Abstract
A suppressor screen using dwarf mutants of barley (Hordeum vulgare L.) led to the isolation of ‘overgrowth’ derivatives, which retained the original dwarfing gene but grew at a faster rate because of a new mutation. The new mutations were in the Slender1 (Sln1) gene (11/13 cases), which encodes the DELLA protein central to gibberellin (GA) signalling, showed 100% genetic linkage to Sln1 (1/13), or were in the Spindly1 (Spy1) gene (1/13), which encodes another protein involved in GA signalling. The overgrowth mutants were characterized by increased GA signalling, although the extent still depended on the background GA biosynthesis capacity, GA receptor function, and DELLA activity. A comparison between two GA responses, α-amylase production and leaf growth rate, revealed degrees of specificity for both the overgrowth allele and the GA response under consideration. Many overgrowth mutants were also isolated in a dwarf line of bread wheat (Triticum aestivum L.) and 19 new alleles were identified in the Rht-B1 gene, one of the ‘Green Revolution’ semi-dwarfing genes and the orthologue of Sln1. The sites of amino acid substitutions in the DELLA proteins of both species provide insight into DELLA function, and included examples where identical but independent substitutions were observed. In both species, the starting lines were too dwarfed to be directly useful in breeding programmes, but new overgrowth derivatives with semidwarf heights have now been characterized. The variation they exhibit in GA-influenced traits identifies novel alleles with perfect markers that are of potential use in breeding.
Key words: barley, DELLA, gibberellins (GA), overgrowth mutant, suppressor screen, wheat.
Introduction
Many gibberellin (GA) mutants, either spontaneous in origin or identified after mutagenesis, have been characterized in a range of plant species. These include distinct dwarf and elongated (‘slender’) phenotypes that typically result from changes in either GA content or GA signalling. Identification of the genes involved and analysis of the proteins they encode have been fundamental in developing an understanding of growth regulation by GA, particularly in the model species of rice and Arabidopsis. Bioactive GAs bind to a GA receptor protein (GID1; Ueguchi-Tanaka et al., 2005; Murase et al., 2008; Shimada et al., 2008) and form a complex that is then bound by DELLA proteins, a subfamily of the GRAS transcription factor family (Griffiths et al., 2006; Willige et al., 2007; Hirano et al., 2010). The GA–GID1–DELLA complex is a target for binding of an SCF E3 ubiquitin ligase via the F-box subunit, resulting in ubiquitination of DELLA and its subsequent proteasome-mediated degradation (Fu et al., 2002; Sasaki et al., 2003; Dill et al., 2004). DELLA proteins are inhibitors of growth, and in one case the evidence suggests that DELLA binding to phytochrome-interacting factor (PIF) transcription factors effectively prevents them from promoting the expression of genes that stimulate hypocotyl growth (de Lucas et al., 2008; Feng et al., 2008).
Della mutants in barley and rice are of particular interest because they exhibit two markedly different phenotypes: highly elongated ‘slender’ types (‘slender DELLA’) and GA-insensitive dwarfs (‘dwarf DELLA’). The former trait is recessive and is characterized by male sterility and an extreme GA response, whereas the latter is dominant or semi-dominant, fully self-fertile, and with dwarfism that is non-responsive to exogenous GA. These radically different phenotypes each result from single-nucleotide substitutions in the Slender1 (Sln1) gene that encodes DELLA (Ikeda et al., 2001; Chandler et al., 2002; Asano et al., 2009). The slender DELLA types involve mutations that abolish the capacity of DELLA to repress growth, whereas the dwarf DELLA types result in DELLA proteins that no longer bind to the GA–GID1 complex, leading to DELLA accumulation.
Semi-dwarfing genes have been prominent in wheat and rice breeding since the Green Revolution because they improve yield. The modest (10–20%) reduction in height of semi-dwarfs is due to a deficiency in growth promotion by endogenous GA, and in wheat this results from mutations at the Rht-1 locus (which encodes DELLA), resulting in growth that is ‘insensitive’ to GA. The original semi-dwarfing mutations were spontaneous in origin, and their agronomic importance is evident from their continuing widespread use in current varieties some 50 years after their first introduction.
Bread wheat is an allo-hexaploid, with DELLA encoded by three homoeologous genes (Rht-A1, Rht-B1, and Rht-D1) on group 4 chromosomes. GA-insensitive semi-dwarfing alleles have been described for the B- and D-genome genes (Peng et al., 1999; Pearce et al., 2011). A more extreme dwarfing allele of the B-genome gene, Rht-B1c, results from insertion of a 2kb DNA element (Wu et al., 2011); all but 90 nt of the insertion are spliced from the transcript, resulting in a predicted 30 aa in-frame insertion in DELLA (Pearce et al., 2011).
‘Suppressor’ screens have been used in many experimental systems to generate near-wild-type (WT) phenotypes. In Arabidopsis, suppressor screens of dwarf mutants have led to the discovery of new genes involved in GA responses, or have confirmed the importance of previously identified genes (Carol et al., 1995; Wilson and Somerville, 1995; Silverstone et al., 1997; Peng et al., 1999). A recent study in rice demonstrated novel properties of a new intragenic suppressor allele of a gid1 mutant (Yamamoto et al., 2010). We have used a similar approach in barley and wheat, with the dual aims of identifying new genes or alleles important in GA responses, and of generating lines closer to a semi-dwarf phenotype that might be useful in breeding.
Materials and methods
Plant material
Himalaya barley and three previously characterized dwarf mutant derivatives are described in Table 1, together with 13 new overgrowth mutants and stocks derived from backcrossing and intercrossing. Grains of tall (Rht-B1a) and dwarf (Rht-B1c) BC7 isolines of the Brazilian breadwheat variety Maringá (Hoogendoorn et al., 1988) were provided by the Australian Winter Cereals Collection, Tamworth, NSW, Australia. Plants were grown in a greenhouse in 20cm pots containing a compost-based mix under natural light (with daylength extension to 14h provided during winter months).
Table 1.
