Abstract
Drought is a major environmental stress limiting growth of perennial grasses in temperate regions. Plant drought tolerance is a complex trait that is controlled by multiple genes. Candidate gene association mapping provides a powerful tool for dissection of complex traits. Candidate gene association mapping of drought tolerance traits was conducted in 192 diverse perennial ryegrass (Lolium perenne L.) accessions from 43 countries. The panel showed significant variations in leaf wilting, leaf water content, canopy and air temperature difference, and chlorophyll fluorescence under well-watered and drought conditions across six environments. Analysis of 109 simple sequence repeat markers revealed five population structures in the mapping panel. A total of 2520 expression-based sequence readings were obtained for a set of candidate genes involved in antioxidant metabolism, dehydration, water movement across membranes, and signal transduction, from which 346 single nucleotide polymorphisms were identified. Significant associations were identified between a putative LpLEA3 encoding late embryogenesis abundant group 3 protein and a putative LpFeSOD encoding iron superoxide dismutase and leaf water content, as well as between a putative LpCyt Cu-ZnSOD encoding cytosolic copper-zinc superoxide dismutase and chlorophyll fluorescence under drought conditions. Four of these identified significantly associated single nucleotide polymorphisms from these three genes were also translated to amino acid substitutions in different genotypes. These results indicate that allelic variation in these genes may affect whole-plant response to drought stress in perennial ryegrass.
Key words: Antioxidant, association mapping, Cyt Cu-ZnSOD, drought, FeSOD, LEA3, leaf water content, Lolium perenne.
Introduction
Perennial ryegrass (Lolium perenne L.) is a cool-season perennial grass species from the family Poaceae. Native to Europe, Asia, and northern Africa, this species is widely cultivated in the temperate regions of the world as a high quality forage and for landscaping, as well as for other elements of ecosystem services such as carbon sequestration, soil formation and protection, and nutrient cycling. Perennial ryegrass is a self-incompatible diploid (2n=2x=14) outcrossing species (Cornish et al., 1980). Among the major perennial grass species, perennial ryegrass is a good model for studying association of gene and drought tolerance because of its diploid genetics, available genetic and genomics resources, rapid stress responses, and available germplasm collections. Knowledge gained from studying this species will facilitate further investigation in other perennial grass species with more complex, polyploid genomes.
Drought stress is an important factor affecting growth and development of cool-season perennial grasses. Plant drought tolerance is a complex quantitative trait, involving multiple metabolic pathways. A number of genes involved in plant drought responses and tolerance have been identified in model plants and crops (Seki et al., 2002; Rabello et al., 2008; Guo et al., 2009; Khowaja et al., 2009; Campo et al., 2012). In rice (Oryza sativa L.) and Arabidopsis thaliana, two groups of genes have been reported to be involved in drought responses: genes encoding functional or structural proteins and genes encoding regulatory proteins (Rabbani et al., 2003; Shinozaki and Yamaguchi-Shinozaki, 2007). Genes in antioxidative metabolic pathways are among the key groups of functional genes, because these antioxidant pathways play an important role in detoxifying reactive oxygen species that can accumulates under drought or other abiotic stress conditions. In perennial ryegrass, ~10% of differentially expressed genes were associated with functions of detoxification (Liu and Jiang, 2010). Transformation of pea (Pisum sativum L.) PsMnSOD encoding manganese superoxide dismutase to rice, and overexpression of Arabidopsis AtAPX encoding cytosolic ascorbate peroxidase and AtMDAR encoding monodehydroascorbate reductase in tobacco (Nicotiana tabacum L.) improved drought tolerance of the targeted plants (Badawi et al., 2004; Wang et al., 2005; Eltayeb et al., 2007). Other functional or regulatory proteins also promote drought tolerance, including late embryogenesis abundant (LEA) proteins (Duan and Cai, 2012) functioning in maintenance of protein and membrane structure (Bartels and Sunkar, 2005), and the aquaporins plasma membrane intrinsic proteins (PIPs) and tonoplast intrinsic proteins (TIPs) that facilitate the diffusion of water and uncharged solutes across membranes (Maurel et al., 2008). Overexpression of OsPIP1 genes in drought-sensitive lowland rice enhanced its drought avoidance by increased root hydraulic conductivity, and higher leaf water potential and relative cumulative transpiration (Lian et al., 2004). Overexpression of BnPIP1 in transgenic tobacco plants showed improved tolerance to drought stress by maintaining normal leaf rigidness after 10 d of water deprivation, while the wild type already showed severe leaf wilting (Yu et al., 2005). These findings suggest that expression of candidate genes encoding functional or regulatory proteins may enhance drought tolerance in plants. At the population level, the relationship between these genes and drought tolerance in a diverse population panel of a plant species is largely unknown; specifically, whether allelic diversity at these genes underlies phenotypic variation associated with whole-plant tolerance to drought stress.
Linkage disequilibrium (LD)-based association mapping is a powerful tool for genetically dissecting complex traits controlled by multiple quantitative trait loci (QTLs) (Yu et al., 2006a; Zhao et al., 2007; Harjes et al., 2008; Tian et al., 2009; Emanuelli et al., 2010; Li et al., 2011; Skøt et al., 2011; Cook et al., 2012). Through exploitation of historical recombination events at a population level, association studies identify marker–trait associations and test large numbers of alleles in a diverse population (Yu and Buckler, 2006). For a species in which LD decays rapidly, candidate gene association mapping is appropriate to relate sequence variations in selected candidate genes to specific traits of interest (Zhu et al., 2008). Rapid LD decay within 300–2000bp has been detected in perennial ryegrass, depending on the population size and individual genes (Skøt et al., 2007; Xing et al., 2007; Brazauskas et al., 2011; Fiil et al., 2011). A candidate gene, FLOWERING LOCUS T, has been found to be associated with changes in flowering time across a range of populations of perennial ryegrass (Skøt et al., 2011). In maize (Zea mays L.), candidate gene association mapping identified single nucleotide polymorphisms (SNPs) in genes that affect abscisic acid levels in floral tissue during drought stress (Setter et al., 2011). These findings indicate that the candidate gene association mapping approach can be effectively used to establish associations between targeted genes with known function and complex traits in outcrossing species.
The objective of this study was to conduct candidate gene association mapping of drought tolerance in perennial ryegrass. It was hypothesized that diverse perennial ryegrass varied greatly in whole-plant physiological traits related to drought tolerance. Given the evidence that genes involved in antioxidant pathways and in other functional or regulatory systems may play a role in drought tolerance, it was also hypothesized that these candidate genes were significantly associated with drought tolerance traits in perennial ryegrass. In this study, whole-plant physiological responses of a global collection of 192 diverse perennial ryegrass accessions to drought stress were examined across six environments. Population structure and relative pairwise kinship of the population were determined by using a set of 109 simple sequence repeat (SSR) markers. Gene–trait associations were analysed after controlling population structure.
Materials and methods
Plant materials
A global collection of 186 perennial ryegrass (L. perenne L.) accessions was obtained from the USDA National Plant Germplasm System at the Western Regional Plant Introduction Station in Pullman, WA, USA, and six turf-type commercial cultivars were obtained from the seed industry (Turf-Seed Company, Gervais, OR, USA; Scotts Inc., Marysville, OH, USA). This collection was based on geographical locations of accessions to maximize ecotype diversity (Supplementary Table S1 available at JXB online). The panel included 72 wild, 66 cultivated, and 54 accessions with uncertain pedigree according to germplasm bank classification. All the accessions were confirmed as diploid by flow cytometry (Y. Wang et al., 2009). A single seed from each accession was initially sown in small plastic pots containing a sandy loam soil with a pH of 6.9. Each accession was then propagated multiple times by tillers to maintain genetic uniformity. The grasses were planted in the field in Wanatah (41°26’N and 86°54’W), West Lafayette (40°25’N and 86°54’W), and Vincennes (38°40’N and 87°31’W) of Indiana, USA in August 2008, respectively. Soil type was sandy loam in Wanatah and Vincennes, and silt loam in West Lafayette. Each location had three replications for each accession with the same genotypes across locations. The maintenance of plants in the field followed the routine practices of perennial ryegrass during the experiment. A total of 179 accessions were alive after winter 2008 and were used for phenotypic data collection. Drought stress was imposed in the plots by withholding water for 7–10 d after initial measurements were taken under well-watered conditions in May at each location in 2009 and 2010. On average, soil water content and mean air temperatures under well-watered and drought conditions were 15.0% and 12.1% and 20.6 ºC and 23.1 ºC across 2 years, respectively. The detailed information on sampling dates, soil water, and air temperatures in all three locations are shown in Supplementary Table S2. All accessions were under drought stress for the same amount of time. The typical climatic pattern at the field site included multiple dry spells each lasting 7–15 d during the growing season, which caused light to severe leaf wilting in the perennial ryegrass accessions.
Phenotypic traits
Data were collected before and after drought stress at three locations in 2009 and 2010 for the following traits: canopy and air temperature difference (CAD), leaf water content (LWC), chlorophyll fluorescence (Fv/Fm), and leaf wilting. CAD was measured using a hand-held infrared thermometer (AGRI-THERM III, Everest Interscience Inc., Tuscon, AZ, USA). LWC was measured according to the following equation: LWC (g g–1)=(FW−DW)/DW, where FW is the leaf fresh weight and DW is the dry weight. FW was measured immediately after leaves were cut in the field. The leaves were then dried in an oven at 75 °C for 72h to obtain the DW. Fv/Fm was measured on 4–5 arbitrarily selected leaves for each accession at 2200h using a fluorescence meter (OS-30P, OPTI-Sciences, Hudson, NH, USA). It allowed a minimum of 45min for leaf dark adaptation prior to sampling, which was sufficient for photoinhibited leaves to recover their fluorescence ability (Demmig and Björkman, 1987). Leaf wilting was assessed by visual rates using a scale of 1 (slight wilting) to 9 (severe wilting). CAD, LWC, and Fv/Fm were measured under both well-watered and drought-stressed conditions, while leaf wilting was only assessed under drought stress. Endophyte Neotyphodium lolii infection in each plant accession was determined using a tissue print immunoblot procedure (Agrinostics Ltd. Co., Watkinsville, GA, USA).
