Abstract
The nucleoskeleton of plants contains a peripheral lamina (also called plamina) and, even though lamins are absent in plants, their roles are still fulfilled in plant nuclei. One of the most intriguing topics in plant biology concerns the identity of lamin protein analogues in plants. Good candidates to play lamin functions in plants are the members of the NMCP (nuclear matrix constituent protein) family, which exhibit the typical tripartite structure of lamins. This paper describes a bioinformatics analysis and classification of the NMCP family based on phylogenetic relationships, sequence similarity and the distribution of conserved regions in 76 homologues. In addition, NMCP1 in the monocot Allium cepa characterized by its sequence and structure, biochemical properties, and subnuclear distribution and alterations in its expression throughout the root were identified. The results demonstrate that these proteins exhibit many similarities to lamins (structural organization, conserved regions, subnuclear distribution, and solubility) and that they may fulfil the functions of lamins in plants. These findings significantly advance understanding of the structural proteins of the plant lamina and nucleoskeleton and provide a basis for further investigation of the protein networks forming these structures.
Key words: Allium cepa, bioinformatics analysis, immunofluorescence microscopy, immunoelectron microscopy, LINC proteins, NMCP1 proteins, nucleoskeleton, phylogenetic analysis, plamina, plant lamina, protein analysis.
Introduction
The lamina is a protein meshwork associated with the inner nuclear membrane and the nuclear pore complexes. In metazoans it consists of a polymeric assembly of lamin filaments and lamin-binding proteins that form the peripheral nucleoskeleton (NSK) (Goldberg et al., 2008). Although lamins are most abundant in the lamina, they also form stable complexes in the nucleoplasm (Dechat et al., 2010b). Lamins are type V intermediate filament proteins that exhibit a typical tripartite structure, featuring a long coiled-coil rod domain flanked by a short N-terminal head domain and a tail domain, the latter containing a nuclear localization signal, an IgG fold, and a C-terminal CAAX box (Dechat et al., 2010a). Lamins are classified as type A and B, which display distinct expression patterns, mitotic behaviour, and biochemical characteristics (Peter and Stick, 2012). At least one B-type lamin is expressed in all somatic metazoan cells, whereas A-type lamins are expressed in differentiated tissues, although they are absent in most invertebrates. Transcripts of the genes encoding lamins are alternatively spliced to create multiple isoforms. Additionally, lamins undergo various post-translational modifications such as farnesylation, phosphorylation, and sumoylation, which determine their retention at the inner nuclear membrane and their state of polymerization (Dittmer and Misteli, 2011).
Lamins are involved in many nuclear functions, including the maintenance of nuclear shape and architecture, the association of NSK to the cytoskeleton (CSK), chromatin organization and positioning, DNA replication, repair, and transcription, cell cycle progression, and mitosis and differentiation (Dechat et al., 2010a; Mejat and Misteli, 2010). Lamins appear to be restricted to metazoans as no clear homologues have been identified in unicellular organisms or plants (Dittmer and Misteli, 2011), suggesting a metazoan origin. Thus, it is of interest to identify functional analogues of lamin in non-metazoans (Peter and Stick, 2012) and indeed, two lamin-like proteins were recently described in unicellular eukaryotes. The Dictyostelium NE81 protein is considered an evolutionary precursor of metazoan lamins (Kruger et al., 2012), while the large coiled-coil nucleoskeletal protein NUP1 of Trypanosoma fulfils lamin functions but it is otherwise unrelated to lamins (Dubois et al., 2012). Plants lack genes that encode lamins but they have a fibrous structure similar to the animal lamina (also called plamina) underlying the inner nuclear membrane (Fiserova et al., 2009; Moreno Díaz de la Espina, 2009). Moreover, there are few lamin-binding proteins that are conserved between plants and animals. Such examples include the SUN proteins, which form part of the LINC (linker of the nucleoskeleton to the cytoskeleton) complex that binds the NSK and CSK (Graumann et al., 2010; Murphy et al., 2010; Oda and Fukuda, 2011) and the nucleoporin Nup136, a functional homologue of animal lamin-binding Nup153 (Tamura and Hara-Nishimura, 2011).
The presence of a structure similar to the lamina and lamin-binding proteins and the fulfilment of the main lamin functions in the plant nucleus suggest that although plant genomes lack obvious homologues, they may express proteins that functionally substitute lamins. These proteins probably share some structural properties of lamins that are essential for their activity rather than specific sequence homology. Early studies of the plant NSK described proteins that are immunologically related to lamins and IFs, with similar molecular weights, pI, solubility, and nuclear distribution in both monocots and dicots (Blumenthal et al., 2004; Moreno Díaz de la Espina, 2009). However, no full sequence has been ascribed to these proteins to date.
Another candidate analogue of lamin in plants is NMCP1 (nuclear matrix constituent protein 1), a residual protein of the nuclear envelope described for the first time in carrot (Masuda et al., 1993). DcNMCP1 has a tripartite structure with a central rod domain that is predicted to mediate dimerization, which is flanked by a head and tail domain (Masuda et al., 1997). Searches against plant genomes have identified genes encoding NMCP homologues (Dittmer et al., 2007; Kimura et al., 2010), implying the existence of several NMCP variants with distinct functions (Kimura et al., 2010).
In Arabidopsis thaliana, four genes encoding proteins related to DcNMCP1 were characterized. These proteins were named LINC (little nuclei), after the phenotype of linc1linc2 double mutants. Mutation of the genes encoding LINC1 and LINC2 not only affected nuclear size but heterochromatin organization as well, demonstrating that these proteins are important determinants of plant nuclear shape and structure, as are lamins in animal nuclei (Dittmer et al., 2007).
To further characterize functional homologues of lamins in plants, this study analysed the phylogenetic relationships, predicted structures, and sequence similarities of NMCP family members, proposing the classification of NMCP proteins into two types. In addition, the sequence and biochemical characteristics of endogenous NMCP1 were investigated for the first time in a monocot (Allium cepa), comparing the subnuclear expression and distribution of AcNMCP1 in nuclei isolated from meristematic and differentiated root cells.