Barley lines, genotypes and mutations. ND, not determined.
| Line | Genotype | Mutationa | Reference | |
|---|---|---|---|---|
| Nucleotide | Amino acid | |||
| Himalaya | WT | – | – | |
| M640 | Sln1d | G137A | G46E | Chandler et al., 2002 |
| TR1 | Sln1d.4 | C1469Tb | S490Fb | This paper |
| TR9 | Sln1d.5 | G839Ab | R280Hb | This paper |
| TR13 | Sln1d.6 | G803Ab | R268H | This paper |
| TR26 | Sln1d, spy1a | G812A (Spy1) | G271D (SPY1) | This paper |
| TR56c | Sln1d.7 | G829Ab | A277Tb | This paper |
| TR60c | Sln1d.8 | G691Ab | V231Mb | This paper |
| TR100c | Sln1d.9 | G1442Ab | R481Hb | This paper |
| TR103c | ND | ND | ND | This paper |
| TR107 | Sln1d.10 | G844Tb | V282Fb | This paper |
| M463 | grd2b | – | – | Wolbang et al., 2004 |
| TR216 | grd2b, sln1s | G829A | A277T | This paper |
| TR261 | grd2b, sln1m | G680A | G227E | This paper |
| TR305 | grd2b, sln1o | C1454T | S485F | This paper |
| M693 | gse1n | – | – | Chandler et al., 2008 |
| TR407 | gse1n, sln1n | G710A | C237Y | This paper |
| M240c | sln1m | See above | See above | This paper |
| M242c | sln1n | See above | See above | This paper |
| M2434 | sln1s | See above | See above | This paper |
| M244segd | sln1s/Sln1d.7 | See above | See above | This paper |
| M247c | spy1a | See above | See above | This paper |
| M248 | grd2b, spy1a | See above | See above | This paper |
| M249 | gse1l, spy1a | See above | See above | This paper |
a Coordinates refer to the positions in the HvSln1 coding sequence or SLN1 amino acid sequence from Himalaya (identical to Genbank accession no. AK372064) starting at ATG and ending at TGA. For TR26, the coordinate refers to the position in the HvSpy1 (AF035820) coding sequence or SPY1 amino acid sequence, starting at ATG and ending at TGA.
b Sln1d.4–Sln1d.10 are derivatives of Sln1d and contain the original Sln1d mutation in addition to the new substitutions indicated.
c Lines established after two backcrosses to Himalaya before selecting for homozygosity of the allele shown.
d Grains are progeny from heterozygotes segregating at the Sln1 locus as indicated.
Mutagenesis
The method for mutagenesis of barley and wheat grains was a simplification of an earlier procedure (Zwar and Chandler, 1995). Grain (1–2kg) of each line was imbibed in twice its mass of water at 4 °C overnight, transferred to 2 l measuring cylinders filled with water, and aerated from the bottom with pressurized air for 8h, with one change of fresh water given after 4h. The water was removed and grain was incubated for 2h in freshly prepared 1mM sodium azide dissolved in 0.1M potassium phosphate buffer (pH 3.0), and then washed extensively in running water for 2h, placed in a fume hood to dry overnight, and sown in the field within several days of treatment.
Construction of derivative lines carrying overgrowth alleles
The barley overgrowth alleles were backcrossed, intercrossed, and outcrossed to generate a set of lines suitable for detailed physiological characterization (Supplementary Fig. S1 at JXB online). Four of the new Sln1 overgrowth alleles occurred in either grd2b or gse1n dwarfing backgrounds and were backcrossed for two generations to the WT allowing overgrowth phenotypes to be compared in tall and dwarf backgrounds. The loss of the original dwarfing allele was confirmed by PCR. The remaining seven new Sln1 overgrowth alleles occurred in the Sln1d dwarf background, and four of these (Sln1d.7, Sln1d.8, Sln1d.9 and TR103) have been through two generations of back-crossing to the WT. Two overgrowth lines (TR56 and TR216) involved identical mutations in Sln1 (G829A), but one occurred in a Sln1d allele and the other in a Sln1 (WT) allele. These lines were intercrossed, and F2 plants were isolated that were heterozygous at Sln1 (Sln1d.7/sln1s) but lacked grd2b (from the TR216 parent). The progeny of such heterozygotes showed a 1:2:1 ratio of genotypes, allowing the effect of sln1s on growth to be assessed in a ‘tall’ (sln1s/sln1s) versus ‘DELLA dwarf’ (Sln1d.7/Sln1d.7) genetic background. The spy1a allele was crossed into GA deficiency and GA receptor dwarfing backgrounds to determine whether it would enhance GA signalling.
Leaf elongation rates and GA dose–response curves
The methods used have been described previously (Chandler and Robertson, 1999). Curves were fitted to data points using a four-parameter Hill equation.
α-Amylase production by endosperm half-grains
Endosperm half-grains were prepared and incubated with or without gibberellic acid (GA3; 1 μM) at 22 °C for 0, 42, or 72h. To each sample, 1.5ml of a solution of 10mM CaCl2 was added, the half-grains were homogenized, and an aliquot of 1ml was clarified by centrifugation (20 000g for 5min). The supernatant was analysed for α-amylase activity using the Megazyme α-amylase (Ceralpha) procedure.
Coleoptile lengths of barley overgrowth lines
Coleoptile lengths were determined on seedlings after 21 d of growth in the dark with a daily temperature programme of 12h at 12 °C and 12h at 8 °C.
PCR amplification of DNA and sequencing
DNA was prepared from leaf material of barley and wheat lines by the method of Ellis et al. (2005). PCR amplification of barley sequences used primers specific for the Sln1, Spy1, and Gse1 genes (the latter as a ‘control’ gene; see Discussion). Amplified fragments were treated with Exosap-IT (Affymetrix) to remove primers and then sequenced using a BigDye Terminator Sequencing kit (Applied Biosystems). Wheat sequences were amplified using primer pairs in which one primer was specific for the Rht-B1 gene (Supplementary Table S1 at JXB online). The 3’ half of the gene in an initial selection of 60 overgrowth mutants was amplified using conserved forward primers but reverse primers that were specific to B gene sequences in the 3’ untranslated region.
Results
Isolation of overgrowth mutants in barley
The starting material comprised three dwarf derivatives of Himalaya barley, each representing a defined single-nucleotide substitution affecting GA biosynthesis (GA 3-oxidase, Grd2), the GID1 GA receptor (Gse1), or DELLA (Sln1). Grains of each dwarf line were treated with sodium azide, sown in the field for harvest of M2 grains, and M2 seedlings growing in soil were screened at the second-leaf stage for those that showed more rapid growth than their dwarf siblings (1×106 grains sown, representing about 50 000 M1 spikes).
