Abstract
Although a great deal of progress has been made toward understanding the role of abscisic acid (ABA) in fruit ripening, many components in the ABA signalling pathway remain to be elucidated. Here, a strawberry gene homologous to the Arabidopsis gene ABI1, named FaABI1, was isolated and characterized. The 1641bp cDNA includes an intact open reading frame that encodes a deduced protein of 546 amino acids, in which putative conserved domains were determined by homology analysis. Transcriptional analysis showed that the levels of FaABI1 mRNA expression declined rapidly during strawberry fruit development as evidenced by real-time PCR, semi-quantitative reverse transcription–PCR, and northern blotting analyses, suggesting that the Ser/Thr protein phosphatase PP2C1 encoded by FaABI1 may be involved in fruit ripening as a negative regulator. The results of Tobacco rattle virus-induced gene silencing and PBI121 vector-mediated overexpression suggested that the down- and up-regulation of FaABI1 mRNA expression levels in degreening strawberry fruit could promote and inhibit ripening, respectively. Furthermore, alteration of FaABI1 expression could differentially regulate the transcripts of a set of both ABA-responsive and ripening-related genes, including ABI3, ABI4, ABI5, SnRK2, ABRE1, CHS, PG1, PL, CHI, F3H, DFR, ANS, and UFGT. Taken together, the data provide new evidence for an important role for ABA in regulating strawberry fruit ripening in the processes of which the type 2C protein phosphatase ABI1 serves as a negative regulator. Finally, a possible core mechanism underlying ABA perception and signalling transduction in strawberry fruit ripening is discussed.
Key words: Abscisic acid (ABA), overexpression, strawberry fruit ripening, Tobacco rattle virus, type 2C protein phosphatase ABI1, virus-induced gene silencing (VIGS).
Introduction
Abscisic acid (ABA) not only plays a central role in the adaptation of plants to environmental challenges, but also regulates many aspects of plant growth and development (Leung and Giraudat, 1998; Finkelstein et al., 2002; Himmelbach et al., 2003; Hirayama and Shinozaki, 2007). In recent years, not only has ABA been shown to play important roles in perception and signal transduction involved in the regulation of Arabidopsis seed germination, seedling growth, and stomatal movement (Fujii et al., 2009; Ma et al., 2009; Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Santiago et al., 2009; Shang et al., 2010), but much progress has also been made toward a better understanding of the molecular mechanisms that underlie the roles of ABA in the regulation of fleshy fruit ripening (Chai et al., 2011; Jia et al., 2011; Li et al., 2011; Sun et al., 2011, 2012). Nevertheless, many ABA signalling components involved in fruit ripening remain to be elucidated.
To date, two core ABA signalling pathways have been proposed in Arabidopsis, namely ABA–PYR/PYL/RCAR–type 2C protein phosphatase (PP2C)– SNF1-related protein kinase 2 (SnRK2) (Fujii et al., 2009) and ABA–ABAR–WRKY40–ABI5 (Shang et al., 2010). Structural biology provides a detailed gate–latch–lock mechanism involved in ABA signal perception and transduction, including ABA–PYR1 perception, PYR1–PP2C interaction, inhibition of PP2C activity, and activation of SnRK2 (Fujii et al., 2009). In this model, the early reported PP2Cs, serving as a central and negatively regulated hub in ABA signalling (Leung et al., 1997; Merlot and Giraudat, 1997; Gosti et al., 1999; Merlot et al., 2001), were first integrated into the canonical ABA signalling network by reversible phosphorylation (Fujii et al., 2009), which is a universal mechanism for regulating diverse biological functions in eukaryotes (Smith, 1996).
Most phosphorylation events involve transfer of phosphate to serine and threonine residues catalysed by kinases, such as Ca2+-dependent protein kinase (CDPK), SnRKs, mitogen-activated protein kinase (MAPK), and a receptor-type kinase (RPK1), while removal of this phosphate is catalysed by Ser/Thr protein phosphatases including type 1 (PP1) and type 2 (PP2), of which PP2 was subsequently divided into three groups based on the metal ion requirements: 2A (not requiring metal ions), 2B (activated by calcium), and 2C (Mg2+ dependent) (Cohen and Cohen, 1989; Sopory and Munshi, 1998; Hirayama and Shinozaki, 2007). Note that the Arabidopsis strong ABA-insensitive loci ABI1 and ABI2 have been demonstrated to encode PP2C enzymes, which play a central functional role in the ABA signalling pathway as negative regulators of plant stress, cell differentiation, and growth (Leung et al., 1994; Meyer et al., 1994; Finkelstein et al., 2002; Hirayama and Shinozaki, 2007; Lu and Wang, 2008). Also, plant PP2Cs are reportedly encoded by a large multigene family with 80 and 78 members in Arabidopsis and rice, respectively (Xue et al., 2008). In addition to Arabidopsis and rice, PP2Cs as negative regulators have been characterized only in several other plants, such as pea, wheat, beech, alfalfa, and maize (MacKintosh et al., 1991, 1992; Meskiene et al., 1998; Sheen, 1998; González-García et al., 2003; Hu et al., 2010).
Following the determination of Arabidopsis ABA core signalling function, a great deal of progress has been made toward understanding the role of ABA in fleshy fruit ripening. The down-regulation of the expression of ABA receptor genes Mg-chelatase H subunit (FaABAR/CHLH) or pyrabactin resistance 1 gene (FaPYR1) can destroy strawberry fruit red colouring, indicating that both FaABAR/CHLH and FaPYR1 proteins are positive regulators of fruit ripening (Chai et al., 2011; Jia et al., 2011; Li et al., 2011). A significant reduction in SINCED1 activity leads to a decline in the transcription of genes encoding major cell wall catabolic enzymes, indicating that ABA affects cell wall catabolism during tomato fruit ripening (Sun et al., 2012). Transcriptional analysis suggested that tomato SlPYL1, SlPYL2, SlPP2C1, SlPP2C5, and SnRK2.3 may be involved in the regulation of fruit ripening (Sun et al., 2011). Although PP2C is an important hub in ABA responses, its defined function in fruit ripening remains unclear. In the present study, silencing and overexpression of the FaABI1 gene in strawberry fruit were performed. The results showed that PP2C1 encoded by FaABI1 is a negative regulator of strawberry fruit ripening.