To examine the consistency and accuracy of phenotypic data, repeatability of phenotypic traits across six environments (2 years×3 locations) as reflected by heritability (h2) was calculated using PROC MIXED (SAS Institute, Version 9.1, Cary, NC, USA). The h2 was calculated as follows: h2=σg2/(σg2+σge2/e+σe2/re), where σg2, σe2, and σge2 represent Type III SS (sums of squares) for genotype (G), environment (E), and G×E, respectively. The ‘e’ is degree of freedom of E and ‘re’ is the degree of freedom of G×E. Least square means were calculated based on the pooled data from six environments×three replications since h2 was >0.5 for all traits (Supplementary Table S3 at JXB online). Analysis of variance (ANOVA) was calculated using SAS PROC MIX with both accessions and drought stress as fixed effects, and environments and G×E as random factors.
Molecular profiling
Young leaves from each accession were collected, frozen in liquid N, and stored at –80 ºC for DNA isolation using a cetyltrimethyl ammonium bromide (CTAB) method (Murray and Thompson, 1980). DNA concentration was quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Rockland, DE, USA) and diluted to 15ng μl–1 for polymerase chain reaction (PCR). A total of 109 published genome-wide SSR markers (Kubik et al., 2001; Jensen et al., 2005; Lauvergeat et al., 2005; Gill et al., 2006; King et al., 2008) across seven chromosomes in perennial ryegrass were screened in all accessions (Supplementary Table S4 at JXB online). The forward primer sequence was modified by adding an M13 tail (5’-ACG ACG TTG TAA AAC GAC) to the 5’ end, and the M13 primers were labelled with four fluorescent dyes of different coloirs [FAM (blue), VIC (green), PET (yellow), and NED (red)] for multiplex analysis in ABI 3730 (Applied Biosystems Inc., Foster City, CA, USA). Each 10 μl PCR reaction consisted of 1× PCR buffer, 2.5mM MgCl2, 0.2mM dNTP mix, 0.05 μM forward tailed primer, 0.1 μM reverse primer, 0.05 μM fluorescent-labelled M13 primer, 1.0U of Taq DNA polymerase, and 60ng of DNA. All PCRs were performed in a 384-well iCycler thermocycler (Bio-Rad Inc., Hercules, CA, USA) using a touch-down program (Yu et al., 2006b). In brief, PCR was started at 95 ºC for 5min, then ran for four cycles at 96 ºC for 1min, 68 ºC for 5min, with a decrease of 2 ºC in each consequent cycle; and 72 ºC for 1min; four cycles of 96 ºC for 1min, 58 ºC for 2min, with a decrease of 2 ºC in each consequent cycle, and 72 ºC for 1min; followed by 24 cycles of 96 ºC for 1min, 50 ºC for 1min, 72 ºC for 1min, with a final extension of 72 ºC for 5min and 4 ºC for 5min. The amplified fragments were separated in an ABI 3730 DNA Sequencer (Applied Biosystems Inc.). Alleles were called using GeneMarker 1.6 software (SoftGenetics, LLC, State College, PA, USA) and manually checked twice for accuracy. Allele size differing in at least 2bp was considered to be polymorphic between accessions. If a primer amplified more than one fragment in an accession and these fragments appeared in different accessions, they were counted as different loci. All confirmed polymorphic alleles were used for structure analysis.
Population structure and relative kinship
Population structure was determined by model-based clustering using STRUCTURE 2.3.1 (Pritchard et al., 2000). The structure was run 10 times by setting pre-defined k (the number of population groups) from 1 to 10 using admixture models, with 10 000 burn-in time and 10 000 iterations of Markov chain convergence for each run. The optimal number of groups was determined when k was set to five. Subsequently, 10 independent runs were performed by setting k from three to six, with burn-in time of 50 000 and replication number of 100 000 for each run. This did not produce different results from those of the preliminary runs. Among the 10 runs, the one with the highest likelihood value (k=5) was selected to assign the proportion of membership for each accession. Relative pairwise kinship was calculated using SPAGeDi (Hardy and Vekemans, 2002) with all 109 polymorphic SSR markers. The coefficient of Loiselle (Loiselle et al., 1995) was used to obtain the pairwise kinship matrix. The distributions of pairwise kinship for 192×192 data points were summarized using Excel’s internal function ‘frequency’.
Candidate gene selection, sequencing. and SNP identification
Fourteen candidate genes were selected for sequencing, including 10 putative genes encoding antioxidant enzymes and additional putative genes of LEA, PIP, TIP, and MAPK (encoding mitogen-activated protein kinase) (Supplementary Table S5 at JXB online). Previous results in perennial ryegrass showed that genes involved in detoxification play a role in drought tolerance (Liu and Jiang, 2010), therefore more genes were selected in the antioxidative pathways. Primers were designed based on various sources, including GenBank accession AJ634002 for CAT encoding catalase, EU054285–EU054295 for MAPK, AB533342.1 for LEA3, FJ472349.1 for TIP1, the perennial ryegrass expressed sequence tag (EST) for PIP1, and the conserved coding region of orthologous genes from closely related species such as rice, Brachypodium distachyon, sorghum (Sorghum bicolor L.), and maize for other genes used in this study. Primers for amplification of PCR fragments for sequencing of the 14 candidate genes are listed in Supplementary Table S5.
Both genomic DNA and reverse-transcribed cDNA were used as PCR amplification templates for synthesis and sequencing of these genes. Direct genomic DNA sequencing often resulted in unclean sequence readings because of the high outcrossing and heterozygous nature of perennial ryegrass; thus, the more problematic introns were excluded for sequencing in this study by doing the following. First, the exon distribution of all these candidate genes was plotted in NCBI Splign service (Kapustin et al., 2008) by using the orthologous genes from rice and Arabidopsis. Secondly, each of the two candidate genes, putative CAT and MnSOD, had one long exon that covered the major part of that gene; therefore, the primers for these two genes were designed based on a single exon, and genomic DNA was used as PCR template. The other 12 genes usually formed by short exons, thus an mRNA reverse-transcribed cDNA was used to design the primer.
Total RNA was isolated from leaf tissues using TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA) and was treated with DNase (TURBO DNA-Free Kit; Ambion, Austin, TX, USA) to remove contaminating genomic DNA. The first-strand cDNA was synthesized from 2 μg of DNase-treated RNA using an iscript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). PCR products with the correct length were confirmed using agarose gel and visualized in gel image system (UVP, Upland, CA, USA). The target fragments were cut and recovered using a 96-well high-throughput gel recovery kit (Zymo Research, Irvine, CA, USA). Sequencing reactions were carried out by using Big Dye Terminator kit version 3.1 (Applied Biosystems Inc.). Sequencing was conducted using an ABI 3730 genetic analyzer according to the manufacturer’s instructions (Applied Biosystems Inc.) in the Genomic Center at Purdue University.
Diploid SNPs were identified using the NovoSNP program 3.0.1 Microsoft Windows Platform version (Weckx et al., 2005). This software can deal with heterozygous sequence data. Multiple alignments were performed automatically, and SNP position and the heterozygous allele were detected using the software. Rare SNPs were excluded for SNP counting when the total non-major allele count was <5% (~8 accessions in this population). After manual verification guided by quality score, diploid SNP information with physical positions was obtained.
Linkage disequilibrium and association mapping analysis
The LD was calculated for each candidate gene using TASSEL 2.1 (Bradbury et al., 2007). The generated r 2 value was plotted against the physical distance among each pair of SNPs. Meanwhile, data from all candidate genes was pooled together to generate the overall LD for perennial ryegrass. Candidate gene association analysis was performed on the 179 perennial ryegrass accessions using the GLM by implementing Q model and simple linear model with the TASSEL 2.1 software package (Bradbury et al., 2007). Each SNP was fit as a fixed effect one at a time to test the association between the SNP and phenotype. Minor allele with frequency (MAF) <5% was filtered prior to association analysis. After this filtering, SNPs that passed the threshold of P-value <10−03 were deemed significant according to procedure of Sukumaran et al. (2012) with modifications. Briefly, the threshold P-value was determined jointly by considering that these are SNPs from candidate genes and their numbers are not very large and by considering the pattern of the quantile–quantile (Q–Q) plot of the selected model and the point at which the observed F-test statistics deviated from the expected F-test statistics (Suppelmentary Fig. S1 at JXB online).
In silico sequence analysis for functional substitution(s)
The sequence variations of putative LpCyt Cu-ZnSOD, LpFeSOD, and LpLEA3 showing significant association with traits were further analysed to identify putative functional amino acid substitution(s). DNA sequences were aligned using the ClustalW2 online service (Larkin et al., 2007). The amino acid sequence of each peptide was directly inferred in NCBI using BlastX and matched with other orthologous proteins in other plant species. The predicted secondary structures for the proteins were obtained using PredictProtein (Rost et al., 2004) and Jpred (Cole et al., 2008). The phylogenetic tree was constructed in ClustalW2- Phylogeny (http://www.ebi.ac.uk/Tools/phylogeny/clustalw2_phylogeny/) by alignment of the sequences of plant species including perennial ryegrass, Italian ryegrass (Lolium multiflorum Lam.), maize, rice, Arabidopsis, sorghum, wheat (Triticum aestivum L.), barley (Hordeum vulgare L.), and cotton (Gossypium arboretum L.), and incorporating into the Neighbor–Joining method with bootstrap analysis of 1000 replicates. The calculated tree file was viewed in TreeView version 1.6.6 software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Results
Variations in drought response
Drought stress caused leaf wilting and increased CAD, reduced LWC, but did not affect Fv/Fm in the panel of perennial ryegrass accessions (Supplementary Table S6 at JXB online). Accessions differed significantly in CAD, Fv/Fm. LWC, and leaf wilting (Table 1; Supplementary Table S1, Supplementary Fig. S2). Among all accessions across six environments, CAD ranged from –2.67 ºC to 2.08 ºC, Fv/Fm ranged from 0.75 to 0.83, and LWC ranged from 2.76 (g g–1) to 6.17 (g g–1) under well-watered conditions (Table 1; Supplementary Fig. S2). Under drought conditions, the minimum and maximum values were 0.53 ºC and 6.53 ºC for CAD, 0.71 and 0.84 for Fv/Fm, 1.80 (g g–1) and 3.93 (g g–1) for LWC, and 2.7–8.0 for leaf wilting, respectively (Table 1; Supplementary Table S1). Significant correlations were found between CAD, Fv/Fm, and LWC under well-watered and drought conditions, respectively (Table 2; Supplementary Fig. S3). Under drought stress, Fv/Fm was negatively correlated with CAD and leaf wilting; LWC was negatively correlated with leaf wilting, whereas CAD was positively correlated with leaf wilting. Under well-watered conditions, such correlations were not observed (Table 2). Twenty-six accessions were endophyte infected. Analysis of similarities indicated no significant differences in the overall drought tolerance of the perennial ryegrass accessions based on endophyte infection (Global R=215, P=0.216, 999 permutations). The lack of any significant pattern of drought tolerance among infected and uninfected accessions was confirmed visually using a two-dimensional rendering resulting from the non-metric multidimensional scaling analysis (stress=0.1).
