Abstract
The effect of iron (Fe) and sulphur (S) deprivation on sulphate uptake and assimilation pathways was investigated in durum wheat by analysing the expression of genes coding for major transporters and enzymes involved in sulphate assimilation and reduction: high-affinity sulphate transporters (TdSultr1.1 and TdSultr1.3), ATP sulphurylase (TdATPSul1 and TdATPSul2), APS reductase (TdAPR), sulphite reductase (TdSiR), O-acetylserine(thiol)lyase (TdOASTL1 and TdOASTL2), and serine acetyltransferase (TdSAT1 and TdSAT2). Further experiments were carried out to detect changes in the activities of these enzymes, together with the evaluation of growth parameters (fresh biomass accumulation, leaf green values, and total S, thiol, and Fe concentrations). Fe shortage in wheat plants under adequate S nutrition resulted in an S deficiency-like response. Most of the genes of the S assimilatory pathway induced by S deprivation (TdATPSul1, TdAPR, TdSir, TdSAT1, and TdSAT2) were also significantly up-regulated after the imposition of the Fe limitation under S-sufficient conditions. However, the differential expression of genes encoding the two high-affinity transporters (TdSultr1.1 and TdSultr1.3) indicates that the mechanisms of sulphate uptake regulation under Fe and S deficiency are different in wheat. Moreover, it was observed that the mRNA level of genes encoding ATPS, APR, and OASTL and the corresponding enzyme activities were often uncoupled in response to Fe and S availability, indicating that most probably their regulation involves a complex interplay of transcriptional, translational, and/or post-translational mechanisms induced by S and/or Fe deficiency.
Key words: Iron, Strategy II, sulphate assimilation, sulphate transporter, sulphur, wheat.
Introduction
Sulphur (S) is an essential nutrient for growth and development of plants and it is required in many metabolites, such as amino acids and proteins (Hesse et al., 2004). Besides biomass production and yield stability (Zhao et al., 1999), in wheat S nutrition affects bread- and pasta-making qualities through its effect on grain protein composition (Lerner et al., 2006). Over recent years, the decrease in atmospheric SO2 emissions and lower S supply through mineral fertilization (McGrath et al., 2002) have led to the occurrence of S deficiency in many European countries, and physiological responses of plants to S deficiency have been recorded (reviewed by Lewandowska and Sirko, 2008).
Plants use inorganic sulphate from the soil as the main source of S. The sulphate uptake into root cells is enabled by a family of transmembrane proton sulphate co-transporters (Hawkesford, 2000; Davidian and Kopriva, 2010), then sulphate can be reduced and assimilated into organic compounds. While roots play an important role in taking up sulphate from the soil, the reduction process is thought to occur mainly in leaves, and most of the enzymes involved in the assimilatory pathway are localized within the plastids (Howarth et al., 2008). The first reaction of the pathway is sulphate activation by the enzyme ATP sulphurylase (ATPS), which forms adenosine phosphosulphate (APS) from ATP and sulphate. Then, APS reductase (APR) reduces APS to produce free sulphite, which is further reduced to sulphide by sulphite reductase and then incorporated into cysteine. The synthesis of cysteine by condensation of O-acetylserine and sulphide, catalysed by O-acetylserine(thiol)lyase (OASTL), represents the final step in the reductive sulphate assimilation pathway (reviewed in Davidian and Kopriva, 2010; Takahashi et al., 2011). Apart from being incorporated directly into proteins, cysteine acts as a donor of reduced S in the synthesis of both methionine, which leads to S-adenosylmethionine (SAM) and S-methylmethionine derivatives, and glutathione.
Beyond its essential role in plant metabolism, S nutrition is also important for iron (Fe) uptake and assimilation. Graminaceous plants, such as maize, barley, and wheat, possess an adaptive mechanism to cope with Fe deficiency, called ‘Strategy II’ (Marschner et al., 1986). In Strategy II plants, members of the mugineic acid family of phytosiderophores (PSs) are secreted by the roots into the rhizosphere, where PSs are able to form stable complexes with cationic micronutrients, such as Fe3+. The Fe3+–PS complex is then taken up by root cells through the action of Yellow Stripe 1 (YS1) proteins (Murata et al., 2006). Mori and Nishizawa (1987) identified the S-containing amino acid methionine as the sole precursor of the mugineic acid family of PSs. Accordingly, in grasses, the importance of adequate S nutrition for Fe assimilation arises from increased demand for reduced S for methionine and, consequently, mugineic acid synthesis induced by Fe deprivation. Indeed, S deficiency resulted in decreased synthesis and secretion of mugineic acids and thus lower tolerance to reduced Fe availability (Astolfi et al., 2006a). Moreover, it is expected that changes in Fe availability would affect the regulation of the S uptake and assimilation pathway. Indeed, the Fe use efficiency in maize (Astolfi et al., 2003), barley (Astolfi et al., 2006a), and durum wheat (Zuchi et al., 2012) increased under adequate S nutrition and was further improved when durum wheat plants were supplied with increased concentrations of S (Zuchi et al., 2012), whereas it declined as S supply was decreased. It is noteworthy that in durum wheat the effect of increased S nutrition seems especially advantageous for plants grown under severe Fe limitation (Zuchi et al., 2012).
Previous studies in barley plants have shown that Fe deficiency affects the partitioning of the reduced S pool between the shoot and the root and can affect the uptake of sulphate in conditions of limited S supply (Astolfi et al., 2006b). It was therefore hypothesized that changes in Fe availability modulate the rate of uptake and assimilation of S in wheat plants and that the effects of Fe availability are related to the plant S status. To test this hypothesis, durum wheat plants were subjected to Fe and S deficiencies and the effects on the S uptake and assimilatory pathways, in particular on the level of transcripts and enzymes encoded by the involved genes, were detected and recorded separately in shoots and roots. It is shown here that Fe limitation in wheat plants under adequate S nutrition produced an S deficiency-like response at the molecular level. Most of the genes of the S assimilatory pathway induced by S deprivation (TdATPSul1, TdAPR, TdSir, TdSAT1, and TdSAT2) were also significantly up-regulated by Fe limitation. However, the differential expression of genes encoding the two high-affinity transporters (TdSultr1.1 and TdSultr1.3) indicated that at least the mechanism of sulphate uptake regulation under Fe and S deficiency might be different. Moreover, the uncoupling of regulation of mRNA levels and activities of ATPS, APR, and OASTL by Fe and S availability indicates that their regulation involves a complex interplay of transcriptional, translational, and/or post-translational mechanisms.
Materials and methods
Growth conditions and sampling
Durum wheat (Triticum durum Desf. cv. Svevo) seeds were germinated on moistened filter paper in darkness at 20 °C. Uniform 3-day-old seedlings were transferred to plastic pots (12 seedlings per pot) filled with 2.2 litres of continuously aerated nutrient solution (NS) (Zhang et al., 1991) at two sulphate levels in the nutrient solution (+S=1.2 and –S=0mM sulphate, optimal supply and severe deficiency, respectively) and two FeIII-EDTA levels (0 and 100 μM). The S-deficient NS was prepared by replacing sulphate salts (K+, Mn2+, Zn2+, and Cu2+) with appropriate amounts of chloride salts (K+, Mn2+, Zn2+, and Cu2+). S application rates for this experiment were chosen on the basis of previous studies. In particular, as adequate supply, the same application rate (1.2mM sulphate) was chosen as for other grasses (Astolfi et al., 2006a; Zuchi et al., 2012). NS was continuously aerated and changed every 2 d. Plants were grown in a climate chamber under 200 μmol m–2 s–1 PAR at leaf level and 14h/10h day/night regime (temperature 26 °C/20 °C; relative humidity 80%). Wheat shoots and roots were sampled on day 11 after sowing.
Identification and cloning of full-length cDNAs of wheat genes involved in sulphur metabolism
The available Arabidopsis sequences of genes coding for high-affinity transporters and enzymes involved in sulphate assimilation and reduction (Supplementary Table S1 available at JXB online) were used as queries in a BLAST search of public databases of wheat expressed sequence tags (ESTs): DFCI wheat gene index database (TaGI, version 12) and NCBI. The identified non-redundant sequences were used as templates for 5’ and 3’ RACE (rapid amplification of cDNA ends) extensions using the 5’/3’ RACE kit from Roche following the manufacturer’s instructions. Total RNA was extracted using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, and its concentration and quality were checked as reported in Paolacci et al. (2009) and described in detail in the Supplementary Appendix S1 at JXB online. RACE products were amplified using 2 μg of a pool of total RNA from roots and shoots of the durum wheat cv Svevo. The 5’ and 3’ RACE products were cloned and validated by sequence analysis; the corresponding full-length cDNAs of each identified wheat gene involved in S metabolism were cloned by reverse transcription–PCR (RT–PCR) using total RNA from roots and shoots of cv. Svevo and specific primers designed on the basis of the 5’- and 3’-untranslated regions (UTRs) (Supplementary Table S2). PCRs were performed using 2 μl of the reverse transcription reaction with the GC-Rich PCR System from Roche following the manufacturer’s instructions.