Materials and methods
Plant material
Strawberry (Fragaria×ananassa cv. Camarosa) was grown in a glasshouse (20–25 °C, relative humidity 70–85%, 14h/10h light/dark cycle) during the spring season in 2009–2010. The fruit samples were sampled from seven developmental stages: small green (SG), large green (LG), degreening (DG), white (Wt), initial red (IR), partial red (PR), and full red (FR) for 7, 15, 20, 23, 25, 27, and 31 d after anthesis, respectively. Thirty uniformly sized fruit were sampled at each stage (one replicate). After removing the achenes (seeds), the receptacle (pulp) was cut into cubes measuring 0.5–0.8cm3, snap-frozen in liquid nitrogen, and quickly stored at –80 °C.
RNA isolation, cloning of the FaABI1 gene, and RT–PCR analysis
Total RNA was extracted from 10g of fresh or treated strawberry fruit using a modified cetyltrimethylammonium bromide (CTAB) protocol (Jia et al., 2008). Genomic DNA was removed by 15min incubation at 37 °C with RNase-Free DNase (TaKaRa, Otsu, Japan) followed by an RNA Clean Purification Kit (BioTeke, Beijing, China). The purity and integrity of RNA were analysed both by agarose gel electrophoresis and by the A 260:A 230 and A 260:A 280 ratios. To generate first-strand cDNA, 3 µg aliquots of total RNA were reverse-transcribed using a universal primer (5’-AAGCAGTGGTATCAACGCAGAGTAC(T)30VN-3’ (where N=A, C, G, or T; V=A, G, or C) supplied with the SMART™ RACE cDNA Synthesis Kit (TaKaRa) according to the manufacturer’s protocol.
To clone the FaABI1 gene, an Arabidopsis PP2C protein AB1I (At4g26080) was used to BLAST in a strawberry gene library (https://strawberry.plantandfood.co.nz/index.html), and a high homology protein with a gene locus 07500 was found. Based on the nucleotide sequence, specific primers (forward, 5’-ATGGAGGA CATGTCTCCAGCAG-3’; reverse, 5’-TCAAGATTTGCTCTTGA ACTTTC-3’) were designed to amplify a coding sequence from ‘Camarosa’ strawberry fruit by reverse transcription–PCR (RT–PCR). PCR was performed under the following conditions: 94 °C for 5min, followed by 35 cycles at 94 °C for 30 s, 58 ºC for 30 s and 72 ºC for 2min, with a final extension at 72 ºC for an additional 10min. The PCR products were ligated into a pUC-T vector and subsequently transformed into Escherichia coli DH5α. Positive colonies were selected, amplified, and sequenced by Invitrogen China (Shanghai, China)
Primers for semi-quantitative RT–PCR (SqRT–PCR) of FaABI1 were designed based on the determined sequences (forward, 5’-CAGCAGGTAAAGTGGGAGAA-3’; reverse, 5’-AGGG CAAGAGTTGAAAGATAAG-3’). The primers for real-time PCR were designed as showed in Table 1. A Bio-Rad iQ Sequence Detector (Bio-Rad, Hercules, CA, USA) was used for real-time PCR amplification. The quantitative PCR experiment was repeated three times.
Table 1.
The primers used for real-time PCR.
| Genes | Sequences for real-time PCR | Sources of primers |
|---|---|---|
| ABI1 | Sense: 5’-CAAGAGCCATTCTTTGTCGT-3’ | Chai et al. (2011); Jia et al. (2011) |
| Antisense: 5’-TGGAATAATCCAGGGTTTCA-3’ | ||
| SnRK2 | Sense: 5’-GCACTTCCGTCCAAGAGTG-3’ | Chai et al. (2011); Jia et al. (2011) |
| Antisense: 5’-AGGATATGTAGTGCTGGTAGATT-3’ | ||
| ABI3 | Sense: 5’-CGGCGCCTGTATTAGTCCC-3’ | Chai et al. (2011); Jia et al. (2011) |
| Antisense: 5’-TGCAGTCTCCAGCGTTTGAT-3’ | ||
| ABI4 | Sense: 5’-TCCTCATCACCACCGTCTT-3’ | Chai et al. (2011); Jia et al. (2011) |
| Antisense: 5’-ACTCTGGCTCGTTTGCTCT-3’ | ||
| ABI5 | Sense: 5’-GGAGCTGGCAATGGTCG-3’ | Chai et al. (2011); Jia et al. (2011) |
| Antisense: 5’-AGGCCCGCCTTTCCTT-3’ | ||
| Actin | Sense: 5’-TGGGTTTGCTGGAGATGAT-3’ | Chai et al. (2011); Jia et al. (2011) |
| Antisense: 5’-CAGTAGGAGAACTGGGTGC-3’ | ||
| PYR1 | Sense: 5’-GGAGCTGGCAATGGTCG-3’ | Chai et al. (2011); Jia et al. (2011) |
| Antisense: 5’-AGGCCCGCCTTTCCTT-3’ | ||
| CHS | Sense: 5’-GCTGTCAAGGCCATTAAGGA-3’ | Chai et al. (2011); Jia et al. (2011) |
| Antisense: 5’-GAGCAAACAACGAGAACACG-3’ | ||
| PG1 | Sense: 5’-CGACAGAGTGAAAAATTCCTTAG-3’ | GenBank accession no. EF441274 |
| Antisense: 5’-AGGACTGGGTTAGCAAAATTATTC-3’ | ||
| PL1 | Sense: 5’-TGACTCCCTTGCTGCTTCTT-3’. | GenBank accession no. EF441273 |
| Antisense: 5’-TCTACTGCGTGCTCATTCCA-3’ | ||
| NCED1 | Sense: 5’-CCCAAACGGCACGAAAT-3’ | GenBank accession no. HQ290318.1 |
| Antisense: 5’-GCATCGCTCGCATTCTT-3’ | ||
| ABRE1 | Sense: 5’-GGTGGTGCGAGAAGATGC-3’ | Gene 28250 locus in https://strawberry.plantandfood.co.nz/index.html |
| Antisense: 5’-TTGGGAAAGATTGGTTGC-3’ | ||
| CHI | Sense: 5’-GTTAAGTGGAAGGGCAAGA-3’ | GenBank accession number AB201755 |
| Antisense: 5’-CCCGTCAGCGGTAGTATCA-3’ | ||
| F3H | Sense: 5’-TTTTCTGAGCAATGGGAGG-3’ | GenBank accession no. AB201760 |
| Antisense: 5’-CTGGGTTCTGGAATGTCG-3’ | ||
| DFR | Sense: 5’-ACGAAGTGATAAAGCCAACA-3’ | GenBank accession no. AF029685 |
| Antisense: 5’-AAACACCAACCTCCGAAC-3’ | ||
| ANS | Sense: 5’-CGTGAGACCCAAAGAGGA-3’ | GenBank accession no. AY695818 |
| Antisense: 5’-ATGCCGTGGTTGATAAGG-3’ | ||
| UFGT | Sense: 5’-GGTAAGCCACAGGAGGACA-3’ | GenBank accession no. AY575056 |
| Antisense: 5’-TATGAGCACCGAACCAAAA-3’ |
Construction of virus vectors, the PBI121 vector, and Agrobacterium-mediated infiltration
The pTRV1 and pTRV2 vectors (Liu et al., 2002) were kind gifts of Dr Liu Yu-le, Qinghua University. A 584bp cDNA fragment near the 5’ end of FaABI1 was amplified using appropriate primers (forward, 5’-GGTAACCATCCCATCACAAG-3’; reverse, 5’-GGGTCAAACTCTGGCTCATTCC-3’), cloned into the pMD19-T vector (TaKaRa), digested with SacI and XbaI, and subsequently cloned into the virus vector pTRV2 cut with the same restriction enzymes. Agrobacterium strain GV3101 containing pTRV1, pTRV2, or the pTRV2 derivative pTRV2-FaABI1584 was used for RNA interference (RNAi). For overexpression, the 1641bp cDNA was amplified using appropriate primers (forward, 5’-TCTAGAATGGAGGACATGTCTCCAGCAG-3’; reverse, 5’-G AGCTCTCAAGATTTGCTCTTGAACTTTC-3’), cloned into the pMD-T simple vector (TaKaRa), digested with XbaI and SacI, and subsequently cloned into the binary expression vector PBI121 cut with the same restriction enzymes. Agrobacterium strain GV3101 containing PBI121 or the PBI121derivative PBI121-FaABI11641 was used for overexpression.