Table 1.
Range of canopy and air temperature difference (CAD), leaf water content (LWC) chlorophyll fluorescence (Fv/Fm), and wilting for 179 diverse perennial ryegrass accessions under control (C) and drought (D) conditions.
| CAD (°C) | Fv/Fm | LWC (g g–1) | Wilting | ||||
|---|---|---|---|---|---|---|---|
| C | D | C | D | C | D | D | |
| Minimum | –2.67a | 0.53 | 0.746 | 0.707 | 2.76 | 1.80 | 2.7 |
| Maximum | 2.08 | 6.53 | 0.826 | 0.836 | 6.17 | 3.93 | 8.0 |
| Mean | –1.08 | 2.75 | 0.808 | 0.798 | 4.18 | 2.57 | 4.6 |
| SDb | 0.92 | 0.99 | 0.010 | 0.016 | 0.53 | 0.32 | 0.94 |
a Data summarized across three locations for 2009 and 2010.
b Standard deviation.
Table 2.
Pearson correlation coefficients among canopy air temperature (CAD), chlorophyll fluorescence (Fv/Fm), leaf water content (LWC), and leaf wilting (wilting) under well-watered conditions (C) and drought stress (D).
| Traits | C-CAD | C-Fv/Fm | C-LWC | D-CAD | D-Fv/Fm | D-LWC | D-wilting |
|---|---|---|---|---|---|---|---|
| C-CAD | 1 | ||||||
| C-Fv/Fm | 0.01 | 1 | |||||
| C-LWC | 0.02 | –0.06 | 1 | ||||
| D-CAD | 0.56*** | –0.06 | –0.14 | 1 | |||
| D-Fv/Fm | –0.10 | 0.50*** | 0.05 | –0.16* | 1 | ||
| D-LWC | –0.06 | –0.06 | 0.34*** | –0.02 | 0.06 | 1 | |
| D-wilting | –0.08 | –0.07 | 0.08 | 0.18* | –0.15* | –0.15* | 1 |
*, **, and *** indicate significant at P < 0.05, P < 0.01, and P < 0.001, respectively.
The overall heritability of various traits was high, ranging from 0.51 to 0.86 among six environments in 2009 and 2010 (Supplementary Table S3 at JXB online), indicating high repeatability over testing environments; thus least square means (lsmeans) of individual traits were calculated across all environments tested for association analysis.
Population structure and relative kinship
A total of 2180 SSR alleles were scored for 109 SSR markers across 192 accessions (Supplementary Table S4 at JXB online). The average number of alleles per marker, major allele frequency, polymorphism information content, and the overall mean gene diversity was 20, 0.34, 0.76, and 0.79, respectively (Supplementary Table S7).
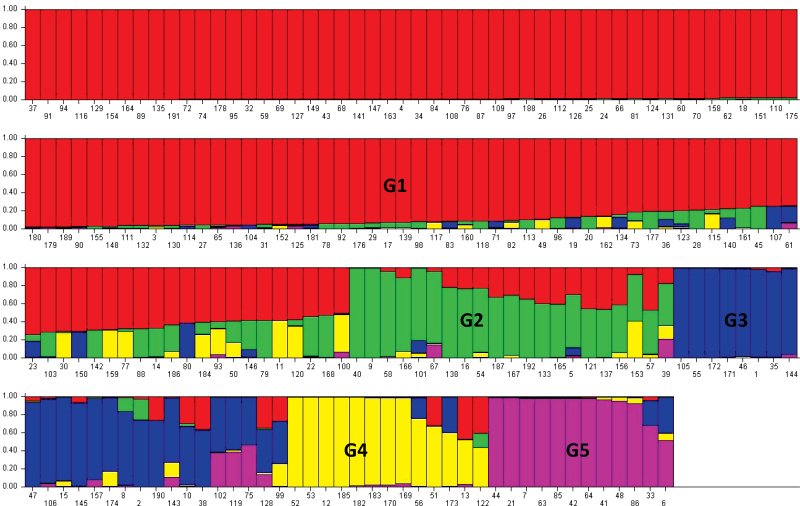
The STRUCTURE analysis identified five groups (G1, G2, G3, G4, and G5) in this panel of perennial ryegrass (Fig. 1) based on likelihood plots of the models, stability of grouping patterns across different runs, and germplasm information (M. Wang et al., 2009). The mean major allele frequency, gene diversity, and observed heterozygosity level ranged from 0.36 to 0.40, 0.71 to 0.76, and 0.42 to 0.53 across the five groups, respectively (Supplementary Table S7 at JXB online). G1 was the largest and most diverse group, with 121 accessions of mixed origins, including all the accessions from Oceania and the majority of the accessions from the USA, Canada, Europe, and South America. Approximately 89.4% of cultivated materials were assigned into G1 including 10 out of 11 turfgrass cultivars. Further distinct subgroups were not detected within G1 after independent structure analysis (data not shown). G2 contained 21 accessions; from Europe (14), Asia (6), and one turf-type commercial cultivar from the USA. G3 was the second largest group, with 25 accessions; mainly from Europe (13), Africa (7), and Asia (3). G4 contained 13 accessions, mainly from North Africa (Algeria, Morocco, and Tunisia). G5 was the smallest, with 12 accessions mainly from Southern Europe and Asia. There was no obvious kinship in this population. More than 55% of the pairwise kinship estimates were zero while ~35% were between zero and 0.05, indicating that 90% of estimates were <0.05. The 10% of estimates of >0.05 represented the minimum relative kinship among samples (Supplementary Fig. S4) and should not cause further complexity in association analysis.
Fig. 1.
Genetic relatedness of 192 perennial ryegrass accessions with 109 simple sequence repeats (SSRs) as analysed by the STRUCTURE program. Numbers on the y-axis indicate the membership coefficient. The colour of the bar indicates the five groups identified through the STRUCTURE program (G1=red, G2=green, G3=blue, G4=yellow, and G5=pink). Accessions with the same colour belong to the same group.
Heterozygous sequences and LD decay
A total of 14 candidate genes involved in antioxidant metabolism, dehydration, and water movement across membrane and signal transduction were sequenced to analyse sequence variation and find SNPs (Table 3). A total of 346 SNPs with an average of one SNP per 21bp were identified in 7288bp of the expressed regions across the 14 candidate genes. The number of SNPs per gene ranged from seven in putative LpMAPK to 50 in LpMDAR, while the average SNP frequency ranged from one SNP per 11bp in LpCAT and LpLEA3 to one SNP per 75bp in MAPK (Table 3). Across the population, heterozygous genotypes occurred up to 21.4% in total, ranging from 1.3% of A/T to 7.9% of A/G (Supplementary Table S8 at JXB online). Multiple alleles (e.g. AA, A/T, A/G, TT, or GG showed up in the same position of different accessions) were observed in some of the polymorphism positions, suggesting a rapid sequence evolution in perennial ryegrass, an outcrossing species.
Table 3.
Summary of genes used in this study and the number of single nucleotide polymorphism (SNP) sites in each gene across a diverse perennial ryegrass population.
| Putative gene | Full name | L1b | L2 | L3 | Ratec | SNP | Freq.d |
|---|---|---|---|---|---|---|---|
| LpCAT a | Catalase | 1478 | 577 | 505 | 93.8 | 45 | 11 |
| LpChl Cu-ZnSOD | Chloroplast Cu-Zn superoxide dismutase | 621 | 428 | 400 | 98.4 | 21 | 19 |
| LpGPX | Glutathione peroxidase | 509 | 545 | 446 | 77.5 | 15 | 30 |
| LpMnSOD | Manganese superoxide dismutase | 695 | 289 | 245 | 90.6 | 15 | 16 |
| LpAPX | Ascorbate peroxidase | 755 | 585 | 555 | 99.5 | >17e | 33 |
| LpMDAR | Monodehydroascorbate reductase | 1307 | 930 | 780 | 95.6 | 50 | 16 |
| LpDHAR | Dehydroascorbate reductase | 641 | 525 | 506 | 98.4 | 13 | 39 |
| LpGR | Glutathione reductase | 1490 | 1130 | 1087 | 99.5 | 44 | 25 |
| LpFeSOD | Iron superoxide dismutase | 767 | 550 | 537 | 90.7 | 24 | 22 |
| LpCyt Cu-ZnSOD | Cytosolic Cu-Zn superoxide dismutase | 459 | 421 | 355 | 91.8 | 17 | 21 |
| LpMAPK | Mitogen-activated protein kinase | 1109 | 612 | 522 | 98.4 | 7 | 75 |
| LpLEA3 | Late embryogenesis abundant, group 3 | 446 | 446 | 350 | 90.1 | 31 | 11 |
| LpPIP1 | Plasma membrane intrinsic protein, type 1 | 866 | 544 | 495 | 98.9 | 21 | 24 |
| LpTIP1 | Tonoplast intrinsic protein, type 1 | 750 | 55 | 505 | 97.8 | 26 | 19 |
| Total 14 | 11 892 | 8087 | 7288 | 94.3 | 346 | 21 |
a DNA template used for LpCAT and LpMnSOD; all others used RNA–cDNA template.
b L1, gene CDS length; L2, total length covered by the primer(s) designed in this study; L3, final readable sequenced length.
c Sequencing rate (%), the proportion of accessions that contain readable sequencing compared with all accessions tested; this was not L3/L2 or L3/L1.
d Frequency of SNP, calculated by L3/SNP, stands for the average SNP per bp length.
e Seventeen SNPs were confirmed, but many uncertain SNPs were found.