RACE and full-length cDNA products were cloned into the pGEMT easy plasmid vector (Promega). Sequencing was performed on both strands by the ABI PRISM 377 capillary sequencer (PE Applied Biosystem) using an ABI Prism Dye Terminator sequencing kit (PE Applied Biosystem) and either vector- or sequence-specific primers. Full-length cDNA sequences were deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases; accession numbers are indicated in Table 1. A two-letter code (Td=Triticum durum) followed by the suffix of the corresponding transporters or enzyme involved in S metabolism was assigned to each sequence. The predicted protein sequences were analysed by searching for conserved motifs in CDD, Pfam HMMs, InterPro, and SMART databases; their subcellular locations were predicted by Target P1.1 and ChloroP 1.1.
Table 1.
Characteristics of the full-length cDNA sequences of wheat genes involved in sulphur metabolism cloned in this study.
| Gene | Accession number | Full-length cDNA | TC sequence (DFCI wheat Gene index) | NCBI wheat equence (GenBank) | Putative orthologous Arabidopsis genes | Predicted subcellular localization | Protein identity (%) | ||
|---|---|---|---|---|---|---|---|---|---|
| 5’ UTR (bp) | 3’ UTR (bp) | ORF (bp) | |||||||
| TdSultr1.1 | JX896648 | 38 | 105 | 1989 | TC387319 | AJ512821 | SULTR1;2 (At1g78000) | 71.1 | |
| SULTR1;1 (At4g08620) | 69.3 | ||||||||
| SULTR1;3 (At1g22150) | 67.9 | ||||||||
| TdSultr1.3 | JX896649 | 55 | 88 | 1971 | TC393732 | BT009249 | SULTR1;2 (At1g78000) | 72.2 | |
| TC430836 | SULTR1;3 (At1g22150) | 69.8 | |||||||
| SULTR1;1 (At4g08620) | 68.8 | ||||||||
| TdATPSul1 | JX896650 | 29 | 65 | 1449 | TC375445 | AK335711 | APS2 (At1g19920) | Plastids | 73.8 |
| APS1 (At3g22890) | 69.8 | ||||||||
| APS3 (At4g14680) | 69.5 | ||||||||
| APS4 (At5g43780) | 68.1 | ||||||||
| TdATPSul2 | JX896651 | 30 | 199 | 1410 | TC399245 | AK334753 | APS3 (At4g14680) | Plastids | 72.4 |
| APS4 (At5g43780) | 72.3 | ||||||||
| APS1 (At3g22890) | 72.0 | ||||||||
| APS2 (At1g19920) | 65.2 | ||||||||
| TdAPR | JX896652 | 20 | 122 | 1386 | TC383251 | BT009195 | APR2 (At1g62180) | Plastids | 73.5 |
| TC416397 | APR1 (At4g04610) | 73.0 | |||||||
| APR3 (At4g21990) | 71.6 | ||||||||
| TdSir | JX896653 | 85 | 120 | 1908 | TC382786 | AK333462 | SIR (At5g04590) | Plastids | 76.0 |
| TdOASTL1 | JX896654 | 17 | 105 | 1179 | TC397724 | – | OASB (At2g43750) | Plastids | 76.8 |
| OASC (At3g59760) | 73.6 | ||||||||
| OASA1 (At4g14880) | 71.7 | ||||||||
| TdOASTL2 | JX896655 | 41 | 49 | 978 | TC369358 | D13153 | OASA1 (At4g14880) | Cytosol | 81.6 |
| BT009041 | OASB (At2g43750) | 72.9 | |||||||
| OASC (At3g59760) | 72.9 | ||||||||
| TdSAT1 | JX896656 | 35 | 164 | 954 | TC444380 | BT008919 | SAT5 (At5g56760) | Cytosol | 72.0 |
| TdSAT2 | JX896657 | 53 | 142 | 1014 | TC426852 | – | SAT2 (At2g17640) | Cytosol | 67.0 |
| SAT4 (At4g35640) | 64.0 | ||||||||
Enzyme extraction and assays
Frozen tissue (~1g fresh weight) was ground to a fine powder in a pre-chilled mortar under liquid N2. Cold extraction buffer containing 50mM HEPES-KOH (pH 7.4), 5mM MgCl2, 1mM EDTA, 10% (v/v) glycerol, 0.1% (v/v) Triton X-100, 5mM dithiothreitol (DTT), 1mM phenylmethylsulphonyl fluoride (PMSF), and 1% (w/v) polyvinylpyrrolidone (PVP) was added in a ratio of 1:7 (w/v) for shoots and of 1:3 (w/v) for roots. The homogenate was filtered through four layers of cheesecloth and then centrifuged at 1000 g for 5min at 4 °C. The resulting supernatant was desalted at 4 °C on a Sephadex G-25 column (PD-10, Pharmacia, Uppsala, Sweden) pre-equilibrated with extraction buffer minus Triton X-100. The desalted extract was centrifuged at 30 000 g for 5min at 4 °C. The supernatant was divided into aliquots which were frozen in liquid N2 and stored at –80 °C until analysis.
ATP sulphurylase (ATPS; EC 2.7.7.4) activity was assayed by the bioluminescence technique (Schmutz and Brunold, 1982; Zuchi et al., 2012).
APS reductase activity was determined as the production of [35S]sulphite, assayed as acid volatile radioactivity formed in the presence of [35S]APS and DTT as reductant (Lee et al., 2011). O-Acetylserine(thiol)lyase (OASTL; EC 4.2.99.8) was determined following the procedures described by Zuchi et al. (2012).
Other biochemical and physiological measurements and statistics
The concentration of chlorophyll per unit area was estimated in attached leaves by a portable Chlorophyll Meter, model SPAD-502 (Soil and Plant Analysis Development) (Minolta Co. Ltd, Osaka, Japan) using the first fully expanded leaf from the top of the plant.
To determine total S concentration, 1g of each shoot or root sample was dried at 105 °C and ashed in a muffle furnace at 600 °C. The ashes were dissolved in 10ml of 3 N HCl and filtered through Whatman No. 42 paper. After mixing with 2.5ml of sieved mixture with 2.5ml of 2% (w/v) BaCl2, the amount of BaSO4 precipitate was determined turbidimetrically (Bardsley and Lancaster, 1962).
Fe concentration in shoots and roots was determined after dry ashing (500 °C) of plant tissue and 1:30 HCl extraction by atomic absorption spectrometry (AAS).
Water soluble non-protein sulphydryl (SH) compounds were determined colorimetrically with DTNB [5,5’-dithiobis-(2-nitrobenzoic acid)] following the procedure described in Astolfi et al. (2003).
Protein concentration was determined according to Bradford (1976) using bovine serum albumin (BSA) as standard.
Each reported value represents the mean ±SD of data from four independent experiments on three biological replicates per experiment. Statistical analyses of data were carried out by analysis of variance (ANOVA) tests with the GraphPad InStat Program (version 3.06). Significant differences were established by post-hoc comparisons (Tukey’s HSD test) at P < 0.001 or P < 0.01.
Expression analyses
The expression of the cloned cDNAs was analysed by quantitative real-time RT–PCR (qRT–PCR) according to Paolacci et al. (2009) and described in detail in the Supplementary Appendix S1 at JXB online. Three biological replicates, resulting from three different RNA extractions, reverse transcription, and qRT–PCRs were used in quantification analysis; three technical replicates were analysed for each biological replicate. The complete set of primer pairs used for qRT–PCR analyses is reported in Supplementary Table S3. The relative expression levels of the analysed genes were computed considering three data sets obtained by the different groupings of the analysed samples: (i) eight samples relative to roots and shoots of seedlings grown with or without 100 μM FeIII-EDTA at two S levels in the nutrient solution (+S=1.2 and –S=0mM sulphate); (ii) four samples including only the shoots of seedlings for the four treatments; and (iii) four samples comprising only the roots of seedlings for the four treatments. The statistical significance of the differences was evaluated by one-way ANOVA followed by Tukey’s test. Different letters denote significant differences at P ≤ 0.01.