Onion is a convenient plant model in which to analyse nuclear structure, as it contains a large and highly structured 2C nucleus with high DNA content (over 90-times that of A. thaliana), little endoploidy in differentiated tissues, and a high proportion of heterochromatin. Moreover, its nuclear and nucleoskeletal structures are well characterized (Moreno Díaz de la Espina, 2009). Taken together with previous findings obtained in Arabidopsis mutants (Dittmer et al., 2007), the present results suggest that NMCPs may be functional homologues of lamins.
Materials and methods
Plant material and culture
A. cepa L. var. francesa bulbs were grown as described previously (Samaniego et al., 2006). Quiescent meristems were excised from unsoaked bulbs.
Cloning and sequencing of cDNAs for AcNMCP1
Cloning and cDNA sequencing was performed as previously described (Kimura et al., 2010) using RNA isolated from the callus of A. cepa and B-degenerate primers AcF2 (5’-GGGGCTKC TTTTGATTGAGA-3’) and AcF3 (5’-ATTGAGAAAAARGARTG GAC-3’) in 3’-RACE, and Ac5RACE-R2 (5’-TAATATGCCTCTG CCCATCAA-3’) and Ac5RACE-R3 (5’-GCAAATGCTCTTTTTGT TCAG-3’) in 5’-RACE.
The cDNAs were ligated into the pGEM T-Easy vector (Promega) using the TA-cloning method and the vectors cloned into Escherichia coli DH5α cells. Plasmid DNA was extracted from the clones and the cDNA sequence was determined. The GenBank/EMBL/DDBJ accession number for AcNMCP1 is AB673103.
Bioinformatics tools
Genome searches were performed using Phytozome version 8.0 (Goodstein et al., 2012), the multiple alignments were carried out using CLUSTAL W2, and the phylogenetic analysis was performed using MEGA5 (Tamura et al., 2011). A search for post-translational modification sites was performed and the molecular weights and isoelectric points (pI) were calculated with ExPASy (http://www.expasy.org/). The nuclear localization signal was localized using NucPred (http://www.sbc.su.se/~maccallr/nucpred/) and MEME used to search for conserved motifs (Bailey et al., 2009). The coiled-coil and polymerization state were predicted using MARCOIL (Delorenzi and Speed, 2002) and Multicoil2 (http://groups.csail.mit.edu/cb/multicoil2/cgi-bin/multicoil2.cgi), respectively.
Antibody production and synthesis of polypeptides with partial AcNMCP1 sequences
The cDNA fragment encoding the 313 N-terminal amino acids of AcNMCP1 was subcloned and expressed using E. coli Rosetta II (Novagen), as described previously (Kimura et al., 2010). Protein expression was induced with 1mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37 °C for 4h, and the cells were harvested and extracted several times with PBS containing 0.2% Triton X-100. The proteins in the insoluble fraction were extracted with 8M urea, 10mM Na-phosphate buffer (pH 8.0), and 1mM 2-mercaptoethanol. The N-terminal region of AcNMCP1 containing a 6 × histidine tag was affinity purified on iMAC resin (Biorad), and the fraction retained by the resin in 10mM imidazole was eluted with 300mM imidazole and dialysed against 6M urea in 10mM Tris-acetate (pH 7.6). The protein in the dialysis solution was then precipitated by adding 1.5 volumes of acetone, dissolved in PBS containing 0.04% SDS, and used for immunization. The anti-AcNMCP1 antibody was generated commercially in rabbits by Sigma Genosys.
Isolation of nuclei and nucleoskeletons
Nuclear and NSK isolations were performed as described previously (Samaniego et al., 2006, Supplementary Text S1, available at JXB online).
PAGE and immunoblotting
Nuclear pellets extracted from onions were dissolved in 400 µl lysis buffer (100mM Tris-HCl pH 7.5, 4.5M urea, 1M thiourea, 2% CHAPS, 0.5% Triton X-100, 10mM DTT) containing protease inhibitor cocktail (Sigma-Aldrich) and 75U benzonase (Sigma-Aldrich). Protein extracts from the root tips of 4-day-old pea, wheat, maize, garlic, and rye seedlings and 3-week-old whole plants of A. thaliana and Nicotiana benthamiana were ground in liquid nitrogen. To each 100 µg of ground tissue 100 µl lysis buffer was added and the samples were incubated for 45min on ice before they were centrifuged at 4 °C for 10 minutes at 1450 g. The protein content was measured using the modified Bradford protein assay (Berkelman, 2008) and then protein extracts were mixed with 6 × Laemmli buffer and resolved by SDS-PAGE on 8% (w/v) polyacrylamide gels or precast 4–15% linear gradient gels (Biorad), as described previously (Samaniego et al., 2006). Two-dimensional electrophoresis (2D-PAGE) was performed using non-linear (pH 3–10) or linear (pH 4–7) gel strips, as described previously (Perez-Munive and Moreno Díaz de la Espina, 2011). The proteins were transferred to nitrocellulose membranes that were probed with an anti-AcNMCP1 antibody (1:1000), as described previously (Samaniego et al. (2006). Molecular weights were determined using Quantity One 1-D analysis software (Biorad).
Treatments with chaotropic agents
Batches of onion nuclear pellets were solubilized in the following buffers: (a) 6M guanidine thiocyanate (GITC) in 100mM Tris-HCl (pH 7.5); (b) 7M urea, 2M thiourea, 4% CHAPS, 18.2mM DTT, 100mM Tris-HCl (pH 7.5); (c) 2 × Laemmli buffer. Samples in GITC or urea were mixed 1:1 with 2 × Laemmli buffer.