Of most interest were 22 plants (Fig. 1), recovered across all three dwarf backgrounds, that were fully fertile and had progeny that were uniform and showed enhanced growth. In each case, the presence of the original dwarfing mutation was confirmed by sequencing the appropriate PCR-amplified fragment. We refer to these as ‘overgrowth’ mutants, as their leaf elongation rate was higher than expected based on the presence of a dwarfing allele. The degree of growth enhancement differed among lines (see below), and their height at maturity ranged from intermediate between the dwarf parent and WT to as tall as the WT.
Fig. 1.
Screening for overgrowth mutants in barley. One tall seedling among dwarf siblings. The pale yellow seedling (front left) is an example of the 2–3% of non-green seedlings that are seen in barley M2 populations.
Identification of mutations and genetic linkage studies
The Sln1 gene was sequenced in DNA from the overgrowth mutants, as this is one of several candidate genes where mutation might lead to an overgrowth phenotype. New mutations in the Sln1 ORF were found in 20 of the 22 lines, and they defined 11 new Sln1 alleles (some lines carried identical mutations and may have been sibs): seven occurred in the Sln1d dwarf background (and were hence intra-allelic mutations and named Sln1d.4–Sln1d.10), three occurred in the grd2b background (sln1m, sln1o, and sln1s), and one occurred in the gse1n background (sln1n). Each of the new alleles (TR lines in Table 1) was a single-nucleotide substitution resulting in a single amino acid substitution in the SLN1 (DELLA) protein. They all occurred in the C-terminal 60% of DELLA, corresponding to the GRAS domain, and they will be described in further detail later, together with equivalent mutations in wheat. There were two identical mutational events, G829A, one being a derivative of Sln1d and the other a derivative of a WT Sln1 gene in the grd2b dwarf background.
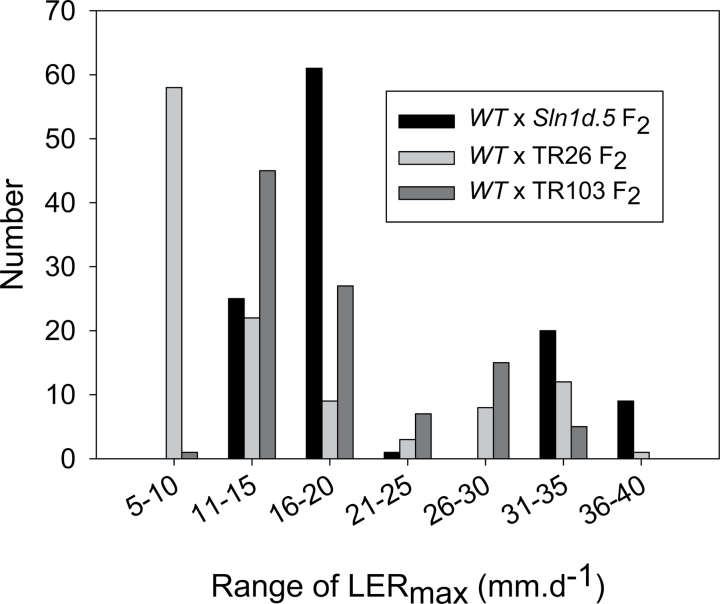
The two remaining overgrowth lines (TR26 and TR103) lacked any new mutation in the Sln1 ORF. Both occurred in a Sln1d dwarf background, and they were crossed to Himalaya, together with Sln1d.5 as a positive control, to assess genetic linkage between the Sln1d dwarfing allele and the new overgrowth allele. The F2 population from the control WT×Sln1d.5 cross showed the expected 3:1 (Sln1d.5:WT) distribution in maximal leaf elongation rates (LERmax; Fig. 2); note the absence of individuals with the slow growth rate of the Sln1d parent, indicating complete linkage between the dwarfing mutation and the intra-allelic overgrowth mutation. A similar result was observed for the TR103×WT F2 population, indicating that the overgrowth mutation in TR103 was 100% linked to Sln1d. By contrast, the TR26×WT F2 population included a majority of seedlings with growth rates the same as Sln1d, about 25% with growth rates the same as WT, and some seedlings with intermediate growth rates (Fig. 2). These results are consistent with the overgrowth mutation in TR26 being in a gene that is genetically unlinked to Sln1. The sequence of Spindly1 in TR26 revealed a single-nucleotide substitution (spy1a; Table 1) that resulted in an amino acid substitution in the sixth TPR motif of SPY1, a region that is important for Arabidopsis SPY activity (Silverstone et al., 2007). SPY1 encodes a negative regulator of GA signalling and was first identified in Arabidopsis, but functionally related genes were later shown to exist in barley (Robertson et al., 1998) and rice (Shimada et al., 2006).
Fig. 2.
Distribution of LERmax values of individual F2 seedlings from different crosses. Values for parents (mean ±standard error) were WT, 35.2±1; Sln1d, 7.6±0.4; Sln1d.5, 13.7±0.4; Sln1d, spy1a, 17.5±0.4; TR103, 16.6±0.6.
Leaf elongation rates of Sln1 overgrowth alleles
The LERmax value achieved by the first leaf under standard conditions is a robust measure of GA response (Chandler and Robertson, 1999) and was determined for all of the overgrowth lines and their parents (Table 2). The 13 independent overgrowth lines all had significantly higher LERmax values than their respective dwarf parent, and the extent of growth enhancement varied in an allele-dependent manner. Three overgrowth alleles (sln1m, sln1n, and sln1s) were compared in both their original dwarfing background and after backcrossing to the tall WT background; growth rates were consistently lower in dwarf backgrounds, indicating that overgrowth alleles are still subject to decreased GA signalling resulting from either impaired GA biosynthesis or GA receptor function. In a WT background, the overgrowth alleles generally enhanced growth rates (additional data presented below). Identical nucleotide substitutions in sln1s and Sln1d.7 allowed us to assess whether impaired DELLA function affected growth promotion by the overgrowth allele sln1s. These two overgrowth alleles are of independent origin, and differ in whether the Sln1d mutation is also present. The M244seg stock segregates tall (sln1s/sln1s) and dwarfed (Sln1d.7/Sln1d.7) lines (see Materials and methods) whose LERmax values were determined as 35.1±0.8 and 12.9±0.4mm d–1, respectively, compared with dwarf Sln1d at 8.3±0.3mm d–1. We concluded that sln1s is subject to impaired DELLA activity. Taken together, these results highlighted a distinctive difference between the new Sln1 overgrowth alleles and typical loss-of-function elongated sln1 alleles such as sln1c; the former showed lower growth rates when GA signalling was impaired by upstream mutations affecting GA biosynthesis or signalling (Chandler and Robertson, 1999), whereas the latter showed constitutive growth rates.