Results
Plant growth parameters
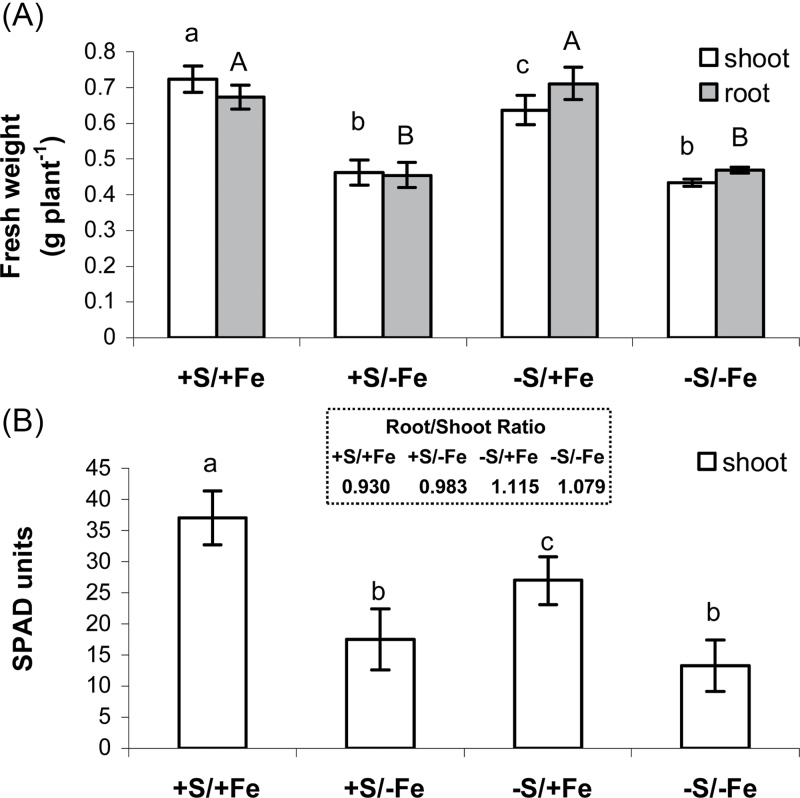
At the end of the experiment, the plants reached the same developmental stage. Plants grown for 11 d on S-free NS reached a shoot biomass (fresh weight basis) 10% lower than that of control plants (Fig. 1A). In contrast, the root biomass was unaffected by S deprivation (Fig. 1A). This adaptive behaviour resulted in a significantly increased root/shoot ratio (+20%), as shown in Fig. 1 (insert). On the other hand, plant growth was significantly more affected by Fe deficiency, which caused a 30% reduction in both shoot and root growth (Fig. 1A).
Fig. 1.
Shoot (white bars) and root (grey bars) fresh weight (A) and chlorosis status (B) of wheat plants grown for 11 d in NS at 0 (–Fe) and 100 (+Fe) μM FeIII-EDTA and under two S concentrations in NS (0 and 1.2mM, –S and +S, respectively). SPAD readings were performed on the first fully expanded leaf from the top of the plant. Insert: root/shoot ratio. Data are means ±SD of four independent replications run in triplicate. Significant differences between samples are indicated by different letters: different upper case letters indicate significant differences among the growth conditions in roots (P < 0.01) (n=4); different lower case letters indicate significant differences among the growth conditions in shoots (P <0.01) (n=4).
Changes in chlorophyll content were also examined to detect stress-induced effects on wheat plants. The leaf chlorophyll concentration of plants grown without S in NS gradually decreased from day 7 onward (the first possible measurement being on day 4) and at day 11 it was at ~70% of the content in control plants, whereas, when plants were grown without Fe, chlorophyll concentration decreased since day 4 and at day 11 it was at 47–49% of the content of S-sufficient and S-deficient plants, respectively (Fig. 1B).
Fe concentration in shoots and roots
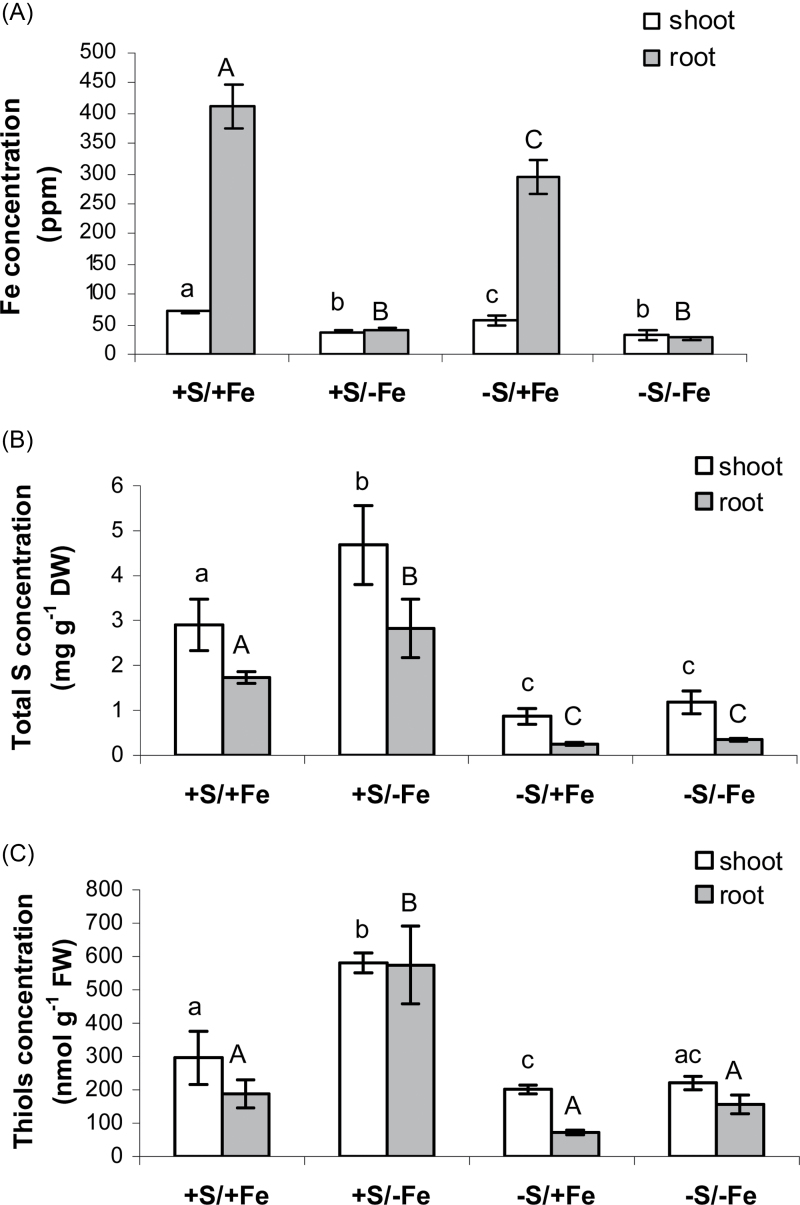
The pattern of Fe accumulation in vegetative plant tissues followed the pattern of chlorophyll measurements. After 11 d of deprivation, the Fe levels were markedly decreased to 53% and 58% of the Fe-sufficient control value in +S and –S shoots, respectively (Fig. 2A). However, in roots from plants exposed to Fe deficiency, Fe concentrations decreased considerably more during deprivation, reaching only 10% of the control value in both +S and –S conditions (Fig. 2A).
Fig. 2.
Iron (ppm) (A), total S (mg g–1 DW) (B), and non-protein thiol (nmol g–1 FW) (C) concentration in shoots (white bars) and roots (grey bars) of wheat plants grown for 11 d in NS at 0 (–Fe) and 100 (+Fe) μM FeIII-EDTA and under two S concentrations in the NS (0 and 1.2mM, –S and +S, respectively). Data are means ±SD of four independent replications run in triplicate. Statistics are as in Fig. 1.
Interestingly, also in S-deficient plants the concentrations of Fe decreased significantly in both shoot and root, although these plants did not experience Fe deficiency (Fig. 2A). This reduction corresponded to a 20% decrease in shoot Fe concentrations (Fig. 2A) and 30% in root concentrations (Fig. 2A). This observation confirms the initial hypothesis and could be explained by a reduced capability of S-deficient plants to produce PSs necessary for Fe uptake.
Plant concentration of total sulphur and non-protein thiols
As expected, S deficiency reduced the accumulation of total S by ~80% in both shoots and roots (Fig. 2B). Interestingly, the highest concentrations of S were observed in shoots and roots of S-sufficient plants exposed to Fe deprivation (about +60% compared with the Fe-sufficient controls) (Fig. 2B).
The concentration of thiols in shoots and roots closely followed the pattern of total S accumulation, with the highest concentrations again being observed in +S/–Fe plants (Fig. 2C). In particular, the thiol concentrations found in Fe-deficient shoots were 2-fold higher than those found in Fe-sufficient shoots, whereas in roots a >3-fold increase compared with control plants was observed (Fig. 2C). The higher increase in roots could be related either to translocation of thiols from shoots to roots or to increased S assimilation rates in roots. Previous studies have shown that the S partitioning from the shoots to the roots through the transport of thiols, mainly cysteine, seems to be one of the adaptive strategies used to cope with Fe deprivation (Astolfi et al., 2006b).