Mass spectrometry (nES-MS/MS)
Scans of 2D-PAGE gels stained with Coomassie brillant blue (G-250, Biorad) were compared with immunoblots of a gel run in parallel, and the spots corresponding to the reactive proteins were excised with EXQuest Spot Cutter (Biorad), destained in 50mM ammonium bicarbonate/50% acetonitrile (ACN), dehydrated with ACN, and dried. The gel spots were rehydrated in 12.5ng/ml trypsin in 50mM ammonium bicarbonate and incubated overnight at 30 °C. Peptides were extracted at 37 °C using 100% ACN followed by 0.5% trifluoroacetic acid, dried by vacuum centrifugation, purified using ZipTip (Millipore), and reconstituted in 0.1% formic acid/2% ACN for injection into the HPLC device. The peptide mixtures from in-gel tryptic digestions were analysed using nLC-MS/MS, and the peptides were scanned and fragmented with an LTQ-Orbitrap Velos (ThermoScientific). Mass spectra ‘raw’ files were compared with AcNMCP1 sequences using the SEQUEST search engine and Thermo Proteome Discoverer.
Flow cytometry analysis
DNA content was estimated by flow cytometry as described previously (Samaniego et al., 2006, Supplementary Text S1).
Immunofluorescence
Immunofluorescence was performed on suspensions of isolated nuclei or NSKs using the anti-AcNMCP1 antibody (1:100) as described previously (Samaniego et al., 2006, Supplementary Text S1).
Electron microscopy
Isolated nuclei were fixed in 0.25% formaldehyde in PBS (pH 7.2) with 0.5% TX-100 for 30min at 4 °C, washed in PBS (2×10min), and blocked in 2% BSA for 30min. The samples were subsequently incubated overnight at 4 °C with the anti-AcNMCP1 antibody (1:50) in blocking buffer and then washed in PBS containing 0.05% Tween 20 (3×15min). The pellets were incubated for 45min at room temperature with a 5nm gold-conjugated secondary anti-rabbit antibody (1:50, Sigma), washed in PBS (2×15min), fixed in 2% formaldehyde in PBS for 1h at 4 °C, washed again in PBS, dehydrated in a graded ethanol series, and embedded in LR White resin (London Resin). Post-embedding immunogold labelling of NSK fractions with anti-AcNMCP1 (1:20) and subsequent analysis was performed as described previously (Perez-Munive and Moreno Díaz de la Espina, 2011). Sections were contrasted in aqueous 5% uranyl acetate 30min.
Results
Sequence analysis, coiled-coil prediction, and phylogeny of NMCP proteins
AcNMCP1 was predicted to contain 1217 amino acids, with a molecular weight of 139kDa and a pI of 5.39. This AcNMCP1 was aligned with previously reported sequences of DcNMCP1, AgNMCP1, LINC1 and OsNMCP1 (Supplementary Fig. S1), indicating features specific to the NMCP family that were revealed by the bioinformatics analysis described below (coiled-coil prediction, conserved motifs, nuclear localization signal, and phosphorylation sites).
The AcNMCP1 sequence was used for BLAST searches using the Phytozome version 8.0 database, and the gene family with the highest score and e-value (2.2e-177 for DNA sequence and 2.1e-123 for amino-acid sequence) was selected. This family was made up of 71 genes and it also produced high scores using the DcNMCP1 and AgNMCP1 sequences. The matches represented 27 out of 31 plant genomes and the following species lacked NMCP homologues: unicellular algae (Volvox carteri, Chlamydomonas reinhardtii), a clubmoss (Selaginella moellendorfii), and a dicot (Medicago truncatula). However, additional BLASTP searches against non-redundant protein sequence databases revealed matches for clubmoss and Medicago. The sequences were shorter than those typical of NMCPs and included highly conserved regions, suggesting that both species express NMCPs but that the sequence entries are incomplete (data not shown).
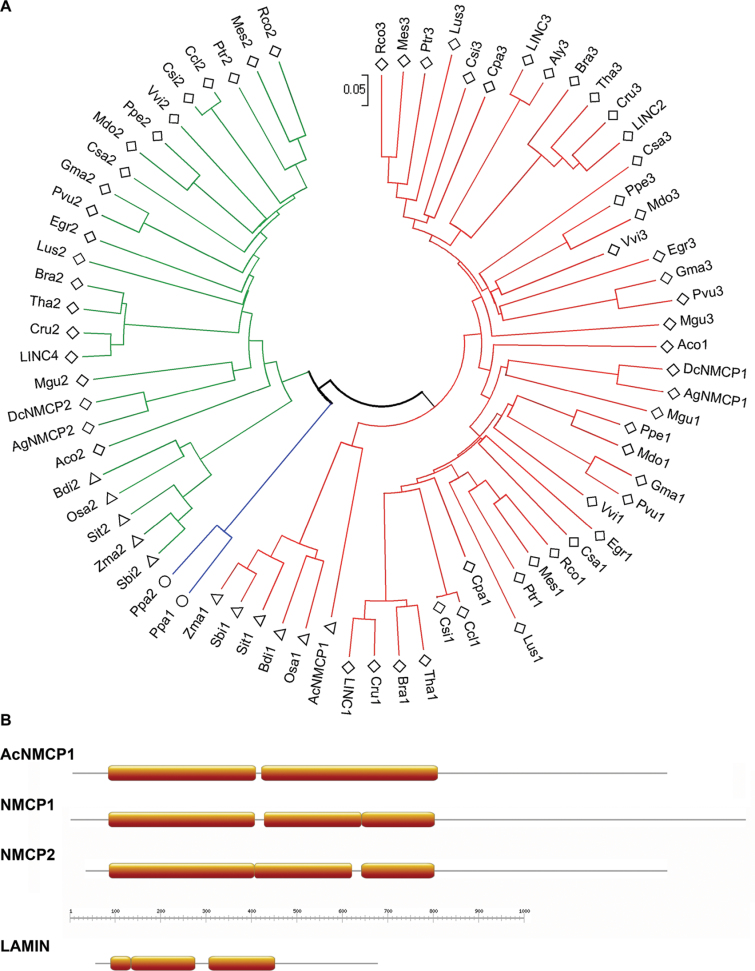
In the selected gene family there were open reading frames from a moss (Physcomitrella patens) and from various monocot and dicot genomes. A phylogenetic tree for all NMCPs was constructed in MEGA5 using the neighbour-joining method, and the distances were computed using the p-distance method. Based on sequence and structure similarities and on phylogenetic relationships, the protein family was classified into two clusters: one containing NMCP1 proteins and a second that contained NMCP2 proteins (Fig. 1A). The moss had two NMCP homologues that evolved from the common NMCP progenitor gene. In vascular plants, NMCP evolved from two genes: the NMCP1 and NMCP2 progenitors. Most dicots have two genes that encode NMCP1, with the exception of A. thaliana which carries three NMCP1 genes (LINC1, LINC2, and LINC3), and all the plants analysed had one NMCP2 gene. In A. thaliana, the LINC4 protein previously described as chloroplast protein was classified as NMCP2.