For the virus-induced gene silencing (VIGS) assay, pTRV1 and pTRV2 or recombinant derivatives (pTRV2-FaABI1) were transformed into Agrobacterium strain GV3101 by the freeze–thaw method (Fire et al., 1998). A 5ml culture of each strain was grown overnight at 28 °C in Luria–Bertani (LB) medium (50mg ml–1 kanamycin and 50mg ml–1 rifampicin, 10mM MES, 20mM acetosyringone). The overnight cultures were inoculated into 50ml of LB medium and grown at 28 °C overnight. The cells were harvested by centrifugation (5000rpm, 5min, 20 °C), resuspended in infiltration buffer (10mM MgCl2, 10mM MES, 200mM acetosyringone), adjusted to an optical density (OD) of 1.0–2.0, and left to stand at room temperature for 3h. About 1ml of Agrobacterium mixture pTRV1, pTRV2, or pTRV2-FaABI1 (1:1 ratio) was infiltrated into every DG fruit with a 50ml syringe. Ten uniformly sized fruit were used in infiltration experiment, and the experiment was repeated three times.
Determination of fruit firmness and soluble solid, anthocyanin, and ABA contents
The fruit firmness was measured after removal of the skin on three sides using a fruit hardness tester (FHM-5, Takemura Electric Work Ltd, Japan). The solid soluble content of flesh was measured using a hand-held sugar measurement instrument (MASTER-100H, ATAGO Master, Japan), onto which fruit juice was applied to obtain a reading. Anthocyanin measurements were performed as described previously by Fuleki and Francis (1968a , b ). ABA measurements were carried out as described by Chai et al. (2011). Ten uniformly sized fruit were used for detection of each parameter, and the experiment was repeated three times.
Probe preparation and northern and siRNA hybridization
DIG-labelled probes were synthesized using a PCR-DIG Probe Synthesis Kit (Roche Diagnostics) according to the manufacturer’s protocol. For northern hybridization analysis, aliquots of RNA (15 µg) were separated by electrophoresis on 1% (w/v) agarose gels containing 2.2M formaldehyde and blotted onto nylon membranes (Hybond N+; Amersham Biosciences). RNA blots were hybridized with a 626bp FaABI1 probe that was amplified using the appropriate primers (forward, 5’-CAGCAGGTAAAGTGGGAGAA-3’; reverse, 5’-AGGGCAAGAGTTGAAAGATAAG-3’). For the small interfering RNA (siRNA) test, siRNA was extracted from 10g of flesh using the miRcute miRNA isolation kit (Tiangen Biotech, Beijing, China), which was then fractionated in a 15% (w/v) polyacrylamide–urea gel and blotted onto a 0.45mm nylon membrane (Whatman, Nytran SPC, Sanford, CA, USA) and hybridized with an ABI1-specific probe corresponding to the FaABI1 region (forward, 5’-GGTAACCATCCCATCACAAG-3’; reverse, 5’-GGGTCAAACTCTGGCTCATT CC-3’). rRNA stained with ethidium bromide was used as a gel loading control. The filters were hybridized overnight with DIG-labelled DNA probes (0.3–1g ml–1) in high stringency hybridization solution (50% formamide, 2× SSPE buffer, 10mM dithiothreitol, 1mg ml–1 herring sperm DNA, 500 µg ml–1 yeast tRNA, and 1mg ml–1 bovine serum) in a shaking water bath at 50 °C. Following hybridization, the filters were washed twice at 50 °C for 15min in each of 2× SSC, 1× SSC, and 0.1× SSC. The membranes were then subjected to immunological detection according to the manufacturer’s instructions using NBT/BCIP stock solution as a chemiluminescent substrate for alkaline phosphatase (Roche Diagnostics).
Southern hybridization
About 0.5g of strawberry leaf tissue was used for genomic DNA isolation using a DNA Extraction Kit (BioTeke, Beijing, China) according to the manufacturer’s protocol. DIG-labelled probes (FaABI1, forward, 5’-CAGCAGGTAAAGTGGGAGAA-3’, reverse, 5’-AG GGCAAGAGTTGAAAGATAAG-3’) were synthesized using a PCR-DIG Probe Synthesis Kit (Roche Diagnostics) according to the manufacturer’s protocol. About 10 µg of DNA was separated by electrophoresis on 0.8% (w/v) agarose gels containing 2.2M formaldehyde and blotted onto nylon membranes (Hybond N+; Amersham Biosciences). The hybridization processes were performed according to the northern analysis above.
Detection of the Tobacco rattle virus (TRV) vector by RT–PCR
Random primers were used to reverse-transcribe RNA for the first strand of infiltrated strawberry fruit to detect the TRV vectors by RT–PCR as described by Chai et al. (2011).