Activities of enzymes involved in sulphate assimilation
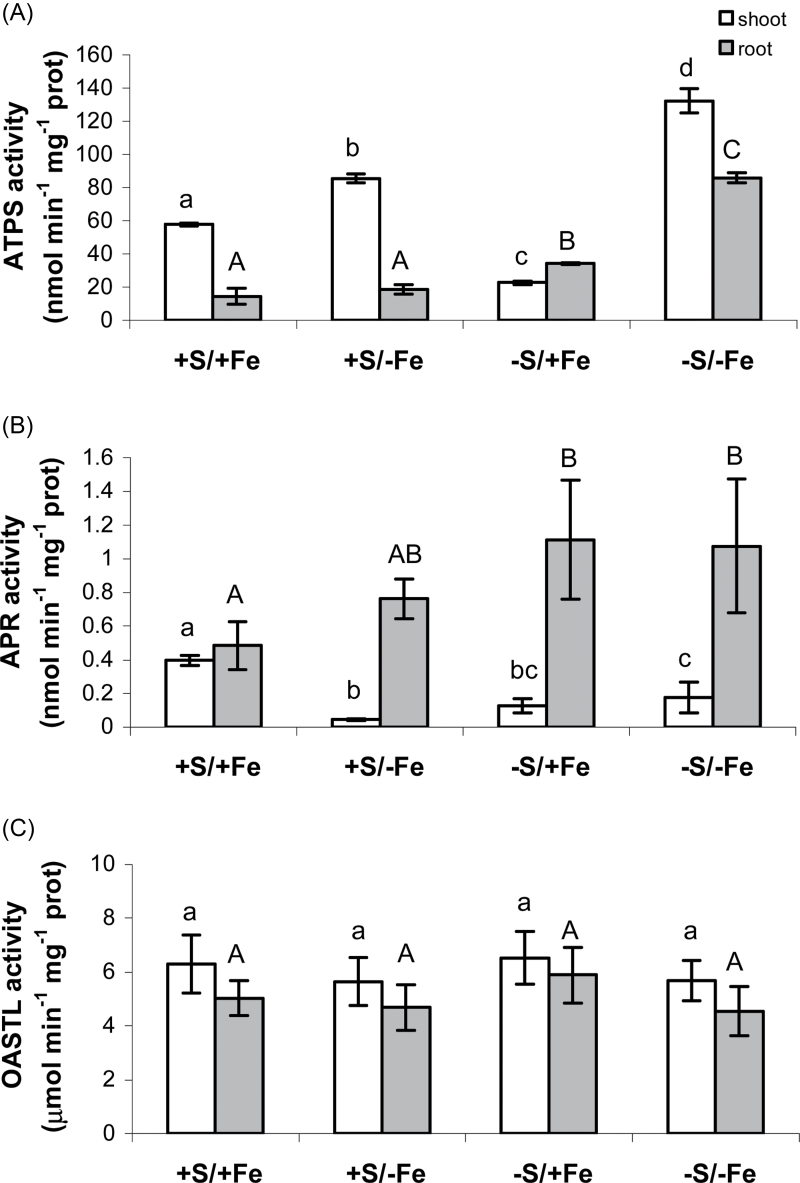
To determine how S and Fe deprivation affects sulphate assimilation, activities of key enzymes of the pathway were measured. The activity of ATPS in roots was generally lower than that found in shoots, except for plants exposed to S deprivation and normal Fe supply. At normal Fe, S deprivation resulted in a decrease in ATPS activity in shoot but an increase in the roots (Fig. 3A). Fe deficiency increased ATPS activity in shoots, by ~50% in S-sufficient plants, but almost 5-fold in S-deficient shoots (Fig. 3A). In roots, the stimulatory effect of Fe deficiency on ATPS was detected only during simultaneous S deficiency (Fig. 3A).
Fig. 3.
Changes in shoot (white bars) and root (grey bars) ATPS (nmol ATP min–1 mg–1 protein) (A), APR (nmol min–1 mg–1 protein) (B), and OASTL (μmol min–1 mg–1 protein) (C) activities of wheat plants grown for 11 d in NS at 0 (–e) and 100 (+Fe) μM FeIII-EDTA and under two S concentrations in the NS (0 and 1.2mM, –S and +S, respectively). Data are means ±SD of four independent replications run in triplicate. Statistics are as in Fig. 1.
Unexpectedly, in contrast to ATPS, APR activity was higher in roots than in shoots of wheat seedlings, undergoing nutrient deficiency (Fig. 3B). APR activity was strongly reduced in shoots by both S and Fe deprivation, whereas withdrawing S and Fe from the NS resulted in an increase in the root APR activity (Fig. 3B). In particular, root APR activity was 60% higher under Fe stress and 2-fold higher under S deprivation or combined S and Fe stress with respect to the control (+S+Fe) roots (Fig. 3B).
On the other hand, levels of OASTL activity measured in shoots and roots did not change significantly after 11 d of both S and Fe deprivation (Fig. 3C).
Expression of durum wheat genes coding for high-affinity transporters and enzymes involved in sulphate assimilation and reduction
In order to analyse the regulation of mRNA levels for genes of sulphate assimilation in durum wheat, the corresponding cDNAs were cloned using BLAST search of the DFCI wheat gene index (TaGI, version 12) and NCBI databases using Arabidopsis sequences (Supplementary Table S1 at JXB online). Ten sequences retrieved from these two databases referred to distinct non-homeologous genes and were used to obtain full-length cDNAs by 5’ and 3’ RACE (Table 1). The durum wheat genes showed 68–74% identity to corresponding Arabidopsis genes on the amino acid level and the same predicted subcellular localizations (Table 1). Their characteristics are described in detail in Supplementary Appendix S2 at JXB online.
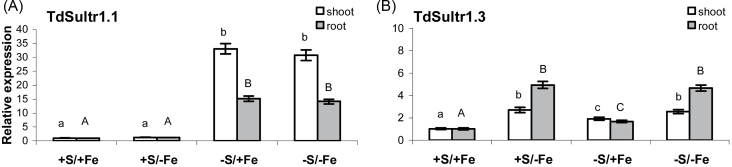
The two high-affinity sulphate transporters TdSultr1.1 and TdSultr1.3 showed very different expression patterns (Fig. 4; Supplmentary Fig. S1 at JXB online). In control plants, TdSultr1.1 was preferentially expressed in roots, with a transcription level ~13 times higher than in shoots; while the transcript of TdSult1.3 was ~15 times higher in shoots than in roots (Supplementary Fig. S1). When plants were grown on S-deprived NS, the transcript level of TdSultr1.1 was ~33 and 15 times increased in shoots and roots, respectively, whereas its expression was unaffected by Fe deprivation in both tissues (Fig. 4A). In contrast, TdSultr1.3 was significantly up-regulated in shoots and roots by Fe and/or S deprivation, with the highest expression level detected in Fe-deficient plants (Fig. 4B).
Fig. 4.
Relative expression levels by quantitative real-time RT–PCR (qRT–PCR) of the genes encoding durum wheat high-affinity sulphate transporters (TdSultr1.1 and TdSultr1.3) in shoots (white bars) and roots (grey bars) of wheat plants grown for 11 d in NS at 0 (–Fe) and 100 (+Fe) μM FeIII-EDTA and under two S concentrations in the NS (0 and 1.2mM, –S and +S, respectively). The 12 cDNA pools (three biological replicates and four seedling samples for each of the two tissues analysed) were tested in triplicate and normalized using the geometric average of the relative expression of the two reference genes ubiquinol-cytochrome c reductase (Unigene Cluster Ta4045; DFCI contig TC350975) and RNase L inhibitor-like protein (Unigene cluster Ta2776; DFCI contig TC278756). Relative expression levels of each sulphate transporter gene were referred to a calibrator set to the value 1, which was represented by the treatment with the lowest expression. Normalized values of relative expression of each gene are given as the average ±SD. Statistical significance of the differences was evaluated by one-way ANOVA followed by Tukey’s test. Different letters denote significant differences at P ≤ 0.01.