Fig. 1.
Classification of NMCPs: evolutionary relationships and predicted protein structures. (A) Phylogenetic relationship of NMCPs inferred using the neighbour-joining method. Evolutionary distances were calculated using the p-distance method and are presented as the number of amino-acid differences per site. The phylogenetic tree is drawn to scale. The sequences classified as type 1 NMCP are marked in red and type 2 are in green, with the two members in Physcomitrella patens in blue. Dicotyledon species are represented by rhombi; monocotyledons by triangles; and moss by circles. Sequence accession data are shown in Supplementary Table S1. (B) Schematic representation of the coiled-coil prediction (MARCOIL) for AcNMCP1, typical NMCP1 and NMCP2, and lamin (orange boxes).
The coiled-coil prediction was performed using MARCOIL, which employs the hidden Markov model and outperforms the popular Multicoil programme. To avoid negative matches and increase reliability, the cut off was set at 0.6, at which MARCOIL is reported to perform best (Gruber et al., 2006). Indeed, a control analysis on a group of lamin sequences confirmed that MARCOIL outperforms Multicoil2 and Multicoil (data not shown). Predictions were generated for 76 NMCP sequences, including the sequences collected in the genome searches and the proteins described previously in carrot, celery, and A. thaliana (Masuda et al., 1993, 1997, 1999; Dittmer et al., 2007; Kimura et al., 2010). These analyses revealed that all NMCPs contained a central coiled-coil domain. The rod domain of NMCP1s contains two coiled coils of similar lengths separated by a short linker, the first from 250 to 300 residues, and the second from 350 to 400. On several occasions MARCOIL analysis revealed a short linker within the second segment that divided it into two coils of 200 and 150 residues, respectively (Fig. 1B). The predicted structures of NMCP2 proteins resembled the latter arrangement, although not all NMCP2 sequences contained the first linker (Fig. 1B). The positions of the linkers in NMCP1 and NMCP2 corresponded, suggesting that the structure of the rod domain is conserved across the NMCP family. The polymerization state predicted by Multicoil2 indicated that all coiled-coil regions have a high probability of forming dimers.
Multiple sequence alignment confirmed that NMCPs share a high degree of sequence similarity in the rod domain. A search for conserved regions using MEME detected multiple conserved motifs within the rod domain and several in the tail domain, although the general sequence similarity in the tail domain was relatively low (Fig. 2A, selected regions with a high e-value and conserved localization are shown in Fig. 2B). While region 3 was absent in moss, region 7 was absent in NMCP2 proteins and region 8, which was preceded by a stretch of acidic amino acids (Supplementary Fig. S1), was absent in dicot NMCP2, although it was present in monocot NMCP2. The search also detected a possible nuclear localization signal conserved across NMCP1 proteins, followed by the conserved region 7. Region 6 was followed by a consensus recognized by the cdc2 kinase SPXK/R. A NucPred prediction indicated that almost all (62 out of 76) NMCPs contained the nuclear localization signal consensus sequence, although its localization and pattern was only conserved in NMCP1 proteins. In the search for possible conserved post-translational modification sites, a few phosphorylation sites for cdc2, PKA, and PKC were identified in the head and tail domains (Fig. 2A).
Fig. 2.
Conserved regions and phosphorylation sites in AcNMCP1, NMCP1, and NMCP2. (A) Schematic representation of conserved regions, predicted nuclear localization signals (green boxes) and phosphorylation sites (red bars, cdk1; grey bar, PKA/PKG). Localization of the conserved regions is indicated by green bars with corresponding numbers. Coiled coils are represented as orange boxes. (B) MEME motifs displayed as ‘sequence LOGOS’. The height of each letter reflects the probability of its localization at this position. Letters are coloured using the same colour scheme as the MEME motifs based on the biochemical properties of the amino acids.
Identification and characterization of AcNMCP1
To identify endogenous AcNMCP, a polyclonal antibody was raised against the N-terminal portion of the protein that includes the highly conserved regions 1 and 2 (Fig. 2A). Cross-reactivity of the antibody was evaluated in the monocots A. cepa, Allium sativum, Triticum aestivum, Secale cereale, and Zea mays, and in the dicots A. thaliana, N. benthamiana, and Pisum sativum. In immunoblots, the antibody specifically recognized bands in all species except for N. benthamiana, and no bands were detected in negative controls. Although NMCP transcripts were similar in size (3300–3600bp for NMCP1 and 2700–3000bp for NMCP2) the molecular weights of the detected bands were highly variable across species (Fig. 3A). In A. thaliana the antibody recognized a major band of 150kDa, which roughly corresponds to the predicted molecular weight of AtNMCP/LINC proteins (120–130kDa, www.arabidopsis.org). In other monocots such as wheat and rye and also garlic that belongs to the genus Allium, the antibody cross-reacted with proteins of 100kDa, while in maize the antibody recognized a triplet of about 80kDa. In pea, a major band of a similar size (70kDa) to a protein of the peripheral nuclear matrix described previously (Blumenthal et al., 2004) was detected. The diversity of molecular weights across species may indicate that NMCPs undergo alternative splicing and/or post-translational modifications.
Fig. 3.