Table 2.
Leaf elongation rates and α-amylase production of overgrowth mutants. For results for TR26, see Table 3. ND, not determined.
| Line | Genotype | LERmax (mm d–1) | α-Amylase activity (Ceralpha units/grain) | ||
|---|---|---|---|---|---|
| 0h (×103) | 42 h | 72 h | |||
| Him | WT | 34.6±0.9 | 1.6 | 0.36±0.08 | 1.7±0.09 |
| Him (GA3) | WT | 52.3±0.1 | ND | 9.81±0.86 | 23.1±2.8 |
| M640 | Sln1d | 9.2±0.2 | 1.6 | 0.21±0.01 | 0.19±0.05 |
| M640 (GA3) | Sln1d | 12.1±0.3 | ND | ND | ND |
| TR1 | Sln1d.4 | 23.8±0.5 | 2.5 | 0.26±0.01 | 2.88±0.27 |
| TR9 | Sln1d.5 | 15.5±0.7 | 1.6 | 0.21±0.01 | 0.16±0.04 |
| TR13 | Sln1d.6 | 15.8±0.6 | 1.8 | 0.20±0.02 | 0.19±0.04 |
| TR56 | Sln1d.7 | 16.7±0.6 | 2.7 | 0.93±0.22 | 3.24±0.47 |
| TR60 | Sln1d.8 | 28.6±0.9 | 3.6 | 0.82±0.13 | 3.76±0.23 |
| TR100 | Sln1d.9 | 20.2±0.6 | 3.2 | 4.34±0.13 | 14.6±0.39 |
| TR103 | ND | 16.1±0.4 | 3.4 | 0.74±0.10 | 0.39±0.1 |
| TR107 | Sln1d.10 | 13.2±0.3 | 2.4 | 0.39±0.07 | 0.17±0.03 |
| M463 | grd2b | 16.9±0.8 | 2.5 | 0.67±0.04 | 1.3±0.2 |
| M463 (GA3) | grd2b | 49.8±0.8 | ND | 7.20±0.73 | 18.9±1.4 |
| TR216 | grd2b, sln1s | 23.2±0.6 | 4.1 | 1.01±0.12 | 4.16±0.55 |
| TR261 | grd2b, sln1m | 38.4±1.5 | 3.8 | 7.50±0.24 | 18.1±0.8 |
| TR305 | grd2b, sln1o | 32.9±0.9 | 3.8 | 9.85±0.43 | 16.9±1.2 |
| M693 | gse1n | 19.7±0.4 | 2.5 | 0.29±0.05 | 0.11±0.01 |
| M693 (GA3) | gse1n | 35.3±1.2 | ND | ND | ND |
| TR407 | gse1n, sln1n | 29.3±0.5 | 9.9 | 0.27±0.03 | 0.26±0.13 |
| M240 | sln1m | 42.2±1.6 | 6.5 | 11.6±1.4 | 22.6±2.3 |
| M242 | sln1n | 44.3±3.1 | 3.0 | 1.07±0.08 | 6.39±1.52 |
| M243 | sln1s | 35.4±0.9 | 3.8 | 3.66±0.53 | 15.4±0.9 |
The spy1a overgrowth allele enhances growth of GA biosynthesis and GA receptor dwarfs
The mutant spy1a overgrowth allele was backcrossed into GA biosynthesis and GA receptor dwarf backgrounds, as well as to the WT (Table 3). The presence of spy1a significantly increased growth rates of the dwarf lines, although not to WT values (Table 3). In a WT background, spy1a caused only marginal increases in growth rate, at the borderline of statistical significance when different experiments and different grain harvests were assessed (Table 3). We concluded that spy1a enhanced growth in all three dwarfing backgrounds but only to a limited extent, indicating that growth rates were still restricted by impaired GA signalling, as found for the Sln1 overgrowth alleles.
Table 3.
Effect of spy1a on growth rates and α-amylase production.
| Line | Genotype | LERmax (mm d–1) | α-Amylase activity (Ceralpha units/grain) | ||
|---|---|---|---|---|---|
| 0h (×103) | 42 h | 72 h | |||
| Himalaya | WT | 34.6±0.9 | 1.6 | 0.36±0.08 | 1.7±0.09 |
| M247 | spy1a | 37.6±0.7 | 4.6 | 3.33±0.22 | 12.5±1.2 |
| M640 | Sln1d | 9.2±0.2 | 1.6 | 0.21±0.01 | 0.19±0.05 |
| TR26 | Sln1d, spy1a | 17.1±0.4 | 2.3 | 0.66±0.10 | 2.84±0.46 |
| M463 | grd2b | 17.7±0.6 | 2.1 | 0.53±0.1 | 3.7±0.3 |
| M248 | grd2b, spy1a | 23.4±0.5 | 3.6 | 10.2±0.9 | 15.0±1.8 |
| M691 | gse1l | 20.6±0.4 | 1.6 | 0.07±0.01 | 0.13±0.03 |
| M249 | gse1l, spy1a | 25.8±0.5 | 2.1 | 1.2±0.2 | 7.2±1.0 |
α-Amylase production by endosperm half-grains of Sln1 and Spy1 overgrowth mutants
The production of α-amylase by endosperm half-grains of barley is typically dependent on the presence of an active GA. Therefore, monitoring α-amylase activity in the absence of an active GA provides a convenient measure of the extent of basal GA signalling. Two control lines with normal GA sensitivity (WT and grd2b) showed a near 15-fold increase in α-amylase activity over basal levels after 72h incubation with GA3 (Table 2). When overgrowth mutants and their dwarf parents were examined, the initial amount of α-amylase activity in mature endosperm half-grains was very low (Table 2, 0 h, note values are ×103), but, with incubation some overgrowth lines (Sln1d.4; Sln1d.7; Sln1d.8; Sln1d.9; grd2b, sln1m; grd2b, sln1o; and grd2b, sln1s) showed enhanced production of α-amylase relative to their dwarf parent, whereas others (Sln1d.5; Sln1d.6; Sln1d.10; and gse1n, sln1n) did not. Among the overgrowth derivatives of Sln1d, the Sln1d.9 derivative was exceptional, accumulating very high levels of α-amylase at both 42 and 72h incubation, despite this line showing only a modest restoration of growth rate (Table 2).