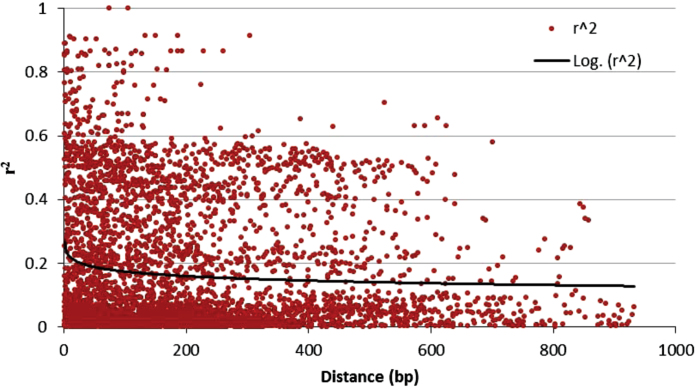
Combined LD analysis of 13 genes (LpAPX was dropped because of many uncertain SNPs) showed LD decay close to 0.1 in <1kb (Fig. 2), although variations in LD patterns were noted for each individual gene (Supplementary Fig. S5 at JXB online). The putative LpCAT, LpChl Cu-ZnSOD encoding chloroplastic copper-zinc superoxide dismutase, LpMnSOD, LpFeSOD encoding iron superoxide dismutase, LpCyt Cu-ZnSOD, LpDHAR encoding dehydroascorbate reductase, LpMAPK, LpLEA3, LpPIP1 encoding type 1 plasma membrane intrinsic protein, and LpTIP1 encoding type 1 tonoplast intrinsic protein showed a rapid LD decay pattern, with r 2 values close to or <0.1 within 200–500bp. LD decay of LpGR encoding glutathione reductase dropped to 0.1 within 1kb. The putative LpMDAR and LpGPX encoding glutathione peroxidase had relatively higher r 2 values from 0.3 to 0.5 at a distance >300–600bp (Supplementary Fig. S5 at JXB online). Diverse LD decay rates in perennial ryegrass might indicate a biased evolution of individual genes within the same species.
Fig. 2.
Linkage disequilibrium (LD) decay in perennial ryegrass. Plots of squared correlations of allele frequencies (r 2) against physical distance between pairs of SNPs in the 13 pooled genes. The LD pattern of individual genes can be found in Supplementary Fig. S5 at JXB online. (This figure is available in colour at JXB online.)
Association between genes and traits
Five population structures and minimum relative pairwise kinship were detected in the perennial ryegrass population used for this study (Fig. 1; Supplementary Fig S4 at JXB online). Therefore, the population structure controlled model (Q model) was selected as the best fit for association mapping analysis of genes and traits, which was also confirmed by quantile–quantile plots of F-test value (Supplementary Fig. S1). After removal of non-polymorphic and low-quality SNPs, a total of 346 SNPs were used for association analysis. By implementing the Q model, three SNPs at loci 134, 220, and 287 from LpCyt Cu-ZnSOD, one at locus 411 from LpFeSOD, and two at loci 237 and 285 from LpLEA3 were significantly associated with Fv/Fm, LWC, and LWC under drought stress, respectively, with a P-value ranging from 9.02×10–5 to 4.94×10–4 (Table 4). Compared with the simple linear model, this was an elimination of ~70% false-positive correlations under drought stress (Supplementary Table S9). Since locus 77 from LpLEA3 had a significant association with D-LWC under a P-value <0.01 level and linkage disequilibrium with locus 237 and 285 from LpLEA3, this locus along with another six significant associated loci were further analysed for amino acid substitutions among different haplotypes. R2 marker values ranged from 0.075 to 0.097 among all those associated SNPs. On average, each marker explained 8.9% (±0.8%) of phenotypic variance for its associated trait (Table 4). Four amino acid substitutions were identified from significant associated LpLEA3 and LpCyt Cu-ZnSOD, and gene-affecting traits were further analysed. In contrast, the significant associated SNP from LpFeSOD resulted in synonymous substitution and no amino acid change was found (Supplementary Fig. S6); therefore, it was not included for further analysis.
Table 4.
Association of candidate genes with traits of perennial ryegrass assessed by using a population structure controlled model (Q model).
| Trait | Putative Gene | Locus | R 2_Marker | P_Marker |
|---|---|---|---|---|
| D-Fv/Fm | LpCyt Cu-ZnSOD | 134 | 0.0969 | 9.02E-05 |
| D-Fv/Fm | LpCyt Cu-ZnSOD | 220 | 0.0828 | 2.48E-04 |
| D-Fv/Fm | LpCyt Cu-ZnSOD | 287 | 0.0752 | 4.94E-04 |
| D-LWC | LpLEA3 | 285 | 0.0905 | 3.92E-04 |
| D-LWC | LpLEA3 | 237 | 0.0914 | 3.63E-04 |
| D-LWC | LpLEA3 | 77 | 0.0625 | 0.0048a |
| D-LWC | LpFeSOD | 411 | 0.0945 | 2.63E-04 |
a Locus 77 was included for further analysis due to its significant association under the P < 0.01 level and linkage disequilibrium with locus 237 and 285.
D-Fv/Fm, chlorophyll fluorescence under drought conditions; D-LWC, leaf water content under drought conditions.
Putative LpLEA3 affecting leaf water content
The LpLEA3 is a putative LEA3 gene in perennial ryegrass, and an orthologue of wheat cold accumulated gene (WCS19) (Chauvin et al., 1993) and Italian ryegrass LEA3 (Lcs19) (Oishi et al., 2010). The total sequenced length of LpLEA3 was ~450bp, varying by accession. Its nucleotide sequence had up to 98% identity to Lcs19 and could align well until the 5’ stop codon. Based on the alignment results, the first 48bp coding sequence of Lcs19 was used to impute the first 48bp of LpLEA3. A 447bp full-length LpLEA3 gene was identified coding a 148 amino acid residue peptide with a mol. wt of 14.5kDa. There was a 22-mer signal peptide at the N-terminus (Supplementary Fig. S7 at JXB online), 18 strongly basic (+) amino acids (K, R), 22 strongly acidic (–) amino acids (D, E), and 38 alanines (A, 25.7%) in this predicted protein.
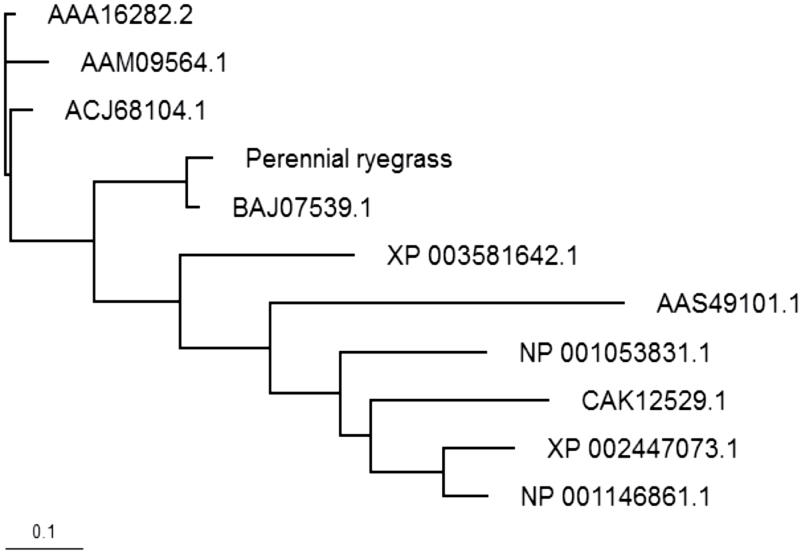
Phylogenetic analysis of ~500 LEA genes from different plant species confirmed that the putative LpLEA3 gene belonged to subfamily three (Supplementary Fig. S8 at JXB online) according to the common features of LEA3 that the gene family was positively charged at the N-terminus and negatively charged at the C-terminus, and was rich in alanine (>20%) and in polar residues. In this study, LpLEA3 showed a very similar structure to WCS19 (Chauvin et al., 1993): the C-terminal half was rich in the acidic amino acids aspartate and glutamate, thus giving this region a net negative charge of –10, whereas the N- terminal half was rich in the basic amino acids lysine and arginine, having a net positive charge of +6, and rich in alanine (25.7%). A high rate of amino acid substitutions was found during the evolution of this gene (Fig. 3). Phylogenetic analysis of LEA3 subfamily members in the selected species revealed that LpLEA3 was more closely related to its orthologue in Italian ryegrass, rye (Secale cereal L.), wheat, barley and B. distachyon than to rice and Arabidopsis (Fig. 3).
Fig. 3.
Phylogenetic tree of the late embryogenesis abundant group 3 protein sequence from plant species. NCBI protein accession numbers were used to indicate homologous proteins except perennial ryegrass (Lolium perenne). Bar=0.1 amino acid substitutions per site. AAA16282.2, wheat (Triticum aestivum); AAM09564.1, rye (Secale cereal); ACJ68104.1, Barley (Hordeum vulgare); BAJ07539.1, Italian ryegrass (Lolium multiflorum); XP_003581642.1, Brachypodium distachyon; AAS49101.1, Arabidopsis thaliana; NP_001053831.1, rice (Oryza sativa); CAK12529.1, Sporobolus stapfianus; NP_001146861.1, maize (Zea mays); and XP_002447073.1, sorghum (Sorghum bicolor).