Characterization of AcNMCP1. (A) Immunoblot detection of proteins using anti-AcNMCP1 in Zma (corn), Psa (pea), Ath (Arabidopsis thaliana), Tae (wheat), Sce (rye), Asa (garlic), and Ace (onion). Ace’, overexposure of Ace; (–), negative control with primary antibody omitted. (B) Detection of AcNMCP1 in onion nuclear fractions extracted in 2 × Laemmli buffer (SDS), 7M urea/2M thiourea (7M U), and 6M guanidine thiocyanate (GITC). (C, D) 2D-immunoblots of A. cepa whole-nuclear extracts (C) and total Arabidopsis protein (D), probed with the anti-AcNMCP1 antibody.
In onion the antibody recognized a major band of 200kDa, although some minor bands of 150 and 100kDa were also observed. The presence and intensity of the lower bands varied between experiments, suggesting that these were proteolytic products. As the predicted molecular weight was much lower than that detected, this study investigated the possibility that the 200-kDa band represents a dimer by denaturing the protein in high concentrations of urea (7M) or guanidine thiocyanate (6M). These treatments had no effect on band mobility (Fig. 3B), suggesting that the 200-kDa band represents the true molecular weight of AcNMCP1. To rule out any possible protein aggregation in the stacking gel, the sample was also resolved in 4–15% gradient gels, with no apparent effect on band migration (data not shown).
In 2D-immunoblots of the onion nuclear fraction, the antibody detected spots of 200kDa with isoelectric points in the range of 3–5.8, with the main spots with a pI of 5.2 and 5.8 (Fig. 3C). In Arabidopsis, a single 150-kDa spot with a pI of 4.9 was detected (Fig. 3D).
Protein identification with nLC-MS/MS
To confirm that the proteins detected by the antibody in A. cepa corresponded to AcNMCP1, the spots separated by 2D-PAGE (Fig. 3C) were excised and identified as AcNMCP1 by nLC-MS/MS. In the first spot, 49 peptides (34.9% coverage) were confirmed by SEQUEST with a score of 174.6, while 61 peptides (41.6% coverage) were identified in the second with a score of 193.4.
Distribution of AcNMCP1 in the nuclei of meristematic cells
Confocal immunofluorescence microscopy of isolated nuclear fractions revealed a consistent pattern of AcNMCP1 staining at the nuclear periphery that showed a punctuate-like distribution. Variable intranuclear staining was also observed in the interchromatin domains revealed by DAPI counterstaining of nuclei depending on the preparation (Fig. 4A, B, B’’, D, E). Sections of isolated membrane-depleted nuclei showed a peripheral structure with associated pore complexes firmly attached to condensed chromatin masses similar to the plant lamina (Moreno Díaz de la Espina et al., 1991). Pre-embedding immunogold labelling for electron microscopy of these nuclei confirmed the distribution of AcNMCP1 and revealed its association with the peripheral plant lamina. AcNMCP1 labelling was abundant in the zones of the plamina closely associated with condensed chromatin masses. The labelling of the fibrillar network in the interchromatin domains was scarce (Fig. 4F).
Fig. 4.
Subnuclear localization of AcNMCP1. (A–E) Confocal sections of meristematic nuclear fractions after incubation with the anti-AcNMCP1 antibody, demonstrating the distribution of the protein along the nuclear periphery (A–E) and in the nucleoplasm on occasion (D, E). (B’’) High magnification of a portion of the nucleus in B showing the punctuate-like distribution of the peripheral labelling. (C) Negative control incubated with the secondary antibody alone. (A’, B’, C’, D’, and E’) Overlay of the corresponding anti-NMCP1- and DAPI-stained images. (F) High-resolution pre-embedding immunogold labelling. Portion of a nucleus that exhibit accumulations of gold particles in the peripheral plant lamina (thick arrows) and scarce labelling in the interchromatin domains (id) (thin arrows). The condensed chromatin masses (chr) and nucleolus (No) showed no labelling. Bar in F = 100nm.
AcNMCP1 is bound to the nucleoskeleton
To investigate the association of AcNMCP1 with the NSK, the NSK was isolated by sequential extraction of nuclear protein fractions. Immunoblotting with the anti-AcNMCP1 antibody revealed that the protein was only present in insoluble fractions and that it was resistant to extraction with non-ionic detergent, DNase, and high salt concentrations. Together, these results demonstrate that AcNMCP1 is a highly insoluble nuclear protein and a component of the NSK (Fig. 5A, B). Indeed, confocal immunofluorescence microscopy and electron microscopy immunogold labelling of nucleoskeletal fractions revealed that AcNMCP1 is mainly associated with the plamina and to a lesser extent with the internal NSK, revealing a similar distribution to that found in isolated nuclei (Fig 5C–E).
Fig. 5.
AcNMCP is a component of the nucleoskeleton (NSK). (A) Detection of AcNMCP1 in the nuclear (N), insoluble (F1, F2, NSK), and soluble (S1, S2, S3) fractions obtained during NSK extraction in immunoblots probed with anti-AcNMCP1. The 200-kDa band of AcNMCP1 was present in all the insoluble fractions but not in the soluble fractions. (B) Coomassie blue staining of a gel run in parallel showing the complex protein composition of the insoluble and soluble fractions. (C, D) Confocal images of NSKs showing the predominant accumulation of AcNMCP1 in the lamina and weaker staining associated with the internal NSK. (C’, D’) Differential interference contrast images of the corresponding fields. (E) Immunogold labelling of NSK showing the association of gold particles with the plant lamina and internal NSK. Bars, 25 µm (C, C’), 10 µm (D, D’), 100nm (E).
Levels and nuclear distribution of AcNMCP1 in root cells at different stages of proliferation
The level and nuclear distribution of AcNMCP1 was analysed in immunoblots and by immunofluorescence in nuclear fractions from cells in the meristem (1–2mm from the root tip), elongation (2–6mm), and mature (≥6mm) root zones, as well as in the non-proliferating meristem of quiescent roots. Flow cytometry analysis revealed that cells in the elongation and mature zones were mostly non-proliferating, while those in the meristematic zone proliferated and had abundant nuclei with a DNA content ranging from 2C to 4C, therefore corresponding to the S-phase. The cells of quiescent meristems were mostly in G1-phase, with no cells in the S-phase (Fig. 6A). In immunoblots, AcNMCP1 was most abundant in meristematic cells, either proliferating or quiescent. Its accumulation decreased slightly in the elongation zone and dramatically in the mature zone, with very weak expression in the cells located 18–20mm from the root tip (Fig. 6B).
