protein Expression and Purification 20, 169–178 [DOI] [PubMed] [Google Scholar]
- Tompa P. 2009. Structure and function of intrinsically disordered proteins. Boca Raton FL: CRC Press; [Google Scholar]
- Tunnacliffe A, Wise MJ. 2007. The continuing conundrum of the LEA proteins. Naturwissenschaften 94, 791–812 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang H, Li R, Ma Y, Wei J. 2011. Expression of a SK2-type dehydrin gene from Populus euphratica in a Populus tremula×Populus alba hybrid increased drought tolerance. African Journal of Biotechnology 10, 9225–9232 [Google Scholar]
- Whitmore L, Wallace BA. 2004. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Research 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski M, Webb R, Balsamo R, Close TJ, Yu XM, Griffith M. 1999. Purification, immunolocalization, cryoprotective, and antifreeze activity of PCA60: a dehydrin from peach (Prunus persica). Physiologiae Plantarum 105, 600–608 [Google Scholar]
- Xing X, Liu Y, Kong X, Liu Y, Li D. 2011. Overexpression of a maize dehydrin gene, ZmDHN2b, in tobacco enhances tolerance to low temperature. Plant Growth Regulation 65, 109–118 [Google Scholar]
- Yin Z, Rorat T, Szabala BM, Ziólkowska A, Malepszy S. 2006. Expression of a Solanum sogarandinum SK3-type dehydrin enhances cold tolerance in transgenic cucumber seedlings. Plant Science 170, 1164–1172 [Google Scholar]
- Zhong L, Johnson WC., Jr 1992. Environment affects amino acid preference for secondary structure. Proceedings of the National Academy of Sciences, USA 89, 4462–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.