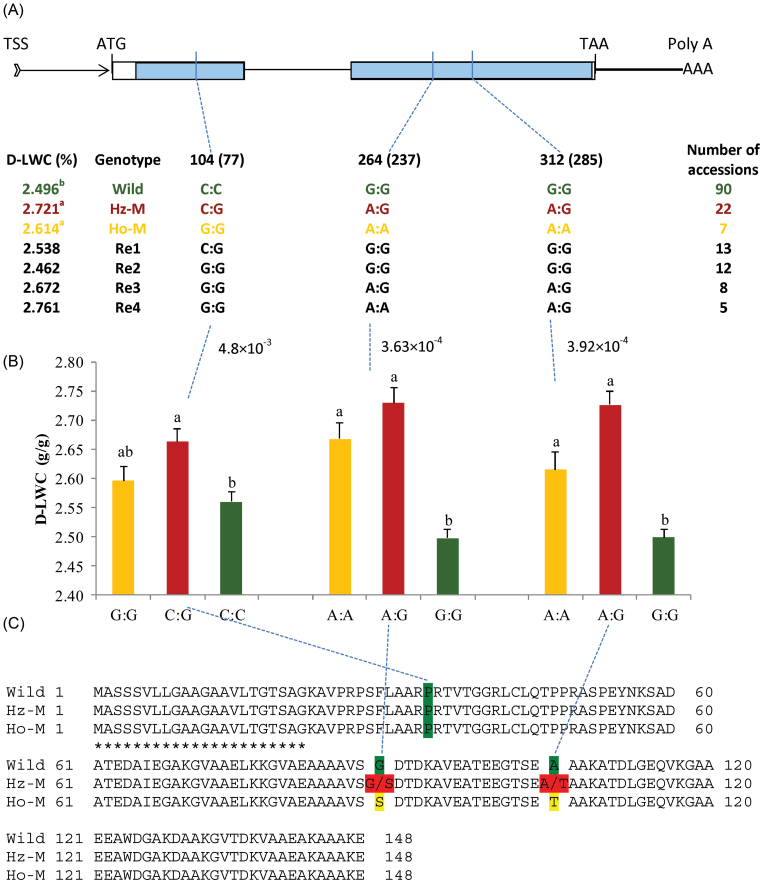
After comparison of sequences from the majority of the exon regions in LpLEA3 (Fig. 4A), D-LWC was found to be significantly associated with three SNPs at position 104, 264, and 312 of LpLEA3 (Fig. 4A), corresponding to loci 77, 237, and 285 in Table 4, respectively. Further analysis of all accessions using these three SNPs identified 90 wild-type, 22 heterozygous mutation (Hz-M), seven homozygous mutation (Ho-M), and 38 recombinant genotypes (R1–R4) (Fig. 4A). The D-LWC values of Hz-M and Ho-M genotypes were significantly higher than those of the wild genotype accessions, which indicated a dominance genetic model of this gene (Fig. 4A). Among these three SNPs, at the synonymous substitution position 104, only the Hz-M (C:G) genotype had significantly higher D-LWC than the wild genotype (C:C), but not the Ho-M (G:G) genotype (Fig. 4B). In contrast, Hz-M (A:G) and Ho-M (A:A) types had significantly higher D-LWC than the wild type (G:G) at two non-synonymous substitution positions 264 and 312 (Fig. 4B). Among these three SNP positions, two amino acid substitutions, from glycine (G) to serine (S) at residue 89 and from alanine (A) to threonine (T) at residue 105, were identified (Fig. 4C). Both these substitutions were from non-polar residues (G, A) to polar residues (S, T), which might improve the hydrophilic feature of this peptide and contribute to water absorption or conservation and eventually drought tolerance of plants.
Fig. 4.
Variant alleles and association mapping at LpLEA3. (A) Gene structure and genotypes analysis of LpLEA3: wild type, heterozygous mutation (Hz-M), homozygous mutation (Ho-M), and recombinants one to four (R1–R4) genotypes were identified on the basis of LpLEA3 sequence from 158 perennial ryegrass accessions. The positions of these three significant SNPs were shown using the full-length LpLEA3 gene sequence as reference, with the start codon designated as position zero. The SNP positions at 104, 264, and 312 were located according to the reference mentioned above, whereas 77, 237, and 285 (in parentheses) were SNP positions relative to the SNP calling reference (accession 120 sequence). The proposed wild-type genotype was highlighted in green, Hz-M in red, Ho-M in yellow, and R1–R4 in black, respectively. Values of leaf water content under drought conditions (D-LWC) with the same letter were not significantly different at P < 0.05 for the wild type, Hz-M, and Ho-M. TSS, transcription start site; arrowed band, untranslated region; thick bar, downstream region after the stop codon; blank box, exon; thin line, intron; ATG, start codon; TAA, stop codon; blue shaded area, region sequenced in this study. (B) Leaf water content under drought stress (D-LWC) against its significant associated SNP markers. Values beside dashed blue lines indicate P-values of association mapping results. The wild-type genotype was highlighted in green, Hz-M in red, and Ho-M in yellow, respectively. Values of D-LWC with the same letter were not significantly different at P < 0.05. (C) Amino acid substitutions in LpLEA3 among wild type, Hz-M, and Ho-M. Underlining with 21 asterisks indicate its signal peptide.
Putative LpCyt Cu-ZnSOD affecting Fv/Fm
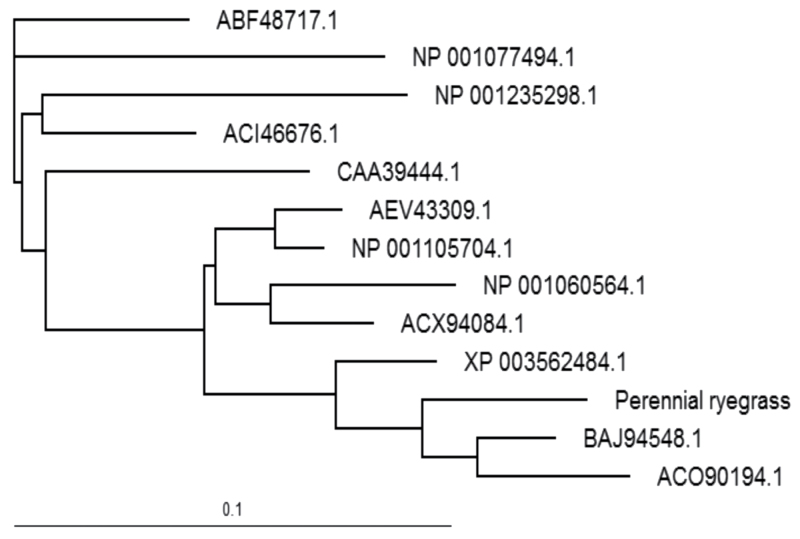
A partial coding sequence (356 out of 459bp) of LpCyt Cu-ZnSOD was identified in perennial ryegrass. Genotype A:A in LpCyt Cu-ZnSOD exhibited a significantly higher Fv/Fm than that of genotype A:T under drought conditions (Supplementary Fig. S9 at JXB online). A low rate of amino acid substitutions was found during the evolution of this gene (Fig. 5). The three SNPs (loci 134, 220, and 287) from LpCyt Cu-ZnSOD significantly associated with Fv/Fm under drought stress (D-Fv/Fm) had two amino acid substitutions with one from alanine (A) to glutamate (E) at residue 40 and the other from E to valine (V) at residue 91 from the N-terminus (Fig. 6). One of the substitutions was from a non-polar, neutral residue to a polar, acidic residue, while the other was from a polar, acidic residue to a non-polar, neutral residue. Moreover, the two non-polar, neutral residues (A and V) had very similar structures, which might indicate a complementary substitution within the same gene. Phylogenetic analysis showed that LpCyt Cu-ZnSOD in perennial ryegrass was more closely related to that in barley, wheat, and B. distachyon than to that in rice, zoysiagrass (Zoysia japonica), bamboo (Bambusa oldhamii), tobacco, cotton, Aradidopsis, soybean (Glycine max L.), and populus (Populus suaveolens) (Fig. 5).
Fig. 5.
Phylogenetic tree of cytosolic copper-zinc superoxide dismutase sequence from plant species. NCBI protein accession numbers were used to indicate homologous protein except perennial ryegrass (Lolium perenne). Bar=0.1 amino acid substitutions per site. ABF48717.1, populus (Populus suaveolens); NP_001077494.1, Arabidopsis thaliana; NP_001235298.1, soybean (Glycine max); ACI46676.1, cotton (Gossypium arboretum), CAA39444.1, tobacco (Nicotiana plumbaginifolia); AEV43309.1, zoysiagrass (Zoysia japonica); NP_001105704.1, maize (Zea mays); NP_001060564.1, rice (Oryza sativa); ACX94084.1, bamboo (Bambusa oldhamii); XP_003562484.1, Brachypodium distachyon; BAJ94548.1, barley (Hordeum vulgare); and ACO90194.1, wheat (Triticum aestivum).
Fig. 6.
Non-synonymous SNPs resulted in amino acid substitutions for putative LpCyt Cu-Zn SOD in perennial ryegrass. WT, wild type; DS, double substitutions. The first amino acid residue substitution from A to E occurred at position 32 (position 40 in reference to Cyt Cu-ZnSOD peptide of other species), which corresponded to an SNP at nucleotide position 287 in reference accession 92; the second amino acid residue substitution from E to V occurred at position 83 (position 91 in reference to Cyt Cu-ZnSOD peptide of other species), which corresponded to SNP at nucleotide position 134 in reference accession 92. This mutation A (E) reversed another mutation from G (A) to C (E) with in the same gene. (This figure is available in colour at JXB online.)
Discussion
Drought responses of perennial ryegrass
Leaf wilting, CAD, Fv/Fm, and LWC provide rapid and easy measurements for whole-plant responses; thus, they have been used to screen drought-tolerant plant materials and characterize drought tolerance at the whole-plant level (O’Neill et al., 2006; Jansen et al., 2009; Jiang et al., 2009; Luo et al., 2011). Relatively low LWC, positive CAD values, and high leaf wilting symptoms potentially indicate the occurrence of drought stress. Large variations in these traits across accessions of perennial ryegrass and further correlation analysis of traits suggest that they are adequate parameters for assessing drought tolerance in the field. High canopy temperature of grass plants may indicate a lower transpiration, and sensitive stomatal closure may act to maintain leaf water status and prevent leaf wilting in the field.