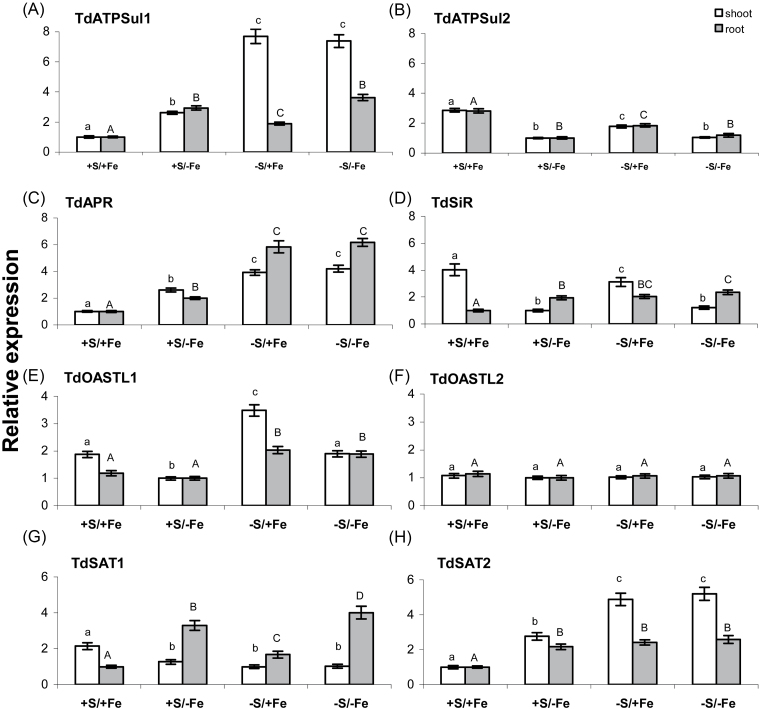
The relative expression of TdATPSul1 was about twice as high in roots than in shoots from control plants (Supplementary Fig. S2 at JXB online) and S deprivation induced significant changes in its expression in both tissues, with the highest expression level detected in shoots (~7 times higher than in control plants) (Fig. 5A). Interestingly, the expression of TdATPSul1 was also significantly up-regulated by Fe deprivation in roots and shoots of plants grown in a non-limiting S concentration in the NS, but in this case the expression level was higher in roots than in shoots (Fig. 5A). The expression pattern of the gene coding for the other durum wheat ATPS isoform, TdATPSul2, was very different; in fact its transcription level was ~28 times higher in shoots than in roots of control plants (Supplementary Fig. S2) and its expression was significantly down-regulated by S and/or Fe deprivation in shoots and roots, with the lowest transcription level observed in Fe-deficient plants (Fig. 5B).
Fig. 5.
Relative expression levels by qRT–PCR of the genes coding for enzymes involved in sulphate assimilation and reduction: ATP sulphurylase (TdATPSul1 and TdATPSul2), APS reductase (TdAPR), sulphite reductase (TdSiR), O-acetylserine(thiol)lyase (TdOASTL1 and TdOASTL2), and serine acetyltransferase (TdSAT1 and TdSAT2) in shoots (white bars) and roots (grey bars) of wheat plants grown for 11 d in NS at 0 (–Fe) and 100 (+Fe) μM FeIII-EDTA and under two S concentrations in the NS (0 and 1.2mM, –S and +S, respectively). The 12 cDNA pools (three biological replicates and four seedling samples for each of the two tissues analysed) were tested in triplicate and normalized using the geometric average of the relative expression of the two reference genes ubiquinol-cytochrome c reductase (Unigene Cluster Ta4045; DFCI contig TC350975) and RNase L inhibitor-like protein (Unigene cluster Ta2776; DFCI contig TC278756). Relative expression levels of the eight genes were referred to a calibrator set to the value 1, which was represented by the treatment with the lowest expression. Normalized values of relative expression of the eight genes are given as the average ±SD. Statistical significance of the differences was evaluated by one-way ANOVA followed by Tukey’s test. Different letters denote significant differences at P ≤ 0.01.
The expression of TdAPR was higher in shoots than in roots (Supplementary Fig. S3 at JXB online) and the S and/or Fe deprivation resulted in a significant increase in the relative amount of its transcripts in both tissues; however, the highest effect was observed in S-deficient plants, rather than in Fe-deficient plants (Fig. 5C). In contrast, the expression pattern of TdSiR was very different in roots and in shoots, although the relative amount of its transcripts in both tissues of control plants was similar (Supplementary Fig. S3). The transcript level of TdSiR was, in fact, significantly down-regulated by S and/or Fe deprivation in shoots and up-regulated in roots (Fig. 5D).
The two durum wheat genes coding for plastidic and cytosolic OASTL enzymes, TdOASTL1 and TdOASTL2 (Table 1; Supplementary Appendix 2 at JXB online), were preferentially expressed in shoots (transcription level 12 times higher in control plants) (Supplementary Fig. S4), although their expression was differentially regulated by S and/or Fe deprivation (Fig. 5E, F). When plants were grown on S-deprived NS, the mRNA level of TdOASTL1 increased ~2-fold in both roots and shoots, whereas its expression was slightly but significantly down-regulated by Fe deprivation in shoots (Fig. 5E). In contrast, the expression of the gene coding for the cytosolic isoform, TdOASTL2, was unaffected in both roots and shoots by the S and Fe availability (Fig. 5F). Finally, the expression pattern and the transcriptional level of the two genes coding for the two cytosolic SAT enzymes were very different. TdSAT1 was preferentially expressed in shoots, with a transcription level ~7 times higher than in roots of control plants, whereas the relative mRNA amount of TdSAT2 was similar in both tissues of the same plants (Supplementary Fig. S5). However, the expression level of TdSAT1 was ~10- and 60-fold higher than that of TdSAT2 in roots and shoots, respectively (Supplementary Fig. S5, and results not shown). Moreover, TdSAT1 was significantly down-regulated in shoots by Fe and/or S deprivation and up-regulated in roots in Fe- and S-deficient plants, with the highest effect observed for the Fe deficiency (Fig. 5G). In contrast, S and/or Fe deprivation induced a significant increment in the relative amount of transcripts of TdSAT2 in both roots and shoots (Fig. 5H).
Discussion
The relationship between S availability in nutrient solution and the ability of a plant to take up Fe has been observed in a range of Strategy II species, including maize (Astolfi et al., 2003), barley (Astolfi et al., 2006a), and durum wheat (Zuchi et al., 2012). In grasses, the importance of an adequate S nutrition for Fe assimilation has been hypothesized to arise from Fe deprivation-induced increased demand for reduced S for methionine and, consequently, synthesis of mugineic acids. Then changes in Fe availability could affect the regulation of the S uptake and assimilation pathway. Moreover, since knowledge of whether and how Fe plays a role in the regulation of S acquisition and assimilation could help to plan possible strategies to enhance the uptake and storage of Fe in wheat, in this study the similarities and differences of the responses to S and Fe deprivation and their effect on sulphate uptake and assimilation were investigated in durum wheat seedlings.
Response to S deficiency in durum wheat
The present results on durum wheat are consistent with studies in other species reporting that plants respond to a limited S supply by increasing the expression of genes encoding components of the uptake and assimilation pathway and by increasing the root/shoot ratio and lateral root proliferation (reviewed in Lewandowska and Sirko, 2008; Takahashi et al., 2011). As expected, complete removal of S from NS resulted in a significant increase in the root/shoot ratio (Hopkins et al., 2004; Astolfi et al., 2006b; Zuchi et al., 2012) (inset in Fig. 1). Leaves from S-deprived plants appeared chlorotic at harvest, as detected by a decrease in SPAD readings (Fig. 1B). The visual symptoms were accompanied by a strong decrease in both total Fe and S concentration in the S-deprived tissues (Fig. 2A, B).
As widely reported, the control of the S assimilatory pathway occurs primarily at the step of sulphate uptake and APS reduction. Indeed, the transcript levels of sulphate transporters (mainly high-affinity sulphate transporters of group I) and APS reductase are regulated by sulphate availability and well correlated with the fluxes through the sulphate assimilation pathway (Scheerer et al., 2010). Accordingly, in the present study the most significant increment of transcript levels in response to S deprivation was detected for TdSultr1.1 and TdAPR in both roots and shoots (Figs 4A, 5C). The up-regulation of TdSultr1.1 by S deprivation in roots indicates an increase in the sulphate uptake capacity, whereas its elevated expression in shoots is probably an indication of increased sulphate re-mobilization and transport within the plant. The spatial expression patterns of the homologous gene from common wheat (TaeSultr1.1) using in situ hybridization (Buchner et al., 2010) suggest, in fact, a general function for high-affinity uptake not only for the initial uptake of sulphate by the roots, as proposed for SULTR1.1 and SULTR1.2 in Arabidopsis (Barberon et al., 2008), but also for a more widespread uptake and distribution into different cell types in multiple tissues, which include parenchymal and vascular tissues. The elevated expression of TdSultr1.1 in shoots could be related to an increased sulphate re-mobilization from the vacuolar sulphate pool and to its interorgan transport via phloem, including its re-distribution from shoots to roots, as observed in Brassica (Abdallah et al., 2010) and poplar (Honsel et al., 2012), to support the enhanced root growth upon S depletion. This process also requires the expression of the sulphate transporters involved in vacuolar efflux (group 4 transporters) and in phloem loading. In common wheat, the increase in the transcript level of TaeSultr1.1 observed in shoots in response to S deprivation is accompanied by a parallel increased expression of TaeSultr4.1, a putative vacuolar sulphate transporter (Buchner et al., 2010). These results could indicate an enhanced phloem loading capacity of sulphate in combination with increased sulphate efflux from the vacuole. In Arabidopsis, a third high-affinity sulphate transporter, SULTR1.3, which is significantly induced in roots and leaves in response to S deprivation, has a function in the loading of sulphate into phloem, facilitating the re-translocation of S sources in plants (Yoshimoto et al., 2003). Although it is difficult to identify the strictly orthologous genes coding for high-affinity transporters between wheat and Arabidopsis (Table 1; Supplementary Appendix S2 at JXB online), it is interesting to note that the other wheat high-affinity sulphate transporter TdSultr1.3, preferentially expressed in shoots, was only weakly induced upon S deprivation (Fig. 4B), indicating that the most responsive gene coding for the high-affinity transporter to the sulphate availability is TdSultr1.1.