Overgrowth alleles backcrossed into a tall background had higher rates of α-amylase accumulation than the original dwarf lines; for instance, the increase in α-amylase production by sln1s was higher in the WT background (M243) than in either Sln1d (TR56) or grd2b (TR216) dwarf backgrounds. Similar results were noted for sln1n (M242 vs TR407) and possibly sln1m (M240 vs TR261). This observation was comparable to those above for growth rates.
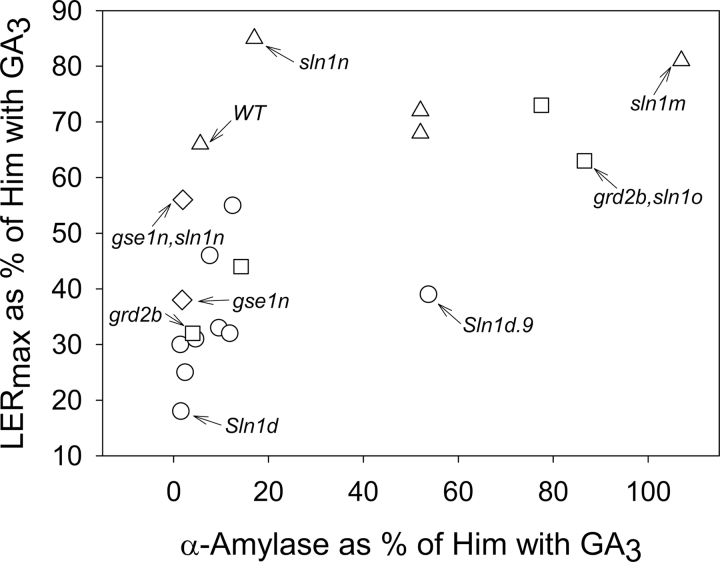
The extent of two different GA responses (growth rate and α-amylase production, both in the absence of GA) was plotted for each overgrowth line as a percentage of the WT+GA value (Fig. 3). Selected examples that illustrate marked differences in the extents of the two responses compared with the parent line are indicated. Sln1d.9 was clearly different from Sln1d and the other Sln1d-derived overgrowth lines (Fig. 3, circles) in its high levels of α-amylase and relatively slow growth rate. Other comparisons revealed similar differences; the sln1o allele in a grd2b background had greatly increased α-amylase accumulation relative to its parent (Fig. 3, squares); the sln1n allele in a gse1n background grew more rapidly than its parent (Fig. 3, diamonds), although α-amylase accumulation was similar; and the backcrossed lines sln1m and sln1n had similar growth rates to WT (triangles) but differed greatly in their extent of α-amylase accumulation. Overall, these results illustrated examples of large differences between Sln1 overgrowth alleles in two defined GA responses, indicating that the effect of an overgrowth allele in enhancing GA signalling depends on both the allele and the GA response being investigated.
Fig. 3.
Relationship between leaf elongation rate and α-amylase production for each overgrowth line. The data from Tables 2 and 3 were plotted as percentages of the GA3-treated Himalaya control, with the α-amylase data being a mean of the 42 and 72h values. Data indicated with circles, squares, diamonds, and triangles refer, respectively, to Sln1d and its overgrowth derivatives, grd2b and its overgrowth derivatives, gse1n and its overgrowth derivative, and WT and its overgrowth derivatives.
The spy1a allele showed much higher levels of α-amylase production in both WT and dwarf backgrounds than in the original Sln1d dwarfing background (Table 3). This behaviour contrasts with its much smaller effects on growth rate (Table 3), and the conclusions reached above on the GA response specificity of Sln1 overgrowth alleles also extend to spy1a.
GA responsiveness of overgrowth lines
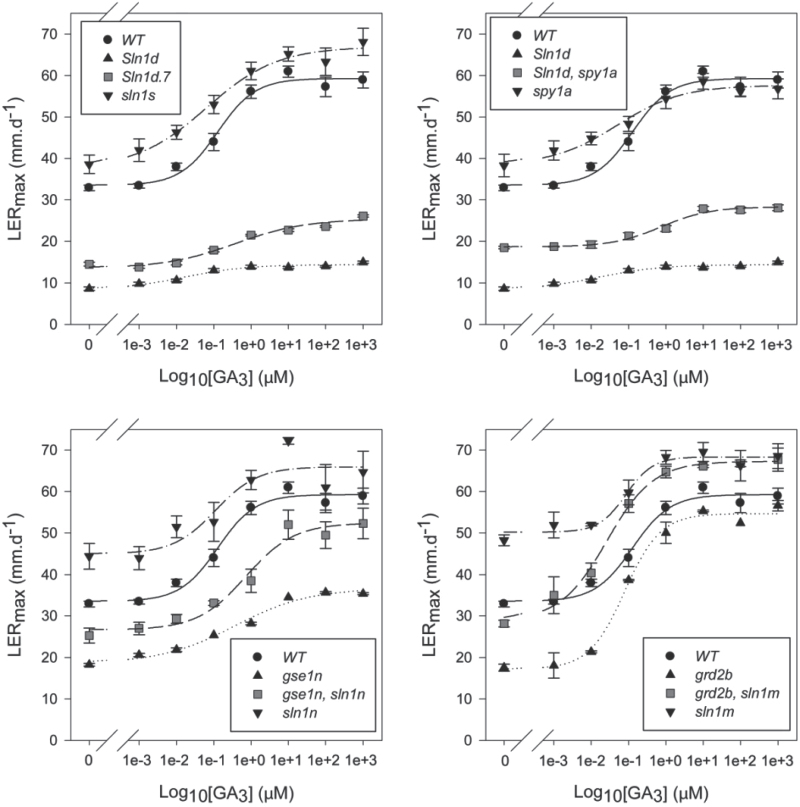
The LERmax values (Tables 2 and 3) indicated that overgrowth alleles enhance leaf elongation rates of both dwarf and tall genotypes but are still influenced by the extent of GA signalling. To investigate this in more detail, we determined GA dose–response growth curves for each overgrowth allele. The curves for four selected alleles are shown in Fig. 4, and three general observations could be made. First, the overgrowth alleles conferred enhanced growth rates across all concentrations of applied GA in dwarf backgrounds, and generally did so in a WT background as well, although at high GA concentrations the effect was small or even non-existent. Secondly, overgrowth derivatives of GA signalling dwarf lines (Sln1d, gse1n) never fully restored their growth rate to that of the WT, even at high GA concentrations (Fig. 4A–C), in contrast to the GA biosynthetic dwarf background (Fig. 4D). Thirdly, most of the transition in growth rates in overgrowth derivatives of lines with normal GA signalling occurred over the GA concentration range of 0.01–1 μM, the same as that identified previously for GA-deficient mutants (Chandler and Robertson, 1999). This indicated that overgrowth alleles do not markedly change their ‘sensitivity’ (H50) for growth responses to GA. These results confirmed that overgrowth alleles enhanced the degree of GA signalling in a manner that was influenced by different genetic backgrounds and by the concentration of exogenous GA.