Fig. 6.
Expression and distribution of AcNMCP1 in nuclei isolated from different root cell types. (A) Localization of the onion root zones used in this analysis and their corresponding DNA content determined by flow cytometry. (B) AcNMCP1 levels detected by immunoblotting with the anti-AcNMCP1 antibody. AcNMCP1 expression was abundant in the proliferating (m) and quiescent (q) meristems, although it decreased significantly in non-meristematic cells (e = elongation zone; d = differentiated zone). H1 histones stained with Coomasie blue were used as loading controls. (C) Peripheral (qP, mP, dP) and central (qC, mC, dC) confocal sections showing the distribution of AcNMCP1 in the periphery and nuclear interior of quiescent (q) and proliferating (m) meristems and in differentiated cells (d). Arrows in qC point to the nucleoplasmic aggregates of the protein in quiescent meristems and arrows in dP to the gaps in the peripheral distribution of the protein in differentiated cells. qP’, qC’, mP’, mC’, dP’, and dC’ show overlays of AcNMCP1 and DAPI staining.
Confocal immunofluorescence revealed a general distribution of AcNMCP1 at the nuclear rim and in the nucleoplasm of all cell types with two peculiarities. Large intranuclear accumulations of AcNMCP1 were frequently observed in the quiescent meristematic nuclei (Fig. 6C). Also, there were large gaps in AcNMCP1 distribution along the nuclear periphery in nuclei isolated from elongation and mature root zones (Fig. 6C). The corresponding differential interference contrast images appeared to rule out nuclear envelope damage (data not shown). Immunofluorescent staining in whole cells was impeded by non-specific cross-reaction of the anti-AcNMCP1 antibody in the cytoplasm. The signal was not caused by non-specific binding of the secondary antibody, as revealed by the negative controls, nor was it observed in immunoblots of cytoplasmic fractions with the anti-AcNMCP1 antibody (data not shown).
Discussion
While no lamin-coding genes have been identified in plant genomes, the presence of a structure similar to the lamina and the fulfilment of the main functions of lamin in plants suggest the presence of plant-specific proteins analogous to lamins. Several proteins have been proposed as lamin analogues in plants, including members of the NMCP protein family. These are conserved nuclear coiled-coil proteins with a tripartite organization similar to that of lamins (Masuda et al., 1993, 1997; Dittmer et al., 2007; Kimura et al., 2010). Functional analysis of A. thaliana has revealed that mutation of two of its four NMCP genes (LINC1 and LINC2) affects nuclear size and morphology and heterochromatin distribution (Dittmer et al., 2007), features that are influenced by lamins in metazoan nuclei (Dechat et al., 2010a).
This study identified members of the NMCP family sharing a high degree sequence similarity in all land plants (embryophytes) analysed, including a moss (P. patens) and vascular plants (tracheophytes), although they are absent in single-cell plants. These proteins were classified into two clusters based on sequence, structural analogies, and phylogenetic relationships, findings that were consistent with previous studies performed in a few species (Dittmer et al., 2007; Kimura et al., 2010). NMCPs have evolved from two genes, the NMCP1 and NMCP2 progenitor, while the two P. patens homologues have evolved from the common NMCP ancestor. Monocots carry one NMCP1 and one NMCP2 gene, while dicots carry an additional gene encoding an NMCP1-related protein, designated NMCP3. The subnuclear distribution of NMCP1 and NMCP2 differs, indicating that they probably mediate different functions (Kimura et al., 2010). The present study found that A. thaliana LINC2, which was thought to encode an NMCP2-related protein, in fact encodes an NMCP1 homologue (NMCP3), while the phylogenetic tree indicated that LINC4 is related to NMCP2 despite its previous annotation as a chloroplast protein in a proteomic study (Kleffmann et al., 2006). The presence of a predicted nuclear localization signal suggests that LINC4 is present in the nucleus (data not shown).
NMCPs have a tripartite structure featuring non-coiled head and tail domains and a central coiled-coil rod domain. The prediction with the MARCOIL programme revealed that the composition of coiled-coil domains between NMCPs is much more similar than that previously suggested by predictions obtained with Multicoil or COILS (based on the Lupas algorithm) (Dittmer et al., 2007; Kimura et al., 2010), which are considered overly restrictive approaches (Gruber et al., 2006). The present prediction reveals that most NMCPs contain two coiled coils separated by a linker of around 20 residues and forming a central rod domain with short linkers inside the coiled-coil segments in some cases. Similar predictions for lamins confirmed that their general structure and organization of coiled-coil domains is similar to that of NMCP1, although the NMCP rod domain is twice as long.
NMCPs exhibit a high degree of sequence similarity in the rod domain, which contains five highly conserved regions at each end and at the positions of the predicted linkers. Lamins exhibit a similar pattern, whereby the highly conserved motifs at either end of the coiled-coil domain are prime candidates to mediate head-to-tail associations (Kapinos et al., 2010). The similar structure and location of conserved motifs in NMCPs and lamins suggest similar mechanisms of oligomerization and protofilament formation. This hypothesis is further supported by the presence of consensus sequences recognized by kinases at each side of the rod domain.
Although the NMCP tail domains do not share strong sequence similarity, several conserved regions were found. Based on a search against the MyHits-PROSITE database, all conserved motifs appeared to be specific to the NMCP family. However, one region of the NMCP1 tail domain (RYNLRR) was found to contain five amino acids identical to a specific region of lamin A (EYNLRSRT, Peter and Stick, 2012) that probably serves as an actin-binding site (Simon et al., 2010). Thus, the conservation of this sequence suggests that this region of NMCP1 may also be a binding site for actin. Like lamins, most NMCPs contain a predicted nuclear localization signal in the tail domain that is conserved in NMCP1 proteins. Although a few sequences lacked a predicted nuclear localization signal, two such sequences (DcNMCP2 and AgNMCP2) still localized in the nucleus, to which they are probably directed via an alternative pathway (Kimura et al., 2010). The retention of lamins in the inner nuclear membrane is mediated by the C-terminal CAAX box, although as seen for lamin C, this motif is not an absolute requirement for inner nuclear membrane association (Dittmer and Misteli, 2011). While NMCPs lack a CAAX box, the C-terminus of all members (except the dicot NMCP2) contains a highly conserved region that may be involved in the inner nuclear membrane association. It is preceded by a stretch of acidic amino acids that is also present in the tail domain of vertebrate lamins (Erber et al., 1999).