The up-regulation at the transcriptional level of TdAPR induced by S deprivation in roots and shoots (Fig. 5C) would probably suggest an increased sulphate reduction capacity in both tissues. However, it was observed that, although the APR activity was significantly higher in roots during S deficiency, it was strongly reduced in shoots (Fig. 3B), indicating that responses of the mRNA level and enzyme activity to S stress were uncoupled in this tissue and that APR could be differentially regulated in wheat shoots and roots by unknown translational and/or post-translational mechanisms in S deficiency conditions.
The genes coding for the two ATPS isoforms, TdATPSul1 and TdATPSul2, showed different expression responses to S depletion; TdATPSul1 was significantly up-regulated in both shoots and roots, with the highest expression level detected in shoots, whereas TdATPSul2 was slightly but significantly down-regulated in both tissues (Fig. 5A, B). Also the transcript levels of the genes coding for the four Arabidopsis ATPS isoforms showed different responses to S deprivation. APS1 mRNA levels have been reported either to decrease slightly (Liang et al., 2010) or to not be affected by sulphate deficiency (Kawashima et al., 2011), whereas the expression of APS4 and APS3 was strongly down- and up-regulated, respectively (Liang et al., 2010; Kawashima et al., 2011). On the other hand, results for APS2, coding for the putatively cytosolic isoform, were inconsistent between different studies (Logan et al., 1996; Takahashi et al., 1997; Kawashima et al., 2011). APS1, APS3, and APS4 were shown to be post-transcriptionally regulated by miR395, a microRNA which is induced upon S depletion (Liang et al., 2010; Kawashima et al., 2011). However, a different cellular or subcellular localization of miR395 and its targets can prevent the post-transcriptional regulation and might result in a discrepancy between gene expression and enzyme activity of ATPS (Kawashima et al., 2011). For instance, Kawashima et al. (2011) showed that despite strong down-regulation of the miR395-targeted APS4, total ATPS activity is only moderately affected by sulphate deprivation. Most probably, the miR395-induced cleavage of APS4 could be counteracted by the increased transcription of the other plastidic isoform APS3 so that the level of total enzymatic ATPS activity is not significantly altered during sulphate deficiency. The predicted amino acid sequence of TdATPSul2 was most similar to that of the three chloroplast isoforms APS1, APS3, and APS4, whereas that of TdATPSul1 was more closely related to the protein sequence of APS2 that, despite the presence of a putative plastid-argeting peptide, has been proposed to be the only cytosolic ATPS in Arabidopsis (Hatzfeld et al., 2000) (Table 1; Supplementary Appendix 2 at JXB online). Thus, on the basis of the sequence homology, it may be speculated that the down-regulation of TdATSul2 observed in this study might be due to a post-transcriptional process mediated by miR395. Expression analysis of miR395 in response to S deprivation in wheat, bioinformatic prediction of its target wheat genes, and their validation by RML 5’ RACE (Llave et al., 2002) will help to address this question in future studies and, in particular, to elucidate whether the mechanism of miR395 regulation during S deficiency is also conserved in wheat. As TdATPSul2 expression was much higher in shoots than in roots, the action of miR395 might have a higher impact in this organ. Indeed, the present results showed that the total ATPS activity in shoots was strongly reduced (–60%) by imposed S deficiency, while it was significantly increased (>2-fold higher than the S-sufficient control) in roots (Fig. 3A). Thus the increase in ATPS activity is probably due to increased expression of TdATPSul1, which may be induced to compensate the reduction in TdATPSul2 levels, and is consistent with increased APS3 levels in S-starved Arabidopsis roots (Kawashima et al., 2011).
Another potentially rate-limiting step in the S assimilatory pathway is the synthesis of cysteine. The activities of the two enzymes involved in cysteine synthesis, SAT and OASTL, are however strongly regulated by protein–protein interactions in the multienzyme complex of cysteine synthase, although transcriptional regulation also takes place (Droux, 2003; Kawashima et al., 2005). However, SAT is strongly activated by association with OASTL, which is inactive and has only a regulatory role in the complex (Droux et al., 1998). The interaction of the two enzymes is strongly affected by the concentration of OAS, which dissociates the complex, and by sulphide which, acting in the opposite manner, strengthens the binding (Berkowitz et al., 2002; Wirtz and Hell, 2006). In the regulation of cysteine synthesis, SAT seems to play the most important role since the cysteine synthesis might be limited by the availability of OAS rather than sulphide (Wirtz et al., 2004) and the cellular activity of OASTL exceeds that of SAT, although the OASTL/SAT activity ratio differs with the compartment (Heeg et al., 2008). In the present experiment, the transcription level of TdSAT2 was strongly increased in both roots and shoots under S-deprived conditions, whereas TdSAT1, encoding the other cytosolic wheat SAT, showed a tissue-specific response to S deficiency; in fact it was down-regulated in shoots and slightly, although significantly, up-regulated in roots (Fig. 5G). Remarkably, out of the five Arabidopsis SAT genes, only SAT2 and SAT4, the two putative orthologous genes to TdSAT2 (Table 1; Supplementary Appendix 2 at JXB online), were induced by S deprivation, indicating a specific role for these genes in response to S availability (Kawashima et al., 2005). It is known that OAS levels increase by S deprivation and that the intermediate of cysteine biosynthesis is probably an important positive regulator of the pathway at the transcriptional level during S deficiency (Ohkama-Otsu et al., 2004; Kawashima et al., 2005). It may be speculated that the induced expression of TdSAT2 in both roots and shoots during S depletion might contribute to the increased level of OAS and, as OAS positively influences sulphate assimilation, it might improve sulphate uptake and reduction capacities and ensure cysteine biosynthesis under S deprivation. With respect to the two OASTL genes analysed in this study, the transcription level of TdOASTL1, coding for a plastidic isoform (Table 1; Supplementary Appendix S2), was slightly but significantly up-regulated by S deprivation in both roots and shoots, whereas the expression of the gene coding for the cytosolic isoform, TdOASTL2, was unaffected in both tissues. Similarly, only slight differences (~1.5-fold) in the mRNA level of a plastidic OASTL isoform were observed in spinach cell culture under S deprivation (Takahashi and Saito, 1996). However, in the present study, no significant alteration was observed upon S deprivation in the level of OASTL activity in both roots and shoots. These results are in agreement with previous studies showing that OASTL activity is not significantly affected by S deprivation in plants (Krueger et al., 2010) and indicate that the change in mRNA level of an OASTL isoform may not necessarily be translated into a change in the OASTL enzyme activity.
Response of S assimilation to Fe deficiency
Wheat belongs to Strategy II plants and therefore copes with Fe deficiency by the secretion of PSs, and by the uptake of Fe–PS complexes (reviewed in Kobayashi et al., 2010; Kobayashi and Nishizawa, 2012). PSs are derived from nicotianamine, whose precursors are methionine and SAM, and the methionine cycle efficiently supplies SAM for PS production (Kobayashi et al., 2010). The genes encoding key enzymes for PS synthesis, the methionine cycle, and transport of the chelated Fe–PS complexes into roots have been isolated from grasses such as barley and rice, and their expression is strongly induced by Fe deficiency (Nagasaka et al., 2009; Kobayashi et al., 2010; Kobayashi and Nishizawa, 2012). In addition to the well documented up-regulation of the genes involved directly in the synthesis of PSs, a general relationship between sulphate assimilation, methionine biosynthesis, and increased production of PSs in response to Fe deficiency may exist, but little research has been focused on this aspect so far. As PSs represent 1–2% of the root dry weight of Fe-deficient barley and durum wheat plants (Nagasaka et al., 2009; Zuchi et al., 2012), increased methionine synthesis may be essential for the proper functioning of the methionine cycle under Fe-deficient conditions.