Fig. 4.
GA3 dose–response curves for growth of the first leaf of barley. Each graph contains four curves, representing the WT (same curve for each panel), the original dwarf line, the overgrowth derivative of the dwarf, and the overgrowth allele in a WT background.
Other traits associated with overgrowth alleles
The overgrowth lines were close to normal in appearance during growth and at maturity, apart from differences in overall height. None of the nine overgrowth derivatives of Sln1d was as tall as the WT at maturity, in contrast to the remaining four overgrowth lines, which were as tall as the WT. Coleoptile lengths of overgrowth lines (Supplementary Table S2 at JXB online) varied in general accordance with LERmax values and with final plant height.
A general feature of overgrowth lines is that they produce larger grains than their dwarf parent. In different harvests from different growing seasons, grain sizes were generally intermediate between the parental dwarfs (grain mass about 40mg) and the tall WT (grain mass about 55mg). In different greenhouse generations, the grains of grd2b,sln1m were on average 40% larger than those of the grd2b dwarf parent. When backcrossed to Himalaya, the sln1m allele resulted in an average 20% increase in grain size. Selected overgrowth alleles have been backcrossed into the commercial barley variety ‘Sloop’, and a full analysis on grain size and other traits will be made on BC3 sister lines.
Overgrowth mutants in wheat
Grains of an Rht-B1c dwarf in a var. ‘Maringá’ background were treated with sodium azide, sown in the field, and M2 plants were screened for overgrowth mutants either during early growth or at maturity in the field. From these two screens (1.6 million M2 plants), approximately 400 plants were selected and characterized either by rapid early leaf elongation or by increased height at maturity. As seen for barley, there was a wide range among different plants in their degree of growth restoration (Fig. 5).
Fig. 5.
Wheat overgrowth mutants. Overgrowth mutants and control isolines were grown to maturity in the greenhouse. Left to right: Maringá Rht-B1c, Maringá Rht-B1a, and three overgrowth mutants that differed in height.
We sequenced the portion of the wheat Rht-B1 gene that corresponds to the region of Sln1 in barley where overgrowth mutations were identified. From an initial selection of 60 wheat overgrowth lines, we identified 19 new derivative alleles of Rht-B1c, which were named using the same convention as for overgrowth derivatives of Sln1d barley, i.e. Rht-B1c.1, Rht-B1c.2, Rht-B1c.3, and so on (Table 4). These included two different classes of mutation responsible for the overgrowth phenotype. Four alleles (Rht-B1c.11, Rht-B1c.13, Rht-B1c.19, and Rht-B1c.20) were premature termination codons, which in barley result in an elongated slender phenotype and male sterility. These four wheat alleles had growth and fertility similar to that of the tall isoline; presumably the expression of the A- and/or D-genome DELLA proteins provides genetic compensation for the B-genome null mutant.
Table 4.
Wheat overgrowth lines, mutations, and stem length at maturity.
| Genotypea | Mutationb | Stem length (% Rht-B1a) | |
|---|---|---|---|
| Nucleotide | Amino acid | ||
| Rht-B1a | 100 | ||
| Rht-B1c | Insertion | Insertion | 42 |
| Rht-B1c.1 | G2715A | G260E | 91 |
| Rht-B1c.2 | G2726A | V264M | 95 |
| Rht-B1c.3 | G2747A | A271T | 74 |
| Rht-B1c.4 | G2829A | G298D | 68 |
| Rht-B1c.5 | G2831A | A299T | 60 |
| Rht-B1c.6 | G2849A | A305T | 60 |
| Rht-B1c.7 | C2865T | A310V | 93 |
| Rht-B1c.8 | C2966T | P344S | 55 |
| Rht-B1c.9 | C2972T | L346F | 85 |
| Rht-B1c.10 | G3065A | G377R | 64 |
| Rht-B1c.11 | G3076A | W380ter | 102 |
| Rht-B1c.12 | C3117T | P394L | 50 |
| Rht-B1c.13 | G3190A | W418ter | 93 |
| Rht-B1c.15 | G3477A | R514H | 95 |
| Rht-B1c.16 | C3507T | T524I | 93 |
| Rht-B1c.17 | C3519T | S528F | 72 |
| Rht-B1c.18 | G3624A | G563D | 92 |
| Rht-B1c.19 | G3697A | W587ter | 92 |
| Rht-B1c.20 | G3874A | W646ter | 96 |
a All genotypes were in the Maringá genetic background, and the stem length of the tall (Rht-B1a) line ranged from 1050 to 1150mm under irrigated field conditions.
b The nucleotide and amino acid sequences are under accession no. KC434134.
The remaining alleles identified 15 new amino acid substitutions in the wheat DELLA protein encoded by the B-genome, which were associated with varying degrees of growth recovery (Table 4). It is of interest to compare these substitutions with those found in barley (Fig. 6). Across the two species, there were 26 single amino acid substitutions, including four sites where identical amino acid changes occurred. We interpreted this to mean that there is a moderately limited set of amino acid substitutions that will lead to an overgrowth phenotype. This conclusion was supported by the observation that within both wheat and barley there were identical but independent mutations observed.
Fig. 6.