While the predicted molecular weights of NMCPs from dicot and monocot species were similar (~130–140kDa for NMCP1 and 110–120kDa for NMCP2), the mobility of the endogenous proteins was very variable across species. In some cases, the molecular weights of the bands detected were higher than the predicted values: 60kDa higher in onion and 20–40kDa higher in A. thaliana, carrot, and celery (Fig. 3: (Kimura et al., 2010). These differences could reflect incomplete denaturation or post-translational modification of the native protein, although the first possibility appears unlikely given the protein’s behaviour in conditions favouring protein denaturation. Moreover, the lower molecular weights detected in monocots suggest the involvement of alternative splicing or post-translational modification.
Confocal microscopy demonstrated a consistent association of AcNMCP1 with the nuclear periphery, as reported for the carrot and celery proteins (Masuda et al., 1997; Kimura et al., 2010). AcNMCP1 also associated with the nucleoplasm, as described for the rice NMCP1a (Moriguchi et al., 2005), Arabidopsis LINC2 (Dittmer et al., 2007) and lamins (Dechat et al., 2010b). Some variability of the staining may have been produced by the reduced accessibility of the internal AcNMCP1 pool to the antibody. Immunogold electron microscopy demonstrated that onion NMCP1 preferentially localizes in the plant lamina, close to condensed heterochromatin masses, which suggests a role in anchoring peripheral heterochromatin to this structure. Indeed, the protein was also detected in the interchromatin domains, which suggests that it is involved in nuclear functions associated with these domains.
AcNMCP1 is an abundant component of the NSK, as witnessed here by the sequential extraction of nuclei and in previous reports of the carrot protein (Masuda et al., 1993). Immunofluorescence and immunogold electron microscopy staining of nucleoskeletons confirmed that the protein is a component of the plant lamina and that it is also present in the internal NSK. These results demonstrate that NMCP1 is a structural protein that may be involved in the organization of multimeric complexes in the plant NSK, a function fulfilled by lamins in metazoans.
In the different root cell populations, the expression of AcNMCP1 is developmentally regulated. This protein was abundant in the proliferating and quiescent meristem, while it was much more weakly expressed in cells of the mature root zones. This expression profile resembles that of lamin B1, which is abundant in proliferating and quiescent cells but that is weakly expressed in differentiated cells (Lehner et al., 1987; Broers et al., 1997; Shimi et al., 2011). The present results also revealed alterations in the distribution of AcNMCP1 in differentiated cells: while AcNMCP1 was distributed along the nuclear envelope in meristematic cells, its distribution in differentiated cells displayed large gaps depleted of AcNMCP1.
In conclusion, plant NMCPs share several important features with metazoan lamins: (1) NMCPs have a similar tripartite structure with a central α-helical rod domain that is predicted to form coiled coils, albeit twice as long as that found in lamins; (2) both ends of the rod domain, which is important for lamin polymerization, are highly conserved in NMCPs; (3) the C-terminus of the protein is highly conserved (except in dicot NMCP2), reflecting important functional conservation, and the stretch of acidic amino acids preceding this region is also present in the tail domain of vertebrate lamins; (4) As lamins in vertebrates, plants have two types of NMCPs, NMCP1 (two genes in dicots, one in monocots) and NMCP2 (one gene); (5) NMCP1 is a nucleoskeletal component in the lamina and the internal NSK, like lamins in animal nuclei; (6) NMCP1 appears to be developmentally expressed, like lamins; (7) NMCPs are expressed in multicellular but not in single-cell plants consistent with the expression of lamins in metazoans alone; and (8) double linc1linc2 mutants of Arabidopsis demonstrate the role of NMCP proteins in the control of nuclear size and shape, and in chromatin organization (Dittmer et al., 2007), as described for lamins (Dechat et al., 2010a). Based on these similarities, this study proposes proposes NMCPs to be candidates to fulfil the functions of lamin in plants. However, to fully elucidate the functions of NMCPs, further studies will clearly be necessary, analysing their roles in different nuclear activities in mutants and identifying their protein partners (such as SUN proteins, Nup136, actin, and other plant-specific proteins). These experiments are currently in progress in the present and other groups.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Text S1. Experimental procedures.
Supplementary Fig. S1. Multiple sequence alignment of AcNMCP1.
Supplementary Table S1. Sequence accession data.
Acknowledgements
The authors thank M Carnota, F García, and MI Fernández for expert technical assistance and Dr M Sefton for English editing. This work was supported by the Spanish Ministry of Science and Innovation (BFU2010-15900) and CSIC (PIE 201020E019). Malgorzata Ciska was supported by a Junta de Ampliacion de Estudios grant (JAEPre_08_00012/JAEPre027).