Expression and purification of FaPYR1 recombinant protein
The coding sequence of FaPYR1 (Chai et al., 2011) was amplified by PCR from a cDNA synthesized above using primers [forward, CGGATCC ATGGAGAAACCATCATCGGC (BamHI site); and reverse, GGCGGCCGC TCAGACCTGGGGAGTTAGCG (NotI site)] and cloned into the expression vector pET28a (Novagen, Germany). The modified fusion protein contains two His-tags at both the N-terminus and the C-terminus. BL21(DE3) cells transformed with the expression plasmid were grown in 1 litre of LB medium containing 50 µg ml–1 kanamycin to an OD600 of 0.5–0.6 at 37 °C. Then, 0.5mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added and induction was carried out overnight at 16 °C. Cells were harvested after ~12h incubation and stored at –80 °C before purification.
Purification was carried out at 4 °C under native conditions. A 1 litre pellet was resuspended in 50ml of lysis buffer (20mM TRIS, pH 8.0, 300mM NaCl, 10mM imidazole, and Merck’s protease cocktail inhibitor) and the cells were sonicated in a Misonix Sonifier in an ice bath. A clear lysate was obtained after centrifugations at 30 000 g for 30min. Proteins were purified on Ni-NTA His Bind Resins (Novagen) in a column procedure. The 8ml of supernatant filtered on a 0.45 µm filter was added to the Ni-NTA His Bind slurry and mixed gently by shaking at 4 °C for 60min. The lysate–resin mixture was loaded onto an empty column. The column was washed with lysis buffer supplemented with 20mM imidazole and 10% glycerol to 20ml, and eluted with elution buffer (20mM TRIS, pH 8.0, 300mM NaCl, 500mM imidazole, and Merck’s protease cocktail inhibitor) for 8ml. The eluted fusion protein was stored at –80 °C before isothermal calorimetric analysis.
Isothermal calorimetry analysis
Before the next step of the analysis, the protein was concentrated using an Amicon Ultra-4 centrifugal 10kDa filter (Millipore) at 3000 g for ~10–40min in a swing bucket rotor (Sigma, USA). Purified FaPYR1 fusion protein was desalted for buffer exchange using a HiTrap Desalting column (GE Healthcare). The HiTrap Desalting column was filled with ITC buffer (20mM phosphate buffer, pH 7.4, 150mM NaCl, 20mM KCl) to remove the ethanol completely in order to equilibrate the column. The sample was applied using a 2–5ml syringe and the eluted buffer was discarded from the column. The buffer was changed to ITC buffer, and then injection was carried out and the eluted buffer was collected.
Binding studies were performed using a calorimeter (Microcal ITC200, USA) at 30 °C. The final concentration of FaPYR1 fusion protein was adjusted to 15mM. Ligand (1mM S-ABA) in the same buffer was injected into the protein solution. The experiment was repeated three times. Data fitting was performed by using the ORIGIN 7.0 software supplied with the instrument.
Results
Cloning, bioinformatics, and Southern analysis of the FaABI1 gene
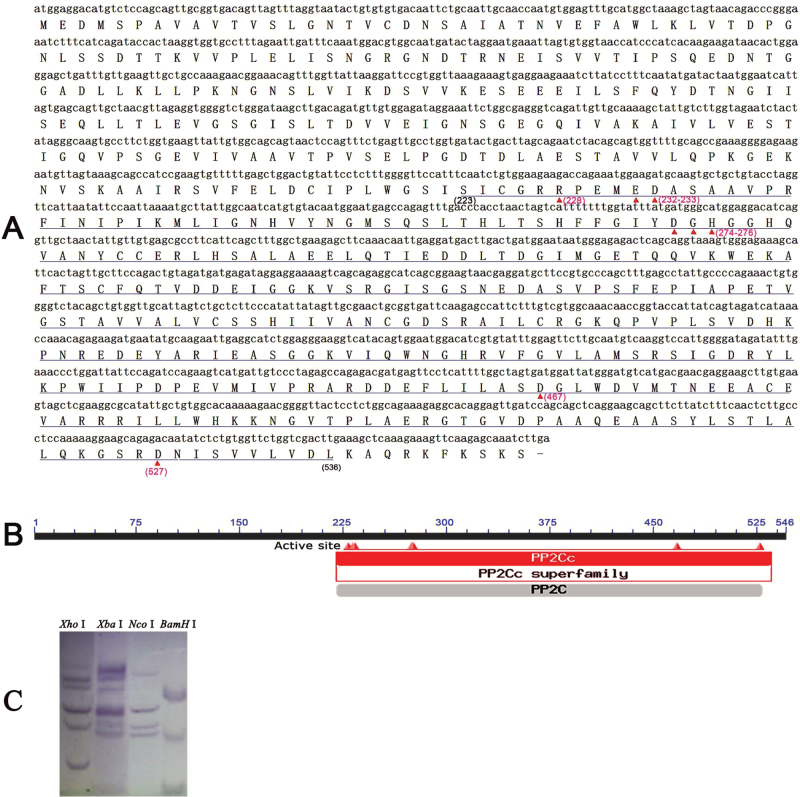
To clone the FaABI1 gene, the Arabidopsis PP2C protein AB1I (At4g26080, GenBank accession no. AY142623) was used for a BLAST search in a strawberry gene library (https://strawberry.plantandfood.co.nz/index.html), which yielded a protein showing a high level of identity with the gene locus 07500, on the basis of which specific primers (forward, 5’- ATGGAGGACATGTCTCCAGCAG-3’; reverse, 5’-TCAAGATTTGCTCTTGAACTTTC-3’) were designed to amplify a coding sequence from ‘Camarosa’ strawberry fruit by RT–PCR. A 1641bp cDNA homologous to Arabidopsis ABI1, named FaABI1, was isolated from strawberry fruit (GenBank accession no. JX989266). The cDNA included an open reading frame encoding a deduced protein of 546 amino acids (Fig. 1A), in which the putative PP2C conserved domains were detected by homology analysis using BLAST for the FaABI1 protein on the NCBI protein blast website (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Fig. 1B), indicating successful isolation of the FaABI1 gene encoding the putative strawberry PP2C.
Fig. 1.
Characterization of the FaABI1 gene sequence. (A) The cDNA and deduced amino acid sequences of the FaABI1 gene. (B) The conserved serine/threonine phosphatase family 2C catalytic domain in the FaABI1 protein. The amino acids underlined with red triangles represent active sites (228, 232–233, 274–276, 467, and 527). Amino acids 223–536 underlined in blue represent the PP2C superfamily domains. (C) A 10 µg portion of strawberry genomic DNA was digested with XhoI, XbaI, NcoI, and BamHI, electrophoresed on a 0.8% agarose gel, and transferred onto a nylon membrane. The membrane was hybridized with a digoxigenin (DIG)-labelled cDNA fragment of FaABI1.