Fv/Fm is an indicator of photochemical efficiency in some species (Baker, 2008), but its value remained unchanged in other perennial grass species such as Kentucky bluegrass (Poa pratensis L.) and prairie junegrass (Koeleria macrantha) under drought stress even when LWC decreased to 68–70% (Bian and Jiang, 2009; Jiang et al., 2010). The present results indicated that overall Fv/Fm was unaffected by drought stress in the field across locations but was significantly different among accessions (Supplementary Tables S1, S6 at JXB online). In addition, Fv/Fm was also correlated with CAD and leaf wilting of perennial ryegrass under drought stress, indicating its potential use for screening drought tolerance of plant populations. Further reductions in Fv/Fm were associated with increasing intensity of drought stress in different plant species (Zulini et al., 2007; Luo et al., 2011).
Collectively, diverse phenotypic responses of perennial ryegrass accessions to drought stress are crucial for gene–trait association. In this study, leaf wilting and other drought response traits were studied in perennial ryegrasses exposed to 7–10 d of drought stress; however, stress could last longer than this duration in some areas where perennial ryegrass are grown, which may further affect phenotypic trait variation in germplasm. The potential variability in short-term versus long-term drought response should be taken into consideration when developing perennial grasses for improved drought tolerance.
SNP frequency and LD decay
The average SNP frequency in the 14 candidate genes was one SNP every 21bp across 192 accessions of perennial ryegrass. This was similar to the average frequency of one SNP per 24–33bp for 11 disease resistance candidate genes in L. perenne (Xing et al., 2007), but higher than one SNP every 94bp in five genes for shoot morphology in perennial ryegrass (Brazauskas et al., 2011) and one SNP per 127bp in nine genes for flowering time in 20 genotypes of perennial ryegrass (Fiil et al., 2011). The variation in SNP frequencies detected in different studies may be attributed to the population size and nature of individual genes in a particular plant species. LD is a measure of non-random association between alleles at different loci. The presence of LD is a prerequisite for association mapping of genes related to phenotypes. Currently, the threshold to indicate LD decay is r 2 < 0.1, r 2 < 0.2, or r 2 reduced to half of its initial value. Generally, LD decays more rapidly in outcrossing species than selfing species (Nordborg et al., 2002; Kim et al., 2007; Brazauskas et al., 2011; Li et al., 2011). A rapid LD decay indicates that more recombination exists within a short distance and that more markers are required to capture the high frequency of recombination (Zhu et al., 2008). In selfing species, LD decayed within 10–50kb on average in Arabidopsis (Nordborg et al., 2002; Aranzana et al., 2005; Kim et al., 2007) and 100–200kb in rice (Mather et al., 2007; McNally et al., 2009; Huang et al., 2010). A rapid decay within a distance of 500–1500bp was found in several outcrossing species such as maize (Remington et al., 2001; Tenaillon et al., 2001), rye (Li et al., 2011), sunflower (Helianthus annuus L.) (Liu and Burke, 2006), Douglas fir (Pseudotsuga menziesii) (Krutovsky and Neale, 2005), and perennial ryegrass (Xing et al., 2007; Brazauskas et al., 2011). Previous studies in perennial ryegrass found that LD decay was at 300bp to 2kb, depending on population size and genes tested (Skøt et al., 2007; Xing et al., 2007; Brazauskas et al., 2011; Fiil et al., 2011). The overall LD decay of all genes close to 0.1 of ~1kb found in this study was consistent with previous studies on candidate genes related to shoot morphology (Brazauskas et al., 2011) and flowering time in perennial ryegrass (Fiil et al., 2011). However, different candidate genes differed in rates of LD decay (relatively rapid or slow) across this diverse population of perennial ryegrass, which may indicate a non-uniformity of plant genome evolution.
Features of putative LpLEA3 and LpCyt Cu-ZnSOD
LEA proteins are extremely hydrophilic proteins that were originally discovered in cotton during seed germination (Dure et al., 1981). Their high content of water-interacting residues facilitates the scavenging of water molecules when a cell experiences changes in water status (Colmenero-Flores et al., 1999). They are thought to be associated with dehydration, cold and salt tolerance (NDong et al., 2002; George et al., 2009; Olvera-Carrillo et al., 2010), acting as protectors of macromolecules or cellular structures and preventing protein aggregation during water deficit (Bray, 1997). However, their physiological and molecular functions still remain largely unknown.
LEA proteins have been classified into between five and nine subfamilies on the basis of sequence similarity (Bies-Ethève et al., 2008) or eight subfamilies as defined by the Pfam database (Punta et al., 2012), the conserved domain, and the InterPro databases (Hunault and Jaspard, 2010). There were only two three-dimensional structures available for this entire LEA gene family (Tolleter et al., 2007), making it challenging to investigate its conserved domain and function–structure relationships. In Arabidopsis, 51 LEA proteins were classified into nine distinct groups with different amino acid lengths (Hundertmark and Hincha, 2008). Although genes within the LEA3 subfamily shared higher similarity than LEA genes from other LEA subfamilies, the diversity was still large within this subfamily, with the length ranging from 147 to 383 amino acids. LEA3 genes have a perfect 11-mer tandem repeats in some cereal crops (Dure, 1993) including sorghum (GQ494000.1), maize (NM_001111828.1), and B. distachyon (XP_003580519.1). Some putative LEA3 proteins such as WCOR14 (ACV60217.1) in common beet (Beta vulgaris L.) have intermediate 11-mer repeats, while others such as Lcs19 in Italian ryegrass (Oishi et al., 2010) have imperfect 11-mer repeats. The results also showed imperfect 11-mer repeats in LpEA3. These results indicate a dynamic evolution of the LEA3 gene family across plant species.
The majority of LEA proteins have been predicted to be highly hydrophilic and natively unstructured (Hundertmark and Hincha, 2008), which indicates a potential high hydrophilic ability with more polar residues sitting on this kind of unstructured peptide. In this study, both substitutions in LEA3 protein of perennial ryegrass were from non-polar residues (G, A) to polar residues (S, T). This might improve the hydrophilic nature of this unstructured peptide, thus enhancing cellular water retention and benefiting drought tolerance. Since the populations of perennial ryegrass were diverse with historical recombination events, the substitutions may be one of the evolutionary patterns of the LpLEA3 gene in perennial ryegrass. Furthermore, these amino acids substitutions were in line with SNPs at positions 237 and 285 in LpLEA3, which were significantly associated with D-LWC. These results indicate that LpLEA3 may facilitate drought tolerance by influencing the water status of plants. LpLEA3 was a homologue of Lcs19, a cold-stimulated gene identified in Italian ryegrass. The distribution of Lcs19 homologues has been found among temperate forage grasses but not in tropical grass species (Oishi et al., 2010). It suggests that LpLEA3 may be a suitable gene for studying the drought response of temperate grasses with more complex genomes.
SODs play a role in protecting against superoxide toxicity (Bowler et al., 1992). The primary sequences of FeSOD and MnSOD apoproteins were related, whereas that of Cu-ZnSOD was distinct (Kliebenstein et al., 1998). Cu-ZnSODs are largely located in the cytoplasm, sharing 80–90% similarity at the amino acids level across species (Crapo et al., 1992; Alscher et al., 2002). The putative LpCyt Cu-ZnSOD in perennial ryegrass had two amino acid substitutions in line with two SNPs associated with Fv/Fm under drought stress; however, the amino acids changed from A (non-polar, non-charged) to E (polar, negatively charged) and from E to V (non-polar, non-charged) with a very similar molecular structure between A and V (Fig. 6). These results suggest that these amino acid substitutions of LpCyt Cu-ZnSOD might not be functionally associated with Fv/Fm of perennial ryegrass. The bBlastx result indicated that this gene had up to 97% sequence similarity compared with 50 other plant species (data not shown). For example, perennial ryegrass and Arabidopsis shared 85% sequence similarity, with the same peptide residue length of 152 amino acids (Supplementary Fig. S10 at JXB online), indicating a highly conserved feature of Cyt Cu-ZnSOD across different species.
Allelic effects on complex traits
Significant allelic effects (8–10%) were found for stress tolerance in association results within perennial ryegrass. The difference in D-LWC between genotypes A:A and G:G in LpLEA3 was 6.8% (Fig. 4B). Similarly, genotype A:A and A:G in LpCyt Cu-ZnSOD had a 2% difference in D-Fv/Fm (Supplementary Fig. S9 at JXB online). In maize, SNPs from lg1 and lg2 explained the leaf angle difference by –0.8 ° and –1.13 ° (Tian et al., 2011). In their study, a total of 203, 287, and 295 SNPs were significantly associated with upper leaf angle, leaf length, and leaf width, respectively; demonstrating that the genetic architecture of the leaf traits was dominated by small allele effects. These allelic effects indicate that association mapping analysis of complex quantitative traits in outcrossing species could be challenging and that larger populations will be needed to uncover alleles with smaller effects. Nevertheless, the advantageous allele from LpLEA3 and LpCyt Cu-ZnSOD discovered in this study could be considered as a target candidate for gene transformation and for further development of markers for breeding programmes to improve drought tolerance of perennial ryegrass and other polyploid perennial grasses. Introgression mapping to introduce genes of chromosome segments from polyploid to diploid grasses may also be effective for improving drought tolerance of forage and turfgrass (Humphreys et al., 2003, 2005). It is expected that the genome sequence of perennial ryegrass will become available in the future, which will undoubtedly impact biology research, gene identification, and cultivar development of perennial ryegrass as well as other grasses with more complex genomes.
Conclusions
Five subpopulations have been identified in 192 global perennial ryegrass accessions. After controlling population structure, six associations were identified between candidate genes involved in antioxidant metabolism and dehydration and traits of Fv/Fm and LWC under drought stress. Two SNPs in the putative LpLEA3 and one SNP in LpFeSOD were significantly associated with variation in LWC and three SNPs in putative LpCyt Cu-ZnSOD were significantly associated with variations in Fv/Fm under drought stress. Molecular characterization of these significantly associated SNPs in LpLEA3 of perennial ryegrass identified two amino acid substitutions, both from a non-polar residue to a polar residue, that may enhance the hydrophilicity of this peptide and contribute to variation in leaf water content under drought stress. The study provided valuable information for candidate gene-based association mapping of drought tolerance in highly heterozygous perennial grasses.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Quantile–quantile plots of F-test value for model comparisons.
Figure S2. Distribution of canopy and air temperature difference (CAD), chlorophyll fluorescence (Fv/Fm), leaf water content (LWC), and leaf wilting under well-watered (C) and drought stress (D) conditions, respectively. The values were least square means calculated for all accessions across six environments.