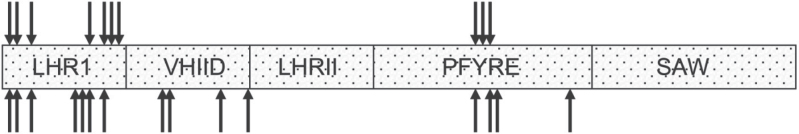
Sites of amino acid substitutions in the DELLA C-terminal GRAS domain for overgrowth mutants of barley (upper arrows) and wheat (lower arrows). Conserved amino acid motifs are indicated.
The sites of amino acid substitutions in the C-terminal half of barley and wheat DELLA proteins are shown in relation to conserved motifs that have been previously identified (Fig. 6). The fact that overgrowth mutations were distributed throughout much of the C-terminal half provides considerable potential for alterations in the binding of DELLA to interacting protein partners.
Discussion
Overgrowth mutants of barley and wheat were isolated following mutagenesis of dwarf lines; they retained the original dwarfing alleles but grew faster than expected because of newly induced mutations. They were characterized by enhanced GA signalling, although the extent was specific for both the allele and the GA response being considered. Previous studies in Arabidopsis employed suppressor screens to identify or confirm four genes with important roles in GA signalling – SPY, SLY, and the two Della genes GAI and RGA. Our results indicated that the Della genes (Sln1 in barley, Rht-1 in wheat) are the most frequent sites of overgrowth mutations. In only a single case was a different gene implicated – one of the barley overgrowth mutants was due to a new mutation in Spy1. Earlier studies on this gene indicated that it acted as a negative regulator of GA signalling in barley (Robertson et al., 1998), although its importance in GA regulation in vivo could not be assessed because no mutants were available. Now, the isolation of the spy1a mutant and the demonstration that it enhances GA signalling in a range of dwarf backgrounds as well as in the WT confirms the earlier findings, and extends the functional importance of SPY1 from Arabidopsis to cereals. The fact that suppressor screens in diverse species have yielded mutations in very similar genes indicates functional conservation of GA responses across a wide taxonomic distance. A similar screen in rice has now extended the list of genes to GID1, the GA receptor (Yamamoto et al., 2010).
Five independent lines of evidence supported the conclusion that the new mutations identified here were responsible for overgrowth phenotypes, rather than being coincidental effects of prior treatment with mutagen. First, for each of the 13 barley mutants, a ‘control’ gene (Gse1) of about the same length as Sln1 was sequenced, and in no case was a nucleotide substitution detected (data not shown). Secondly, the mutations identified were almost (30/31) exclusively G→A (assuming C→T represents G→A in the opposite strand). The redundancy in the genetic code allows 33% of random G→A changes to have no corresponding amino acid substitution; however we did not observe a single case of silent substitution. Thirdly, the mutations we identified involved amino acid residues that are very highly conserved; in all but two cases (both involving very conservative amino acid substitutions), the DELLA mutants involved amino acid residues that are identical between cereal species and the taxonomically distant Arabidopsis. Substitution of highly conserved amino acid residues is much more likely to result in disruption of protein function than changes in poorly conserved residues. Fourthly, independent mutagenesis treatments gave examples of identical mutations induced within barley, between barley and wheat, and within wheat (see Results). This is highly unlikely to occur by chance, and instead suggests that there is a moderately limited spectrum of mutations that will generate an overgrowth phenotype. Fifthly, we always observed 100% linkage between mutant phenotypes and gene sequences following crossing and subsequent segregation in both barley and wheat.
Overgrowth alleles enhance GA signalling and so are most likely to cause reductions in either the amount of DELLA protein or its ‘activity’, the latter probably involving interactions with other proteins. We think it most likely that mutant DELLA proteins have reduced affinity for interacting protein partners, resulting in a greater promotion of ‘GA’ responses. The barley and wheat DELLA proteins share 95% amino acid sequence identity, and in both species a considerable number of amino acid substitutions were in the LHR1 motif, a region that in Arabidopsis is involved in interactions with PIF4 and PIL5 (de Lucas et al., 2008; Feng et al., 2008). Mutations in this region will presumably show reduced affinity for PIF orthologues in cereals. There are also mutations elsewhere in the DELLA protein, including the VHIID and PFYRE motifs, which have substantial effects on growth but which are far removed from the currently identified LHR1 motifs important for PIF interaction. Very little is known about protein partners that interact with the PFYRE motif, or with DELLA proteins in general for other GA responses such as α-amylase production and grain dormancy. The differential effects of particular overgrowth alleles on growth versus α-amylase production in barley suggest that different regions of the DELLA protein interact with different protein partners to regulate these two responses.
The starting lines used in this study are too dwarfed for breeding purposes and were selected for basic/strategic studies on the role of GA in growth regulation. The overgrowth lines generated from these mutants are much closer to ideotypes (e.g. semi-dwarfs) that might be useful in breeding. These are more difficult to isolate in a direct forward screen because typically they fall within the range of variation (largely non-genetic) present in the original tall parent. The identification of new Sln1 overgrowth alleles that are fully fertile and with a range of growth rates both faster and slower than the WT indicates that the previous designation of Sln1 mutants into slender DELLA and dwarf DELLA was an oversimplification, reflecting extremes at the ends of a continuum.
Barley has advantages over wheat for mutational studies, being diploid instead of hexaploid. Typical loss-of-function phenotypes are expected in barley, but not in wheat if homoeologous genes are equally expressed. Our success in isolating many new mutants in wheat stems from the fact that the dwarfism due to semi-dominant alleles such as Rht-B1c is effectively a diploid trait, involving only one of the three genomes, which allows selection of a range of loss-of-function intra-allelic mutations.
In both species new alleles were recovered for an agronomically important trait, plant height. The large collection of wheat mutants is expected to reveal many new semi-dwarfing alleles in the Rht-B1 gene, with the potential to extend the range of dwarfism (more or less dwarfed) beyond that of the existing Rht-1 semi-dwarfing genes. These might be useful in targeting specific alleles to specific environments (Flintham et al., 1997); for instance, stronger (more dwarfing) alleles in cool, moist, high-input environments and weaker alleles in warm, dry, low-input environments. But in addition, variation was also observed in other GA-influenced traits, such as grain size and grain dormancy, which are of practical interest. The dwarf Rht-B1c line has considerably higher grain dormancy than either the tall or semi-dwarf (Rht-B1b) isolines. Initial experiments on the new semi-dwarf overgrowth derivatives indicate that some have higher grain dormancy than Rht-B1b and may therefore be of value in contributing to sprouting tolerance. Furthermore, it has recently been shown that DELLA proteins influence the resistance of both wheat and barley to fungal pathogens (Saville et al., 2012). Among the many new Della alleles, we hope to find examples where suitable height is associated with other useful traits. As a single gene character, and with a perfect molecular marker, such alleles can readily be introduced into breeding programmes.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. Primers for sequencing the 3’ half of wheat Rht-B1c.