To investigate further the members of the FaABI1 gene family in the strawberry genome, Southern blot analysis was performed using a probe corresponding to the FaABI1-coding cDNA sequence. Genomic DNA was digested with XhoI, XbaI, NcoI, and BamHI restriction enzymes, respectively. The probe generated multiple hybridizing bands using every restriction enzyme (Fig. 1C). The results suggest that the strawberry genome indeed contains a FaABI1 gene family with multiple members.
Expression analysis of the FaABI1 gene during strawberry fruit development
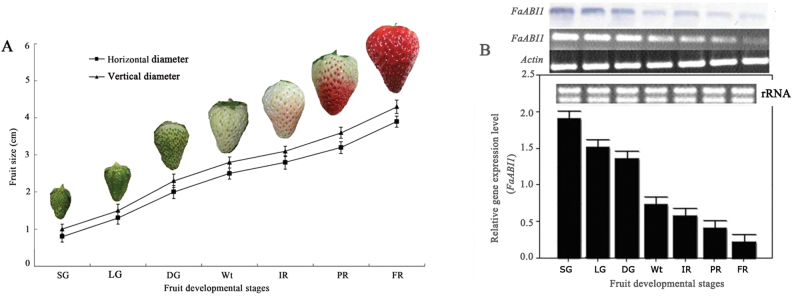
Based on our previous report (Chai et al., 2011) and changes of fruit size and colour, the fruit development processes of strawberry cultivar ‘Camarosa’ was divided into seven different visual stages: SG, LG, DG, Wt, IR, PR, and FR for 7, 15, 20, 23, 25, 27, and 31 d after anthesis, respectively (Fig. 2A).
Fig. 2.
Variations in transcripts of the FaABI1 gene in strawberry fruit during the seven developmental stages. (A) Development was divided into seven stages based on changes in fruit size and colour: small green (SG), large green (LG), degreening (DG), white (Wt), initial red (IR), partial red (PR), and full red (FR). (B) Changes in FaABI1 transcript levels during the seven developmental stages were determined by northern blotting, semi-quantitative RT–PCR, and real-time PCR analyses (from top to bottom). rRNA indicates the loading control of the RNA samples stained with ethidium bromide. Actin mRNA was used as an internal control. The error bars represent the standard error (n=3).
To investigate whether the FaABI1 gene is involved in strawberry fruit ripening, the mRNA expression levels of FaABI1 were determined by real-time PCR, northern blotting, and SqRT–PCR using the seven-stage fruit (Fig. 2A). The results showed that the mRNA expression levels of FaABI1 were extremely high in SG fruit, declined rapidly during strawberry fruit development, and finally remained at extremely low levels at the FR stage (Fig. 2B). The opposite trend between FaABI1 transcripts and fruit size and colour suggested that FaABI1 might negatively regulate strawberry fruit ripening.
Silencing of the FaABI1 gene promotes strawberry fruit ripening
To confirm the function of the FaABI1 gene in strawberry fruit development, a 584bp cDNA fragment of the FaABI1 gene (from 207bp to 790bp) was cloned based on the coding sequence isolated above and inserted into the TRV2 virus vector using the restriction enzymes XbaI and SacI. A mixture of Agrobacterium tumefaciens strain GV3101 cultures containing pTRV1 and pTRV2 or pTRV2 carrying the 584bp fragment of the FaABI1 gene (pTRV2-FaABI1584) in a 1:1 ratio was inoculated into DG fruit using a syringe (Fig. 3A). Control fruit were inoculated with TRV alone (empty vector). Unexpectedly, the surface of the inoculated RNAi fruit developed a red colour 5 d after injection (Fig. 3C), while the surface of the control fruit remained at the Wt stage (Fig. 3B).
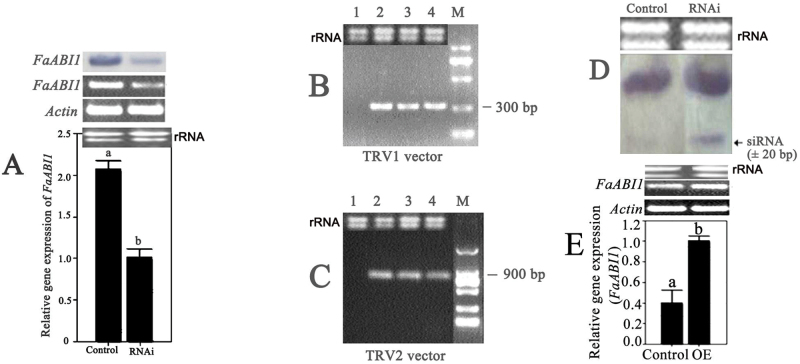
Fig. 3.
The phenotypes of virus-induced FaABI1 gene silencing and FaABI1 overexpression in strawberry fruit. (A) Degreening fruit attached to the plant were infiltrated with Agrobacterium containing TRV alone (control fruit for RNAi), TRV carrying a fragment of FaABI1 (RNAi fruit), PBI121 alone (control fruit for OE), or PBI121 carrying the coding sequence of FaABI1 (overexpressing fruit). (B and C) Photographs of fruit were taken for control (B) and RNAi (C) 5 d after infiltration. (D and E) Photographs were taken for control (D) and overexpressing fruit (E) 12 d after infiltration.
To validate the suppression of FaABI1 at the molecular level, a series of analyses were performed by SqRT–PCR, real-time PCR, northern blotting, and siRNA analyses. The results showed that the FaABI1 transcripts were markedly down-regulated in RNAi fruit compared with the control fruit (Fig. 4A); 300bp TRV-RNA1 and 900bp TRV-RNA2 were both detected in Agrobacterium-mediated TRV-inoculated fruit (Fig. 4B, C, lanes 2–4), but not in fruit inoculated with Agrobacterium alone (Fig. 4B, C, lane 1); FaABI1-related siRNA for specific RNA silencing was also detected in RNAi fruit but not in control fruit (Fig. 4D). Taken together, the FaABI1 gene was silenced successfully in strawberry fruit and thus led to the promotion of red colour development.
Fig. 4.
Silencing and overexpression of the FaABI1 gene in strawberry fruit at the molecular level. (A) Analysis of the FaABI1 transcripts in control and RNAi fruit by northern blotting, semi-quantitative RT–PCR, and real-time PCR (from top to bottom). (B and C) Analysis of the transcripts of TRVs using RT–PCR. Five days after infiltration, the expression of virus vector genes [300bp pTRV1 (B) and 900bp pTRV2 (C)] was detected in fruit infiltrated with Agrobacterium containing TRV in white control fruit (lane 2, empty vector) and in two RNAi fruit (lanes 3 and 4), but not in fruit infiltrated with Agrobacterium alone (lane 1). (D) Detection of siRNA (~20bp) specific to the FaABI1 gene in control and RNAi fruit. (E) Analysis of the transcripts of FaABI1 in control and overexpressing fruit using semi-quantitative RT–PCR and real-time PCR (from top to bottom). rRNA was the loading control for the RNA samples stained with ethidium bromide. Actin mRNA was used as an internal control. The error bars represent the standard error (n=3). Different letters indicate statistically significant differences at P < 0.05 as determined by Duncan’s test.