Figure S3. Two-dimensional plots of significant correlations (P < 0.05 level) among canopy and air temperature difference (CAD), chlorophyll fluorescence (Fv/Fm), leaf water content (LWC), and leaf wilting under well-watered (C) and drought stress (D) of perennial ryegrass.
Figure S4. Distribution of pairwise relative kinship estimates across 192 accessions of perennial ryegrass.
Figure S5. LD decay patterns of individual candidate genes of LpCAT, LpChl Cu-ZnSOD, LpGPX, LpMnSOD, LpMDAR, LpDHAR, LpGR, LpFeSOD, LpCyt Cu-ZnSOD, LpMAPK, LpLEA3, LpPIP1, and LpTIP1 related to plant drought response.
Figure S6. Synonymous SNPs did not cause a change in amino acid sequence for putative LpFeSOD in perennial ryegrass.
Figure S7. Cleavage site prediction of putative LpLEA3 gene in perennial ryegrass.
Figure S8. Phylogenetic tree of ~500 late embryogenesis abundant group 3 (LEA3) protein sequences from plants.
Figure S9. Relationship between chlorophyll fluorescence (Fv/Fm) and SNP markers in LpCyt Cu-ZnSOD.
Figure S10. The sequence similarity of Cyt Cu-ZnSOD between perennial ryegrass and Arabidopsis.
Table S1. Origin, status information, and drought response traits of diverse perennial ryegrass accessions.
Table S2. Sampling date, soil moisture, and air temperatures in three locations before and after drought in 2009 and 2010.
Table S3. Repeatability of phenotypic traits of perennial ryegrasses across six environments under well-watered and drought conditions.
Table S4. Information on 109 simple sequence repeat (SSR) primers across seven linkage group of perennial ryegrass.
Table S5. Primers used for amplification of 14 candidate genes in perennial ryegrass.
Table S6. Analysis of variance (ANOVA) of phenotypic traits of perennial ryegrasses across six environments under well-watered and drought conditions.
Table S7. Summary statistics for all accessions and individual groups (G1–G5) within the panel of perennial ryegrass accessions identified by STRUCTURE analysis.
Table S8. Number of genotypes found in 12 candidate genes, LpChl Cu-ZnSOD, LpGPX, LpMnSOD, LpMDAR, LpDHAR, LpGR, LpFeSOD, LpCyt Cu-ZnSOD, LpMAPK, LpLEA3, LpPIP1, and LpTIP1, across 192 accessions of perennial ryegrass.
Table S9. Number of associations among 14 genes and traits under drought conditions using a simple linear model (S) and population structure (Q) controlled model in perennial ryegrass.
Acknowledgements
The authors thank Dr Torben Asp and Dr Stephen Byrne for kindly providing the reference full-length sequence of the LpLEA3 gene from their unpublished data. The authors also thank Dr Leif Skøt for consultation on perennial ryegrass exon sequencing, and Dr Jeff Holland for statistical advice. This work was supported by the O.J. Noer Research Foundation, The Midwest Regional Turfgrass Foundation, United State Golf Association, Mary S. Rice Farm Grant, and Purdue University Research Foundation.
References
- Alscher RG, Erturk N, Heath LS. 2002. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. Journal of Experimental Botany 53, 1331–1341 [PubMed] [Google Scholar]
- Aranzana MJ, Kim S, Zhao K, et al. 2005. Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genetics 1, e60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badawi GH, Kawano N, Yamauchi Y, Shimada E, Sasaki R, Kubo A, Tanaka K. 2004. Over-expression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit. Physiologia Plantarum 121, 231–238 [DOI] [PubMed] [Google Scholar]
- Baker NR. 2008. Chlorophyll fluorescence: a probe of photosynthesis in vivo . Annual Review of Plant Biology 59, 89–113 [DOI] [PubMed] [Google Scholar]
- Bartels D, Sunkar R. 2005. Drought and salt tolerance in plants. Critical Reviews in Plant Sciences 24, 23–58 [Google Scholar]
- Bian S, Jiang Y. 2009. Reactive oxygen species, antioxidant enzyme activities and gene expression patterns in leaves and roots of Kentucky bluegrass in response to drought stress and recovery. Scientia Horticulturae 120, 264–270 [Google Scholar]
- Bies-Ethève N, Gaubier-Comella P, Debures A, Lasserre E, Jobet E, Raynal M, Cooke R, Delseny M. 2008. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana . Plant Molecular Biology 67, 107–124 [DOI] [PubMed] [Google Scholar]
- Bowler C, Montagu MV, Inze D. 1992. Superoxide dismutase and stress tolerance. Annual Review of Plant Physiology and Plant Molecular Biology 43, 83–116 [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635 [DOI] [PubMed] [Google Scholar]
- Bray EA. 1997. Plant responses to water deficit. Trends in Plant Science 2, 48–54 [Google Scholar]
- Brazauskas G, Lenk I, Pedersen MG, Studer B, Lubberstedt T. 2011. Genetic variation, population structure, and linkage disequilibrium in European elite germplasm of perennial ryegrass. Plant Science 181, 412–420 [DOI] [PubMed] [Google Scholar]
- Campo S, Peris-Peris C, Montesinos L, Peñas G, Messeguer J, San, Segundo B. 2012. Expression of the maize ZmGF14-6 gene in rice confers tolerance to drought stress while enhancing susceptibility to pathogen infection. Journal of Experimetnal Botany 63, 983–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin LP, Houde M, Sarhan F. 1993. A leaf-specific gene stimulated by light during wheat acclimation to low temperature. Plant Molecular Biology 23, 255–265 [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Research 36, W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenero-Flores JM, Moreno LP, Smith CE, Covarrubias AA. 1999. Pvlea-18, a member of a new late-embryogenesis-abundant protein family that accumulates during water stress and in the growing regions of well-irrigated bean seedlings. Plant Physiology 120, 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JP, McMullen MD, Holland JB, Tian F, Bradbury P, Ross-Ibarra J, Buckler ES, Flint-Garcia SA. 2012. Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels. Plant Physiology 158, 824–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish MA, Hayward MD, Lawrence MJ. 1980. Self-incompatibility in ryegrass. Heredity 44, 55–62 [Google Scholar]
- Crapo JD, Oury T, Rabouille C, Slot JW, Chang LY. 1992. Copper,zinc superoxide dismutase is primarily a cytosolic protein in human cells. Proceedings of the National Academy of Sciences, USA 89, 10405–10409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig B, Björkman O. 1987. Comparison of the effect of excessive light on chlorophyll fluorescence (77K) and photon yield of O2 evolution in leaves of higher plants. Planta 171, 171–184 [DOI] [PubMed] [Google Scholar]
- Duan J, Cai W. 2012. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS One 7 e45117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dure L. 1993. A repeating 11-mer amino acid motif and plant desiccation. The Plant Journal 3, 363–369 [DOI] [PubMed] [Google Scholar]
- Dure L, Greenway SC, Galau GA. 1981. Developmental biochemistry of cottonseed embryogenesis and germination: changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry 20, 4162–4168 [DOI] [PubMed] [Google Scholar]
- Eltayeb A, Kawano N, Badawi G, Kaminaka H, Sanekata T, Shibahara T, Inanaga S, Tanaka K. 2007. Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 225, 1255–1264 [DOI] [PubMed] [Google Scholar]
- Emanuelli F, Battilana J, Costantini L, Le, Cunff L, Boursiquot JM, This P, Grando MS. 2010. A candidate gene association study on muscat flavor in grapevine (Vitis vinifera L.). BMC Plant Biology 10, 241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiil A, Lenk I, Petersen K, Jensen CS, Nielsen KK, Schejbel B, Andersen JR, Lubberstedt T. 2011. Nucleotide diversity and linkage disequilibrium of nine genes with putative effects on flowering time in perennial ryegrass (Lolium perenne L.). Plant Science 180, 228–237 [DOI] [PubMed] [Google Scholar]
- George S, Usha B, Parida A. 2009. Isolation and characterization of an atypical LEA protein coding cDNA and its promoter from drought-tolerant plant Prosopis juliflora . Applied Biochemistry and Biotechnology 157, 244–253 [DOI] [PubMed] [Google Scholar]
- Gill GP, Wilcox PL, Whittaker DJ, Winz RA, Bickerstaff P, Echt CE, Kent J, Humphreys MO, Elborough KM, Gardner RC. 2006. A framework linkage map of perennial ryegrass based on SSR markers. Genome 49, 354–364 [DOI] [PubMed] [Google Scholar]
- Guo P, Baum M, Grando S, Ceccarelli S, Bai G, Li R, von Korff M, Varshney RK, Graner A, Valkoun J. 2009. Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. Journal of Experimental Botany 60, 3531–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. 2002. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes 2, 618–620 [Google Scholar]
- Harjes CE, Rocheford TR, Bai L, et al. 2008. Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319, 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wei X, Sang T, et al. 2010. Genome-wide association studies of 14 agronomic traits in rice landraces. Nature Genetics 42, 961–967 [DOI] [PubMed] [Google Scholar]
- Humphreys MW, Canter PJ, Thomas HM. 2003. Advances in introgression technologies for precision breeding within the Lolium–Festuca complex. Annals of Applied Biology 143, 1–10 [Google Scholar]
- Humphreys J, Harper JA, Armstead IP, Humphreys MW. 2005. Introgression-mapping of genes for drought resistance transferred from Festuca arundinacea var. glaucescens into Lolium multiflorum . Theoretical and Applied Genetics 110, 579–587 [DOI] [PubMed] [Google Scholar]
- Hunault G, Jaspard E. 2010. LEAPdb: a database for the late embryogenesis abundant proteins. BMC Genomics 11, 221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M, Hincha D. 2008. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana . BMC Genomics 9, 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M, Gilmer F, Biskup B, et al. 2009. Simultaneous phenotyping of leaf growth and chlorophyll fluorescence via GROWSCREEN FLUORO allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Functional Plant Biology 36, 902–914 [DOI] [PubMed] [Google Scholar]
- Jensen LB, Muylle H, Arens P, et al. 2005. Development and mapping of a public reference set of SSR markers in Lolium perenne L. Molecular Ecology Notes 5, 951–957 [Google Scholar]
- Jiang Y, Liu H, Cline V. 2009. Correlations of leaf relative water content, canopy temperature, and spectral reflectance in perennial ryegrass under water deficit conditions. Hortscience 44, 459–462 [Google Scholar]