References
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research 37, W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkelman T. 2008. Quantitation of protein in samples prepared for 2-D electrophoresis. Methods in Molecular Biology 424, 43–49 [DOI] [PubMed] [Google Scholar]
- Blumenthal SS, Clark GB, Roux SJ. 2004. Biochemical and immunological characterization of pea nuclear intermediate filament proteins. Planta 218, 965–975 [DOI] [PubMed] [Google Scholar]
- Broers JL, Machiels BM, Kuijpers HJ, Smedts F, van den Kieboom R, Raymond Y, Ramaekers FC. 1997. A- and B-type lamins are differentially expressed in normal human tissues. Histochemistry and Cell Biology 107, 505–517 [DOI] [PubMed] [Google Scholar]
- Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. 2010a. Nuclear lamins. Cold Spring Harbor Perspectives in Biology 2, a000547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Gesson K, Foisner R. 2010b. Lamina-independent lamins in the nuclear interior serve important functions. Cold Spring Harbor Symposia on Quantitative Biology 75, 533–543 [DOI] [PubMed] [Google Scholar]
- Delorenzi M, Speed T. 2002. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18, 617–625 [DOI] [PubMed] [Google Scholar]
- Dittmer TA, Misteli T. 2011. The lamin protein family. Genome Biology 12, 222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. 2007. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. The Plant Cell 19, 2793–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois KN, Alsford S, Holden JM, et al. 2012. NUP-1 is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS Biology 10, e1001287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erber A, Riemer D, Hofemeister H, Bovenschulte M, Stick R, Panopoulou G, Lehrach H, Weber K. 1999. Characterization of the Hydra lamin and its gene: a molecular phylogeny of metazoan lamins. Journal of Molecular Evolution 49, 260–271 [DOI] [PubMed] [Google Scholar]
- Fiserova J, Kiseleva E, Goldberg MW. 2009. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. The Plant Journal 59, 243–255 [DOI] [PubMed] [Google Scholar]
- Goldberg MW, Fiserova J, Huttenlauch I, Stick R. 2008. A new model for nuclear lamina organization. Biochemical Society Transactions 36, 1339–1343 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann K, Runions J, Evans DE. 2010. Characterization of SUN-domain proteins at the higher plant nuclear envelope. The Plant Journal 61, 134–144 [DOI] [PubMed] [Google Scholar]
- Gruber M, Soding J, Lupas AN. 2006. Comparative analysis of coiled-coil prediction methods. Journal of Structural Biology 155, 140–145 [DOI] [PubMed] [Google Scholar]
- Kapinos LE, Schumacher J, Mucke N, Machaidze G, Burkhard P, Aebi U, Strelkov SV, Herrmann H. 2010. Characterization of the head-to-tail overlap complexes formed by human lamin A, B1 and B2 ‘half-minilamin’ dimers. Journal of Molecular Biology 396, 719–731 [DOI] [PubMed] [Google Scholar]
- Kimura Y, Kuroda C, Masuda K. 2010. Differential nuclear envelope assembly at the end of mitosis in suspension-cultured Apium graveolens cells. Chromosoma 119, 195–204 [DOI] [PubMed] [Google Scholar]
- Kleffmann T, Hirsch-Hoffmann M, Gruissem W, Baginsky S. 2006. plprot: a comprehensive proteome database for different plastid types. Plant and Cell Physiology 47, 432–436 [DOI] [PubMed] [Google Scholar]
- Kruger A, Batsios P, Baumann O, Luckert E, Schwarz H, Stick R, Meyer I, Graf R. 2012. Characterization of NE81, the first lamin-like nucleoskeleton protein in a unicellular organism. Molecular Biology of the Cell 23, 360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner CF, Stick R, Eppenberger HM, Nigg EA. 1987. Differential expression of nuclear lamin proteins during chicken development. Journal of Cell Biology 105, 577–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Haruyama S, Fujino K. 1999. Assembly and disassembly of the peripheral architecture of the plant cell nucleus during mitosis. Planta 210, 165–167 [DOI] [PubMed] [Google Scholar]
- Masuda K, Takahashi S, Nomura K, Arimoto M, Inoue M. 1993. Residual structure and constituent proteins of the peripheral framework of the cell nucleus in somatic embryos from Daucus carota L. Planta 191, 532–540 [Google Scholar]
- Masuda K, Xu ZJ, Takahashi S, Ito A, Ono M, Nomura K, Inoue M. 1997. Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Experimental Cell Research 232, 173–181 [DOI] [PubMed] [Google Scholar]
- Mejat A, Misteli T. 2010. LINC complexes in health and disease. Nucleus 1, 40–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Díaz de la Espina S. 2009. The plant nucleoskeleton. In: Meier I, ed, Functional organization of the plant nucleus. Berlin, Heidelberg: Springer; pp 79–100 [Google Scholar]
- Moreno Díaz de la Espina S, Barthellemy I, Cerezuela MA. 1991. Isolation and ultrastructural characterization of the residual nuclear matrix in a plant cell system. Chromosoma 100, 110–117 [Google Scholar]
- Moriguchi K, Suzuki T, Ito Y, Yamazaki Y, Niwa Y, Kurata N. 2005. Functional isolation of novel nuclear proteins showing a variety of subnuclear localizations. The Plant Cell 17, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SP, Simmons CR, Bass HW. 2010. Structure and expression of the maize (Zea mays L.) SUN-domain protein gene family: evidence for the existence of two divergent classes of SUN proteins in plants. BMC Plant Biology 10, 269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Fukuda H. 2011. Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. The Plant Journal 66, 629–641 [DOI] [PubMed] [Google Scholar]
- Perez-Munive C, Moreno Díaz de la Espina S. 2011. Nuclear spectrin-like proteins are structural actin-binding proteins in plants. Biology of the Cell 103, 145–157 [DOI] [PubMed] [Google Scholar]
- Peter A, Stick R. 2012. Evolution of the lamin protein family: what introns can tell. Nucleus 3, 44–59 [DOI] [PubMed] [Google Scholar]
- Samaniego R, Jeong SY, de la Torre C, Meier I, Moreno Díaz de la Espina S. 2006. CK2 phosphorylation weakens 90kDa MFP1 association to the nuclear matrix in Allium cepa. Journal of Experimental Botany 57, 113–124 [DOI] [PubMed] [Google Scholar]
- Shimi T, Butin-Israeli V, Adam SA, Hamanaka RB, Goldman AE, Lucas CA, Shumaker DK, Kosak ST, Chandel NS, Goldman RD. 2011. The role of nuclear lamin B1 in cell proliferation and senescence. Genes and Development 25, 2579–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DN, Zastrow MS, Wilson KL. 2010. Direct actin binding to A- and B-type lamin tails and actin filament bundling by the lamin A tail. Nucleus 1, 264–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Hara-Nishimura I. 2011. Involvement of the nuclear pore complex in morphology of the plant nucleus. Nucleus 2, 168–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.