Overexpression of the FaABI1 gene inhibits strawberry fruit ripening
To confirm further the negative regulatory role of FaABI1 in strawberry fruit ripening, the 1641bp coding sequence of FaABI1 was cloned into the plant binary expression construct PBI121 using XbaI and a SacI restriction enzyme named PBI121-FaABI11641. A mixture of A. tumefaciens strain GV3101 cultures containing PBI121 (control) or PBI121-FaABI11641 was inoculated into DG fruit using a syringe (Fig. 3A). Twelve days after inoculation, the surface of control fruit turned full red (Fig. 3D), whereas the inoculated sector on the surface of the overexpressing (OE) fruit remained white (Fig. 3E). SqRT–PCR and real-time PCR analyses indicated that the FaABI1 gene mRNA level was up-regulated by 60% in OE fruit compared with control FR fruit (Fig. 4E). These results indicated that overexpression of the FaABI1 gene inhibited strawberry fruit ripening.
Alteration of FaABI1 expression affects a set of ABA-responsive and ripening-related gene transcripts and ABA levels
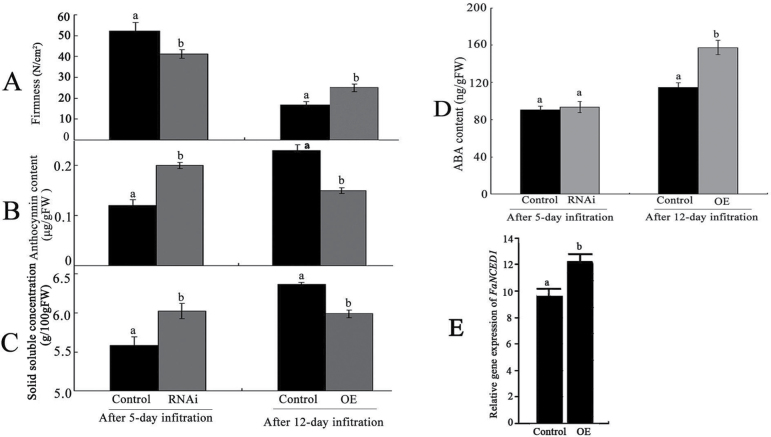
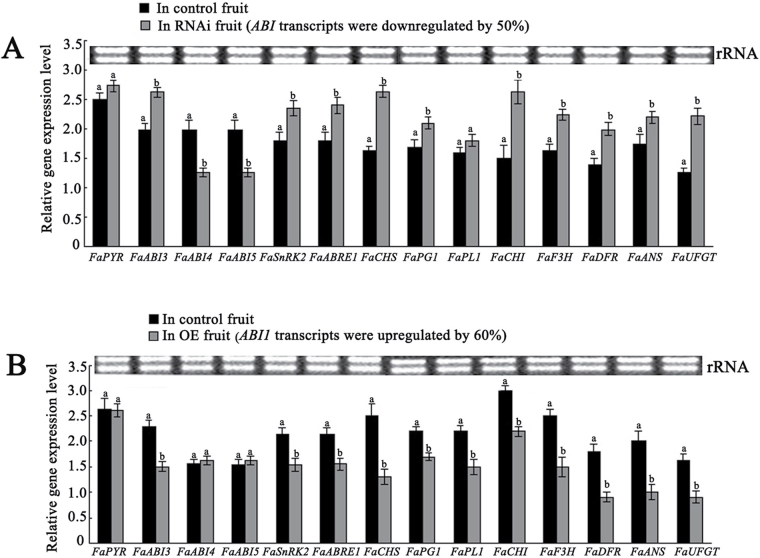
To clarify the mechanism of action of FaABI1 in the regulation of strawberry fruit ripening, several ripening-related physiological parameters were measured, including fruit firmness, solid soluble concentrations, anthocyanin contents, and ABA levels, and ABA-responsive and ripening-related genes, such as ABI3, ABI4, ABI5, PYR1, SnRK2, ABRE1, chalcone synthase (CHS), polygalacturonase (PG1), pectate lyase (PL1), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT), were also examined (Salentijn et al., 2003; Fujii et al., 2009; Chai et al., 2011; Jia et al., 2011; Wei et al., 2011) in both RNAi and OE fruit. The results showed that firmness (Fig. 5A) declined in RNAi fruit but increased in OE fruit; both anthocyanin (Fig. 5B) and solid soluble contents (Fig. 5C) were up-regulated in RNAi but down-regulated in OE fruit, compared with the corresponding control fruit. Gas chromatography–mass spectrometry analysis showed that ABA content was not significantly altered in RNAi fruit but increased in OE fruit compared with the corresponding control fruit (Fig. 5D). Transcriptional analysis of the FaNCED1 gene key to ABA biosynthesis showed that the mRNA expression level of the NCED1 gene was up-regulated in OE fruit compared with that in control fruit (Fig. 5E). Real-time PCR analysis showed that the majority of genes, including ABI3, SnRK2, AREB1, PG1, CHS, CHI, F3H, DFR, ANS, and UFGT, were significantly up-regulated in RNAi fruit (Fig. 6A); in contrast, together with the PL1 gene, they were all markedly down-regulated in OE fruit (Fig. 6B). Notably, a small fraction of genes, such as ABI4 and ABI5, were markedly down-regulated in RNAi fruit (Fig. 6A) but showed no significant alterations in expression level in OE fruit (Fig. 6B). Taken together, the observed regulation of expression of firmness (PG1 and PL1)-, sugar (SnRK2)-, and pigment (CHS, CHI, F3H, DFR, ANS, and UFGT)-related genes through the FaABI1-mediated signalling pathway was consistent with a negative role for PP2C in ABA signalling during fruit ripening.
Fig. 5.
Alteration of FaABI1 expression affects several physiological parameters and FaNCED1 transcripts in RNAi and OE fruit. The physiological parameters included (A) fruit firmness, (B) anthocyanin contents, (C) solid soluble concentrations, and (D) ABA contents. (E) FaNCED1 transcripts in control and OE fruit. RNAi, FaABI1-silenced fruit, in which FaABI1 was down-regulated by 50%; OE, FaABI1-overexpressing fruit, in which FaABI1 was up-regulated by 60%. The error bars represent the standard error (n=3). Different letters indicate statistically significant differences at P < 0.05 as determined by Duncan’s test.