- Jiang Y, Watkins E, Liu S, Yu X, Luo N. 2010. Antioxidative responses and candidate gene expression in prairie Junegrass under drought stress. Journal of the American Society for Horticultural Science 135, 303–309 [Google Scholar]
- Kapustin Y, Souvorov A, Tatusova T, Lipman D. 2008. Splign: algorithms for computing spliced alignments with identification of paralogs. Biology Direct 3, 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khowaja FS, Norton GJ, Courtois B, Price AH. 2009. Improved resolution in the position of drought-related QTLs in a single mapping population of rice by meta-analysis. BMC Genomics 10, 276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M. 2007. Recombination and linkage disequilibrium in Arabidopsis thaliana . Nature Genetics 39, 1151–1155 [DOI] [PubMed] [Google Scholar]
- King J, Thorogood D, Edwards KJ, Armstead IP, Roberts L, Skot K, Hanley Z, King IP. 2008. Development of a genomic microsatellite library in perennial ryegrass (Lolium perenne) and its use in trait mapping. Annals of Botany 101, 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Monde R-A, Last RL. 1998. Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiology 118, 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutovsky KV, Neale DB. 2005. Nucleotide diversity and linkage disequilibrium in cold-hardiness- and wood quality-related candidate genes in Douglas fir. Genetics 171, 2029–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubik C, Sawkins M, Meyer WA, Gaut BS. 2001. Genetic diversity in seven perennial ryegrass (Lolium perenne L.) cultivars based on SSR markers. Crop Science 41, 1565–1572 [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. 2007. Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lauvergeat V, Barre P, Bonnet M, Ghesquiere M. 2005. Sixty simple sequence repeat markers for use in the Festuca–Lolium complex of grasses. Molecular Ecology Notes 5, 401–405 [Google Scholar]
- Li Y, Bock A, Haseneyer G, Korzun V, Wilde P, Schon CC, Ankerst DP, Bauer E. 2011. Association analysis of frost tolerance in rye using candidate genes and phenotypic data from controlled, semi-controlled, and field phenotyping platforms. BMC Plant Biology 11, 146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H-L, Yu X, Ye Q, Ding X-S, Kitagawa Y, Kwak S-S, Su W-A, Tang Z-C. 2004. The role of aquaporin RWC3 in drought avoidance in rice. Plant and Cell Physiology 45, 481–489 [DOI] [PubMed] [Google Scholar]
- Liu A, Burke JM. 2006. Patterns of nucleotide diversity in wild and cultivated sunflower. Genetics 173, 321–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Jiang Y. 2010. Identification of differentially expressed genes under drought stress in perennial ryegrass. Physiologia Plantarum 139, 375–387 [DOI] [PubMed] [Google Scholar]
- Loiselle BA, Sork VL, Nason J, Graham C. 1995. Spatial genetic-structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). American Journal of Botany 82, 1420–1425 [Google Scholar]
- Luo N, Liu J, Yu X, Jiang Y. 2011. Natural variation of drought response in Brachypodium distachyon . Physiologia Plantarum 141, 19–29 [DOI] [PubMed] [Google Scholar]
- Mather KA, Caicedo AL, Polato NR, Olsen KM, McCouch S, Purugganan MD. 2007. The extent of linkage disequilibrium in rice (Oryza sativa L.). Genetics 177, 2223–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Verdoucq L, Luu D-T, Santoni V. 2008. Plant aquaporins: membrane channels with multiple integrated functions. Annual Review of Plant Biology 59, 595–624 [DOI] [PubMed] [Google Scholar]
- McNally KL, Childs KL, Bohnert R, et al. 2009. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proceedings of the National Academy of Sciences, USA 106, 12273–12278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8, 4321–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NDong C, Danyluk J, Wilson KE, Pocock T, Huner NP, Sarhan F. 2002. Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins. Molecular characterization and functional analyses. Plant Physiology 129, 1368–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Borevitz JO, Bergelson J, et al. 2002. The extent of linkage disequilibrium in Arabidopsis thaliana . Nature Genetics 30, 190–193 [DOI] [PubMed] [Google Scholar]
- O’Neill PM, Shanahan JF, Schepers JS. 2006. Use of chlorophyll fluorescence assessments to differentiate corn hybrid response to variable water conditions. Crop Science 46, 681–687 [Google Scholar]
- Oishi H, Takahashi W, Ebina M, Takamizo T. 2010. Expression and gene structure of the cold-stimulated gene Lcs19 of Italian ryegrass (Lolium multiflorum Lam.). Breeding Science 60, 330–335 [Google Scholar]
- Olvera-Carrillo Y, Campos F, Reyes JL, Garciarrubio A, Covarrubias AA. 2010. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis . Plant Physiology 154, 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, et al. 2012. The Pfam protein families database. Nucleic Acids Research 40, D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2003. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiology 133, 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabello A, Guimaraes C, Rangel P, da Silva F, Seixas D, de Souza E, Brasileiro A, Spehar C, Ferreira M, Mehta A. 2008. Identification of drought-responsive genes in roots of upland rice (Oryza sativa L.). BMC Genomics 9, 485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doeblay J, Kresovich S, Goodman MM, Buckler ES. 2001. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proceedings of the National Academy of Sciences, USA 98, 11479–11484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu JF. 2004. The PredictProtein server. Nucleic Acids Research 32, W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, et al. 2002. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal 31, 279–292 [DOI] [PubMed] [Google Scholar]
- Setter TL, Yan JB, Warburton M, Ribaut JM, Xu YB, Sawkins M, Buckler ES, Zhang ZW, Gore MA. 2011. Genetic association mapping identifies single nucleotide polymorphisms in genes that affect abscisic acid levels in maize floral tissues during drought. Journal of Experimental Botany 62, 701–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. 2007. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany 58, 221–227 [DOI] [PubMed] [Google Scholar]
- Skøt L, Humphreys J, Humphreys MO, Thorogood D, Gallagher J, Sanderson R, Armstead IP, Thomas ID. 2007. Association of candidate genes with flowering time and water-soluble carbohydrate content in Lolium perenne (L.). Genetics 177, 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skøt L, Sanderson R, Thomas A, Skot K, Thorogood D, Latypova G, Asp T, Armstead I. 2011. Allelic variation in the perennial ryegrass FLOWERING LOCUS T gene is associated with changes in flowering time across a range of populations. Plant Physiology 155, 1013–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran S, Xiang W, Bean SR, Pedersen JF, Kresovich S, Tuinstra MR, Tesso TT, Hamblin MT, Yu J. 2012. Association mapping for grain quality in a diverse sorghum collection. Plant Genome 5:126–135 [Google Scholar]
- Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS. 2001. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays L.). Proceedings of the National Academy of Sciences, USA 98, 9161–9166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES. 2011. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nature Genetics 43, 159–162 [DOI] [PubMed] [Google Scholar]
- Tian Z, Qian Q, Liu Q, et al. 2009. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proceedings of the National Academy of Sciences, USA 106, 21760–21765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolleter D, Jaquinod M, Mangavel C, Passirani C, Saulnier P, Manon S, Teyssier E, Payet N, Avelange-Macherel MH, Macherel D. 2007. Structure and function of a mitochondrial late embryogenesis abundant protein are revealed by desiccation. The Plant Cell 19, 1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F-Z, Wang Q-B, Kwon S-Y, Kwak S-S, Su W-A. 2005. Enhanced drought tolerance of transgenic rice plants expressing a pea manganese superoxide dismutase. Journal of Plant Physiology 162, 465–472 [DOI] [PubMed] [Google Scholar]
- Wang M, Zhu C, Barkley N, Chen Z, Erpelding J, Murray S, Tuinstra M, Tesso T, Pederson G, Yu J. 2009. Genetic diversity and population structure analysis of accessions in the US historic sweet sorghum collection. Theoretical and Applied Genetics 120, 13–23 [DOI] [PubMed] [Google Scholar]
- Wang Y, Bigelow CA, Jiang Y. 2009. Ploidy level and DNA content of perennial ryegrass germplasm as determined by flow cytometry. Hortscience 44, 2049–2052 [Google Scholar]
- Weckx S, Del-Favero J, Rademakers R, Claes L, Cruts M, De Jonghe P, Van, Broeckhoven C, De, Rijk P. 2005. novoSNP, a novel computational tool for sequence variation discovery. Genome Research 15, 436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Frei U, Schejbel B, Asp T, Lubberstedt T. 2007. Nucleotide diversity and linkage disequilibrium in 11 expressed resistance candidate genes in Lolium perenne . BMC Plant Biology 7, 43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Buckler ES. 2006. Genetic association mapping and genome organization of maize. Current Opinion in Biotechnology 17, 155–160 [DOI] [PubMed] [Google Scholar]
- Yu J, Bai GH, Cai SB, Ban T. 2006. b Marker-assisted characterization of Asian wheat lines for resistance to Fusarium head blight. Theoretical and Applied Genetics 113, 308–320 [DOI] [PubMed] [Google Scholar]
- Yu J, Pressoir Gael, Briggs WH. 2006. a A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genetics 38, 203–208 [DOI] [PubMed] [Google Scholar]
- Yu Q, Hu Y, Li J, Wu Q, Lin Z. 2005. Sense and antisense expression of plasma membrane aquaporin BnPIP1 from Brassica napus in tobacco and its effects on plant drought resistance. Plant Science 169, 647–656 [Google Scholar]
- Zhao K, Aranzana MJ, Kim S, et al. 2007. An Arabidopsis example of association mapping in structured samples. PLoS Genetics 3, e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Gore M, Buckler ES, Yu J. 2008. Status and prospects of association mapping in plants. Plant Genome 1, 5–20 [Google Scholar]
- Zulini L, Rubinigg M, Zorer R, Bertamini M. 2007. Effects of drought stress on chlorophyll fluorescence and photosynthetic pigments in grapevine leaves (Vitis vinifera cv. ‘White Riesling’). Acta Horticulturae 754, 289–294 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.