Therefore, the large amount of PSs produced in Fe-deficient plants may represent an additional requirement for reduced S, that can be somewhat temporary but possibly significant, generating a typical demand-driven coordinated transcriptional regulation of genes involved in sulphate uptake and assimilation, somehow similar to that observed in S-deprived plants. Accordingly, in this study, most of the genes of the S assimilatory pathway induced by S deprivation (TdATPSul1, TdAPR, TdSir, TdSAT1, and TdSAT2) were also significantly up-regulated after the imposition of the Fe shortage under adequate S nutrition (Figs 4, 5), indicating a probably increased rate of sulphate reduction and assimilation to cope with Fe deficiency. Moreover, the present study demonstrates that the need for maintaining a high rate of methionine synthesis to support PS production under Fe deficiency may be met by increasing the sulphate uptake capacity of the roots, probably by the up-regulation of TdSultr1.3, a gene encoding a putative high-affinity transporter (Fig. 4B). This was supported by Fe-deficient roots showing a higher level of total S in S-sufficient conditions (Fig. 2B). The regulation of sulphate uptake may thus represent the first step of the adaptive process required to ensure an adequate S supply to meet the increased demand for methionine in order to sustain an increased production of PSs. Interestingly, Fe deprivation in the growth medium also resulted in higher accumulation of total S in shoots of wheat plants (Fig. 2B), indicating that both S uptake and translocation were enhanced by Fe deprivation. On the other hand, the up-regulation of TdSultr1.3 in shoots of Fe-deficient plants suggests that this high-affinity transporter may also be involved in the modulation of sulphate fluxes throughout the whole plant during Fe deprivation. It is worth noting that the expression of TdSultr1.1, strongly up-regulated by S deprivation, was unaffected in Fe-deprived plants, indicating that the mechanisms of sulphate uptake regulation under Fe and S deficiency are different.
A greater availability of S in Fe-deficient wheat plants resulted in an increased accumulation of thiol compounds, although different S supply significantly influenced the root and shoot concentrations of non-protein thiols (Fig. 2C). This effect is most probably achieved in two different ways: (i) by an enhanced uptake (up-regulation of TdSultr1.3 in roots) and assimilation (increased ATPS activity in shoots) of sulphate and by translocation of thiols from shoot to root, wherein they can be easily utilized as source of reduced S for the synthesis of methionine and its derivatives (PSs) (Nakanishi et al., 1999); and (ii) by sustained sulphate reduction in roots, as APR activity was reduced by Fe deficiency in shoots but not in roots. The decrease in APR activity is independent from the regulation of transcription levels and can be explained by the presence of the Fe–S cluster as a cofactor of this enzyme (Kopriva et al., 2001). This reduced availability of Fe for synthesis of the Fe–S cluster probably limits APR activity in the shoots, which is at least partly compensated by the increase in ATPS, which is not dependent on the cofactor.
Taking into account the wide variety of soils and environmental conditions experienced by plants, it is plausible that they have evolved multiple signal pathways and mechanisms of regulation modulating S metabolism in response to both S requirement and soil sulphate level. However, the nature of these signals and the mechanisms of regulation involved need to be further investigated, since only indirect evidence has been obtained in a few studies. In this study, it was demonstrated that fluctuations in Fe availability other than sulphate are translated into modulation of plant sulphate metabolism at both the transcriptional and physiological level by means of both separate and shared pathways. The first was the case for the sulphate uptake system: S deprivation induced the high-affinity sulphate transporter TdSultr1.1, while Fe depletion up-regulated the high-affinity sulphate transporter TdSultr1.3. A key example of the second mentioned case was the increased mRNA level of most of the genes coding for enzymes involved in the S assimilatory pathway induced by S and Fe deprivation. However, the evidence that the mRNA level of genes encoding ATPS, APR, and OASTL and the corresponding enzyme activities were often uncoupled in response to Fe and S availability most probably suggests that their regulation involves a complex interplay of transcriptional, translational, and/or post-translational mechanisms under S and Fe deficiency. The present research suggests that the molecular mechanism involved in the regulation of sulphate uptake may be related to its induction by S deficiency or by internal metabolic demand, and indicates that the regulation of the enzymes involved in S assimilation cannot be wholly attributed to transcriptional regulation. This has important implications for the analysis of plant sulphate metabolism in response to different nutritional and abiotic stresses because many studies up to now have concentrated on transcriptome analysis to deduce the effects of these stimuli on sulphate uptake and assimilation pathways. It is clear that enzyme activities must be considered together with transcriptional information to gain more physiologically relevant insight into plant responses to their environment.
Supplementary data
Supplementary data are available at JXB online.
Appendix S1. Expression analysis by qRT–PCR of wheat genes involved in sulphur metabolism.
Appendix S2. Cloning and characterization of wheat genes involved in sulphur metabolism.
Fiure S1. Relative expression levels by quantitative real-time RT–PCR (qRT–PCR) and semi-quantitative RT–PCR of the genes encoding durum wheat high-affinity sulphate transporters (TdSultr1.1 and TdSultr1.3) in shoots and roots of seedlings grown with or without 100 μM FeIII-EDTA at two S levels in the nutrient solution (+S=1.2 and –S=0mM sulphate).
Figure S2. Relative expression levels by quantitative real-time RT–PCR (qRT–PCR) and semi-quantitative RT–PCR of the genes encoding durum wheat ATP sulphurylase (TdATPSul1 and TdATPSul2) in shoots and roots of seedlings grown with or without 100 μM FeIII-EDTA at two S levels in the nutrient solution (+S=1.2 and –S=0mM sulphate).
Figure S3. Relative expression levels by quantitative real-time RT–PCR (qRT–PCR) and semi-quantitative RT–PCR of the genes encoding durum wheat adenosine 5-phosphosulphate reductase (APR) (TdAPR) and sulphite reductase (TdSiR) in shoots and roots of seedlings grown with or without 100 μM FeIII-EDTA at two S levels in the nutrient solution (+S=1.2 and –S=0mM sulphate).
Figure S4. Relative expression levels by quantitative real-time RT–PCR (qRT–PCR) and semi-quantitative RT–PCR of the genes encoding durum wheat O-acetylserine(thiol)lyases (TdOASTL1 and TdOASTL2) in shoots and roots of seedlings grown with or without 100 μM FeIII-EDTA at two S levels in the nutrient solution (+S=1.2 and –S=0mM sulphate).
Figure S5. Relative expression levels by quantitative real-time RT–PCR (qRT–PCR) and semi-quantitative RT–PCR of the genes encoding durum wheat serine acetyltransferase (TdSAT1 and TdSAT2) in shoots and roots of seedlings grown with or without 100 μM FeIII-EDTA at two S levels in the nutrient solution (+S=1.2 and –S=0mM sulphate).
Table S1. Arabidopsis genes involved in sulphur metabolism.
Table S2. Primer pairs used for the isolation of the full-length cDNA of wheat genes involved in sulphur metabolism.
Table S3. Primer pairs used in qRT–PCR analyses.
Acknowledgements
This research was financially supported by grants from the Italian M.I.U.R.-PRIN 2009 and by a MAPPA5A project (CRA-MIPAAF, DM 7398/7303/2008).