Fig. 6.
Alteration of FaABI1 expression affects transcripts of a set of ABA-responsive and ripening-related genes in RNAi and overexpressing fruit. (A) The mRNA expression levels of ABA signalling and ripening-related genes in RNAi fruit, in which the FaABI1 transcript was down-regulated by 50%. (B) The mRNA expression levels of ABA signalling and ripening-related genes in overexpressing fruit, in which the FaABI1 transcript was up-regulated by 60%. Actin mRNA was used as an internal control. The error bars represent the standard error (n=3). Different letters indicate statistically significant differences at P < 0.05 as determined by Duncan’s test.
The molecular basis of a negative regulator of FaABI1: FaPYR1 is an ABA receptor
The PYR1–PP2C–SnRK2 core signaling pathway of ABA action has been established in Arabidopsis (Fujii et al., 2009). Given that FaPYR1 acts as a positive regulator in strawberry fruit ripening (Chai et al., 2011), further determination of the receptor nature of FaPYR1 protein contributes to understanding the molecular basis of the negative regulator FaABI1.
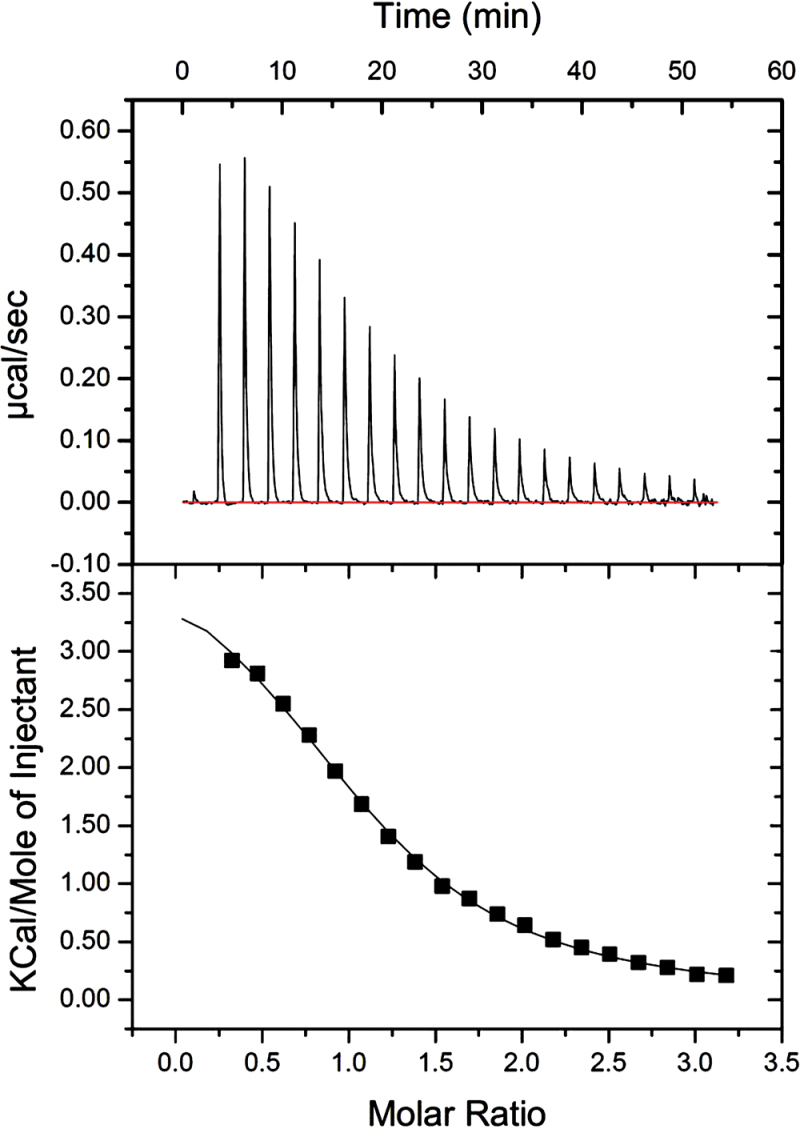
Isothermal calorimetry analysis using 15mM recombinant FaPYR1 protein and 1mM S-ABA showed that each purified FaPYR1 protein molecule could bind approximately one molecule of ABA, and the dissociation constant (K d) was 87.5 µM (Fig. 7). The stoichiometry (N) with a binding ratio of 1:1 between FaPYR1 and ABA demonstrated that strawberry fruit FaPYR1 could be an ABA receptor.
Fig. 7.
Measurement of binding affinity between (+)-ABA and purified FaPYR1 protein using isothermal titration calorimetry (ITC). A typical and specific saturation curve with stoichiometry (N) ~1:1 was obtained, suggesting that one ABA molecule could bind per purified protein molecule with a dissociation constant (K d) of 87.5 µM.
Discussion
The type 2C protein phosphatase ABI1 is a negative regulator of strawberry fruit ripening
In plants, PP2C protein, serving as a negatively regulated hub in ABA signalling, was first integrated into the canonical ABA signalling network by genetics and structural biology in Arabidopsis (Fujii et al., 2009). Although several studies previously reported that ABA plays an important role in strawberry fruit ripening (Jia et al., 2011) and PP2C may be involved in the fruit maturation based on the results of transcriptional analyses (Gambetta et al., 2010; Sun et al., 2011), the defined function of PP2C in fruit ripening remains unclear.
In the present study, it was found that the levels of FaABI1 mRNA expression decrease rapidly during strawberry fruit development (Fig. 2), suggesting that this PP2C protein may negatively regulate fruit development. Importantly, the down- and up-regulation of FaABI1 mRNA expression levels in DG strawberry fruit promotes and inhibits ripening, respectively (Fig. 3), indicating that the PP2C protein is a negative regulator of fruit ripening. Furthermore, this notion is also confirmed by transcriptional analysis of a set of both ABA signalling and ripening-related genes such as ABI3, SnRK2, ABRE1, PG1, PL1, and CHS in FaABI1 RNAi and OE fruit (Fig. 6). The analysis showed that the transcripts of the positive ABA signalling genes, including ABI3, SnRK2, and ABRE1, and firmness/pigment-related genes, including CHS and PG1, are up-regulated significantly in the RNAi fruit, in which the mRNA expression level of FaABI1 is down-regulated by 50%; while they are down-regulated remarkably in the OE fruit, in which the mRNA expression level of FaABI1 is up-regulated by 60% (Fig. 6). These results were consistent with a negative role for PP2C in ABA signalling during fruit ripening. Taken together, this study has provided physiological and molecular evidence to demonstrate that the type 2C protein phosphatase FaABI1 is a negative regulator of strawberry fruit ripening.