Supplementary Table S2. Coleoptile lengths of barley overgrowth lines.
Supplementary Fig. S1. Barley seedlings of different genotypes at the same age.
Acknowledgements
We thank CSIRO and the Grains Research and Development Corporation (project CSP00126) for financial support, and Steve Swain, Tesfamichael Kebrom, Rod King, Rudy Dolferus, Bob McIntosh and Sonja Chandler for comments on the manuscript.
Glossary
Abbreviations:
- GA
gibberellin
- GA3
gibberellic acid
- LER
leaf elongation rate
- PIF
phytochrome-interacting factor
- WT
wild type.
References
- Asano K, Hirano K, Ueguchi-Tanaka M, Angeles-Shim RB, Komura T, Satoh H, Kitano H, Matsuoka M, Ashikari M. 2009. Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Molecular Genetics and Genomics 281, 223–231 [DOI] [PubMed] [Google Scholar]
- Carol P, Peng J, Harberd NP. 1995. Isolation and preliminary characterization of gas1-1, a mutation causing partial suppression of the phenotype conferred by the gibberellin-insensitive (gai) mutation in Arabidopsis thaliana (L.) Heyhn. Planta 197, 414–417 [DOI] [PubMed] [Google Scholar]
- Chandler PM, Harding CA, Ashton AR, Mulcair MD, Dixon NE, Mander LN. 2008. Characterization of gibberellin receptor mutants of barley (Hordeum vulgare L.). Molecular Plant 1, 285–294 [DOI] [PubMed] [Google Scholar]
- Chandler PM, Marion-Poll A, Ellis M, Gubler F. 2002. Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiology 129, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler PM, Robertson M. 1999. Gibberellin dose-response curves and the characterization of dwarf mutants of barley. Plant Physiology 120, 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S. 2008. A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP. 2004. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16, 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MH, Rebetzke GJ, Azanza F, Richards RA, Spielmeyer W. 2005. Molecular mapping of gibberellin-responsive dwarfing genes in bread wheat. Theoretical and Applied Genetics 111, 423–430 [DOI] [PubMed] [Google Scholar]
- Feng SH, Martinez C, Gusmaroli G, et al. 2008. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flintham JE, Borner A, Worland AJ, Gale MD. 1997. Optimizing wheat grain yield: Effects of Rht (gibberellin-insensitive) dwarfing genes. Journal of Agricultural Science 128, 11–25 [Google Scholar]
- Fu X, Richards DE, Ait-Ali T, Hynes LW, Ougham H, Peng J, Harberd NP. 2002. Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14, 3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, et al. 2006. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18, 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Asano K, Tsuji H, Kawamura M, Mori H, Kitano H, Ueguchi-Tanaka M, Matsuoka M. 2010. Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 22, 2680–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogendoorn J, Pfeiffer WH, Rajaram S, Gale MD. 1988. Adaptive aspects of dwarfing genes in CIMMYT germplasm. In: Miller TE, Koebner RMD, eds. Proceedings of the Seventh International Wheat Genetics Symposium. Cambridge: Institute of Plant Science Research, 1093–1100 [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. 2001. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13, 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun TP, Hakoshima T. 2008. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463 [DOI] [PubMed] [Google Scholar]
- Pearce S, Saville R, Chandler PM, Wilhelm EP, Boulton MI, Phillips AL, Hedden P, Nicholson P, Thomas SG. 2011. Molecular characterisation of Rht-1 dwarfing genes in hexaploid wheat. Plant Physiology 157, 1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, et al. 1999. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261 [DOI] [PubMed] [Google Scholar]
- Robertson M, Swain SM, Chandler PM, Olszewski NE. 1998. Identification of a negative regulator of gibberellin action, HvSpy, in barley. Plant Cell 10, 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, et al. 2003. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299, 1896–1898 [DOI] [PubMed] [Google Scholar]
- Saville RJ, Gosman N, Burt CJ, Makepeace J, Steed A, Corbitt M, Chandler E, Brown JKM, Boulton MI, Nicholson P. 2012. The ‘Green Revolution’ dwarfing genes play a role in disease resistance in Triticum aestivum and Hordeum vulgare. Journal of Experimental Botany 63, 1271–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, Kato H, Matsuoka M. 2008. Structural basis for gibberellin recognition by its receptor GID1. Nature 456, 520–544 [DOI] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Sakamato T, Fujioka S, Takatsuto S, Yoshida S, Sazuka T, Ashikari M, Matsuoka M. 2006. The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. The Plant Journal 48, 390–402 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Mak PYA, Martínez EC, Sun TP. 1997. The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146, 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Tseng TS, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun TP. 2007. Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiology 143, 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, et al. 2005. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698 [DOI] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EMN, Maier A, Schwechheimer C. 2007. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19, 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Somerville CR. 1995. Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiology 108, 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbang CM, Chandler PM, Smith JJ, Ross JJ. 2004. Auxin from the developing inflorescence is required for the biosynthesis of active gibberellins in barley stems. Plant Physiology 134, 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Kong X, Wan J, et al. 2011. Dominant and pleiotropic effects of a GAI gene in wheat results from lack of interaction between DELLA and GID1. Plant PhysiologyDOI:10.1104/pp.111.185272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Hirai T, Yamamoto E, Kawamura M, Sato T, Kitano H, Matsuoka M, Ueguchi-Tanaka M. 2010. A rice gid1 suppressor mutant reveals that gibberellin is not always required for interaction between its receptor, GID1, and DELLA proteins. Plant Cell 22, 3589–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwar JA, Chandler PM. 1995. α-Amylase production and leaf protein synthesis in a gibberellin-responsive dwarf mutant of ‘Himalaya’ barley (Hordeum vulgare L.). Planta 197, 39–48 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.