References
- Abdallah M, Dubousset L, Meuriot F, Etienne P, Avice JC, Ourry A. 2010. Effect of mineral sulphur availability on nitrogen and sulphur uptake and remobilization during the vegetative growth of Brassica napus L. Journal of Experimental Botany 61, 2635–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astolfi S, Cesco S, Zuchi S, Neumann G, Roemheld V. 2006. a Sulphur starvation reduces phytosiderophores release by Fe-deficient barley plants. Soil Science and Plant Nutrition 52, 80–85 [Google Scholar]
- Astolfi S, Zuchi S, Cesco S, Sanità di Toppi L, Pirazzi D, Badiani M, Varanini Z, Pinton R. 2006. b Fe deficiency induces sulphate uptake and modulates redistribution of reduced sulphur pool in barley plants. Functional Plant Biology 33, 1055–1061 [DOI] [PubMed] [Google Scholar]
- Astolfi S, Zuchi S, Passera C, Cesco S. 2003. Does the sulphur assimilation pathway play a role in the response to Fe deficiency in maize (Zea mays L.) plants? Journal of Plant Nutrition 26, 2111–2121 [Google Scholar]
- Barberon M, Berthomieu P, Clairotte M, Shibagaki N, Davidian JC, Gosti F. 2008. Unequal functional redundancy between the two Arabidopsis thaliana high-affinity sulphate transporters SULTR1;1 and SULTR1;2 . New Phytologist 180, 608–619 [DOI] [PubMed] [Google Scholar]
- Bardsley CE, Lancaster JD. 1962. Determination of reserve sulphur and soluble sulphate in soils. Soil Science Society of America Proceedings 24, 265–268 [Google Scholar]
- Berkowitz O, Wirtz M, Wolf A, Kuhlmann J, Hell R. 2002. Use of biomolecular interaction analysis to elucidate the regulatory mechanism of the cysteine synthase complex from Arabidopsis thaliana . Journal of Biological Chemistry 277, 30629–30634 [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analitycal Biochemistry 72, 248–254 [DOI] [PubMed] [Google Scholar]
- Buchner P, Parmar S, Kriegel A, Carpentier M, Hawkesford MJ. 2010. The sulfate transporter family in wheat: tissue-specific gene expression in relation to nutrition. Molecular Plant 3, 374–389 [DOI] [PubMed] [Google Scholar]
- Davidian JC, Kopriva S. 2010. Regulation of sulfate uptake and assimilation—the same or not the same. Molecular Plant 3, 314–325 [DOI] [PubMed] [Google Scholar]
- Droux M. 2003. Plant serine acetyltransferase: new insights for regulation of sulphur metabolism in plant cells. Plant Physiology and Biochemistry 41, 619–627 [Google Scholar]
- Droux M, Ruffet ML, Douce R, Job D. 1998. Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants—structural and kinetic properties of the free and bound enzymes. European Journal of Biochemistry 255, 235–245 [DOI] [PubMed] [Google Scholar]
- Hatzfeld Y, Lee S, Lee M, Leustek T, Saito K. 2000. Functional characterization of a gene encoding a fourth ATP sulfurylase isoform from Arabidopsis thaliana. Gene 248, 51–58 [DOI] [PubMed] [Google Scholar]
- Hawkesford MJ. 2000. Plant responses to sulphur deficiency, the genetic manipulation of sulphate transporters to improve S-utilization efficiency. Journal of Experimental Botany 51, 131–138 [PubMed] [Google Scholar]
- Heeg C, Kruse C, Jost R, Gutensohn M, Ruppert T, Wirtz M, Hell R. 2008. Analysis of the Arabidopsis O-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. The Plant Cell 20, 168–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse H, Kreft O, Maimann S, Zeh M, Hoefgen R. 2004. Current understanding of the regulation of methionine biosynthesis in plants. Journal of Experimental Botany 55, 1799–1808 [DOI] [PubMed] [Google Scholar]
- Honsel A, Kojima M, Haas R, Frank W, Sakakibara H, Herschbach C, Rennenberg H. 2012. Sulphur limitation and early sulphur deficiency responses in poplar: significance of gene expression, metabolites, and plant hormones. Journal of Experimental Botany 63, 1873–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins L, Parmar S, Bouranis DL, Howarth JR, Hawkesford MJ. 2004. Coordinated expression of sulfate uptake and components of the sulfate assimilatory pathway in maize. Plant Biology 6, 408–414 [DOI] [PubMed] [Google Scholar]
- Howarth JR, Parmar S, Jones J, et al. 2008. Co-ordinated expression of amino acid metabolism in response to N and S deficiency during wheat grain filling. Journal of Experimental Botany 59, 3675–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima CG, Berkowitz O, Hell R, Noji M, Saito K. 2005. Characterization and expression analysis of a serine acetyltransferase gene family involved in a key step of the sulfur assimilation pathway in Arabidopsis. Plant Physiology 137, 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima CG, Matthewman CA, Huang S, et al. 2011. Interplay of SLIM1 and miR395 in the regulation of sulphate assimilation in Arabidopsis. The Plant Journal 66, 863–876 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nakanishi H, Nishizawa NK. 2010. Recent insights into iron homeostasis and their application in graminaceous crops. Proceedings of Japan Academy 86, 900–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK. 2012. Iron uptake, translocation, and regulation in higher plants. Annual Review of Plant Biology 63, 1–22 [DOI] [PubMed] [Google Scholar]
- Kopriva S, Jones S, Koprivova A, Suter M, von Ballmoos P, Brander K, Flückiger J, Brunold C. 2001. Influence of chilling stress on the intercellular distribution of assimilatory sulfate reduction and thiols in Zea mays. Plant Biology 3, 24–31 [Google Scholar]
- Krueger S, Donath A, Lopez-Martin MC, Hoefgen R, Gotor C, Hesse H. 2010. Impact of sulfur starvation on cysteine biosynthesis in T-DNA mutants deficient for compartment-specific serine-acetyltransferase. Amino Acids 39, 1029–1042 [DOI] [PubMed] [Google Scholar]
- Lee B-R, Koprivova A, Kopriva S. 2011. The key enzyme of sulfate assimilation, adenosine 5’-phosphosulfate reductase, is regulated by HY5 in Arabidopsis. The Plant Journal 67, 1042–1054 [DOI] [PubMed] [Google Scholar]
- Lerner SE, Seghezzo ML, Molfese ER, Ponzio NR, Cogliatti M, Rogers WJ. 2006. N- and S-fertiliser effects on grain composition, industrial quality and end-use in durum wheat. Journal of Cereal Science 44, 2–11 [Google Scholar]
- Lewandowska M, Sirko A. 2008. Recent advances in understanding plant response to sulfur-deficiency stress. Acta Biochimica Polonica 55, 457–471 [PubMed] [Google Scholar]
- Liang G, Yang F, Yu D. 2010. MicroRNA395 mediates regulation of sulphate accumulation and allocation in Arabidopsis thaliana. The Plant Journal 62, 1046–1057 [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056 [DOI] [PubMed] [Google Scholar]
- Logan HM, Cathala N, Grignon C, Davidian JC. 1996. Cloning of a cDNA encoded by a member of the Arabidopsis thaliana ATP sulfurylase multigene family. Journal of Biological Chemistry 271, 12227–12233 [DOI] [PubMed] [Google Scholar]
- Marschner H, Römheld V, Kissel M. 1986. Different strategies in higher plants in mobilization and uptake of iron. Journal of Plant Nutrition 9, 695–713 [Google Scholar]
- McGrath SP, Zhao FJ, Blake-Kalff M. 2002. History and outlook for sulphur fertilisers in Europe York: International Fertiliser Society; [Google Scholar]
- Mori S, Nishizawa N. 1987. Methionine as a dominant precursor of phytosiderophores in Gramineae plants. Plant and Cell Physiology 28, 1081–1092 [Google Scholar]
- Murata Y, Ma JF, Yamaji N, Ueno D, Nomoto K, Takashi I. 2006. A specific transporter for iron(III)–phytosiderophore in barley roots. The Plant Journal 46, 563–572 [DOI] [PubMed] [Google Scholar]
- Nagasaka S, Takahashi M, Nakanishi-Itai R, Bashir K, Nakanishi H, Mori S, Nishizawa NK. 2009. Time course analysis of gene expression over 24h in Fe-deficient barley roots. Plant Molecular Biology 69, 621–631 [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Bughio N, Matsuhashi S, Ishioka NS, Uchida H, Tsuji A, Osa A, Sekine T, Kume T, Mori S. 1999. Visualizing real time [11C]methionine translocation in Fe-sufficient and Fe-deficient barley using a Positron Emitting Tracer Imaging System (PETIS). Journal of Experimental Botany 50, 637–643 [Google Scholar]
- Ohkama-Ohtsu N, Kasajima I, Fujiwara T, Naito S. 2004. Isolation and characterization of an Arabidopsis mutant that overaccumulates O-Acetyl-L-Ser. Plant Physiology 136, 3209–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M. 2009. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Molecular Biology 10, 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheerer U, Haensch R, Mendel RR, Kopriva S, Rennenberg H, Herschbach C. 2010. Sulphur flux through the sulphate assimilation pathway is differently controlled by adenosine 5-phosphosulphate reductase under stress and in transgenic poplar plants overexpressing γ-ECS, SO, or APR. Journal of Experimental Botany 61, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz D, Brunold C. 1982. Rapid and simple measurement of ATP sulphurylase activity in crude plant extracts using an ATP meter for bioluminescence determination. Analytical Biochemistry 121, 151–155 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. 2011. Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annual Review of Plant Biology 62, 157–184 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Saito K. 1996. Subcellular localization of spinach cysteine synthase isoforms and regulation of their gene expression by nitrogen and sulfur. Plant Physiology 112, 273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Yamazaki M, Sasakura N, Watanabe A, Leustek T, Engler JA, Engler G, Van Montagu M, Saito K. 1997. Regulation of sulphur assimilation in higher plants: a sulphate transporter induced in sulphate-starved roots plays a central role in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 94, 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz M, Droux M, Hell R. 2004. O-Acetylserine(thiol)lyase: an enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana . Journal of Experimental Botany 55, 1785–1798 [DOI] [PubMed] [Google Scholar]
- Wirtz M, Hell R. 2006. Functional analysis of the cysteine synthase protein complex from plants: structural, biochemical and regulatory properties. Journal of Plant Physiology 163, 273–286 [DOI] [PubMed] [Google Scholar]
- Yoshimoto N, Inoue E, Saito K, Yamaya T, Takahashi H. 2003. Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiology 131, 1511–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FS, Römheld V, Marschner H. 1991. Role of the root apoplasm for iron acquisition by wheat plants. Plant Physiology 97, 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao FJ, Hawkesford M.J, McGrath SP. 1999. Sulphur assimilation and effects on yield and quality of wheat. Journal of Cereal Science 30, 1–17 [Google Scholar]
- Zuchi S, Cesco S, Astolfi S. 2012. High S supply improves Fe accumulation in durum wheat plants grown under Fe limitation. Environmental and Experimental Botany 77, 25–32 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.