The receptor nature of FaPYR1 lays the molecular foundation for FaABI1 serving as a negative regulator in strawberry fruit ripening
In Arabidopsis, the ABA–PYR1–PP2C–SnRK2 core signalling pathway of ABA action has been established, by which ABA signal transduction consists of a double-negative regulatory mechanism, namely ABA-bound PYR inhibits PP2C activity and PP2Cs inactivate SnRK2s (Fujii et al., 2009). According to this ABA action model, in the absence of ABA, the signalling transduction is blocked by PP2C activity; in the presence of ABA, the signalling transduction is relayed by ABA-bound PYR-inhibited PP2C activity. In a previous study and the present study, it is shown that in green strawberry fruit, low ABA levels (Chai et al., 2011) and the resultant relatively high levels of PP2C expression might block ABA signalling transduction and, as a result, inhibit the burst of the ripening process at these stages. In red fruit, high ABA levels (Chai et al., 2011) and the resultant relatively low levels of PP2C expression might evoke ABA signalling transduction and thus promote the ABA-regulated ripening-related genes such as CHS, PG1, PL1, CHI, F3H, DFR, ANS, and UFGT, and finally lead to the fruit ripening.
The committed step of ABA action is initiated by ABA perception with its receptor. Thus, determination of the receptor nature of FaPYR1 contributes to understanding the negative mechanism of the FaABI1 protein. It was not only demonstrated previously that FaPYR is a positive regulator of strawberry fruit ripening (Chai et al., 2011), but the present study also validated that the binding of one ABA molecule with one purified FaPYR1 protein molecule was determined by stoichiometry (N) with a ratio of ~1:1 as determined by isothermal titration calorimetry analysis (Fig. 7), demonstrating that strawberry FaPYR1 is also an ABA receptor. It is interesting to note that Chai et al. (2011) found that down-regulation of FaPYR1 expression significantly promotes ABA accumulation in the white section of RNAi fruit, and the ABA increased by feedback results from the blocked FaPYR1 signalling. Similarly, in the present study, up-regulation of FaABI1 expression repressed ABA signalling in the white section of OE fruit, and consequently led to the feedback accumulation of ABA by an increase in NCED1 expression. These results were consistent with a positive and negative role for PYR1 and PP2C, respectively, in ABA signalling during fruit ripening. The positive role of FaPYR1 together with the negative role of FaABI1 in regulation of fruit ripening may suggest that ABA perception and signalling transduction underlying PYR1–PP2C–SnRK2 might be a core mechanism in non-climacteric fruit. Reconstitution of an ABA–FaPYR1–FaABI1–FaSnRK2 signalling pathway in vitro will be an important task for future studies.
Acknowledgements
We would like to thank Professor Liu Yu-le for pTRV vectors. This work was supported financially by the National Key Basic Research ‘973’ Program of China (grant no. 2012CB126306), the National Science Foundation of China (grant nos 30971977 and 31272144), and the Newstar of Science and Technology supported by Beijing Metropolis (grant no. Z111105054511048).
References
- Chai YM, Jia HF, Li CL, Dong QH, Shen YY. 2011. FaPYR1 is involved in strawberry fruit ripening. Journal of Experimental Botany 62, 5079–5089 [DOI] [PubMed] [Google Scholar]
- Cohen P, Cohen PT. 1989. Protein phosphatases come of age. Journal of Biological Chemistry 264, 21435–21438 [PubMed] [Google Scholar]
- Finkelstein R, Gampala S, Rock C. 2002. Abscisic acid signaling in seeds and seedlings. The Plant Cell 14, S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas S A, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature 391, 806–811 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Aatoni R, Park SY, Cutler SR, Sheen J, Rotrigues PL, Zhu JK. 2009. In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuleki T, Francis FJ. 1968. a Quantitative methods for anthocyanins. 1. Extraction and determination of total anthocyanin in cranberries. Journal of Food Science 33, 72–78 [Google Scholar]
- Fuleki T, Francis FJ. 1968. b Quantitative methods for anthocyanins. 2. Determination of total anthocyanin and degradation index for cranberry juice. Journal of Food Science 33, 78–82 [Google Scholar]
- Gambetta GA, Matthews MA, Shaghasi TH, McElrone AJ, Castellarin SD. 2010. Sugar and abscisic acid signaling orthologs are activated at the onset of ripening in grape. Planta 232, 219–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García MP, Rodríguez D, Nicolás C, Rodríguez PL, Nicolás G, Lorenzo O. 2003. Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiology 133, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J. 1999. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. The Plant Cell 11, 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E. 2003. Relay and control of abscisic acid signaling. Current Opinion in Plant Biology 6, 470–479 [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. 2007. Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends in Plant Science 12, 343–351 [DOI] [PubMed] [Google Scholar]
- Hu X, Liu L, Xiao B, Li D, Xing X, Kong X, Li D. 2010. Enhanced tolerance to low temperature in tobacco by over-expression of a new maize protein phosphatase 2C, ZmPP2C2. Journal of Plant Physiology 167, 1307–1315 [DOI] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY. 2011. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiology 157, 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HF, Zhu XQ, Jin XS, Shen YY. 2008. An effective method and its modifications for isolation of high-quality total RNA from fruit pulps. Journal of Agricultural Science and Technology 2, 58–62 [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. 1994. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264, 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J. 1998. Abscisic acid signal transduction. Annual Review of Plant Physiology and Plant Molecular Biology 49, 199–222 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. 1997. The Arabidopsis ABSCISIC ACIDINSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatase 2C involved in abscisic acid signal transduction. The Plant Cell 9, 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CL, Jia HF, Chai YM, Shen YY. 2011. Abscisic acid perception and signaling transduction in strawberry: a model for non-climacteric fruit ripening. Plant Signaling and Behavior 6, 1950 –1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Schiff M, Dinesh-Kumar SP. 2002. Virus-induced gene silencing in tomato. The Plant Journal 31, 777–786 [DOI] [PubMed] [Google Scholar]
- Lu G, Wang Y. 2008. Functional diversity of mammalian type 2C protein phosphatase isoforms: new tales from an old family. Clinical and Experimental Pharmacology and Physiology 35, 107–112 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068 [DOI] [PubMed] [Google Scholar]
- MacKintosh C, Coggins J, Cohen P. 1991. Plant protein phosphatase: subcellular distribution, detection of protein phosphatase 2C and identification of protein phosphatase 2A as the major quinate dehydrogenase phosphatase. Biochemical Journal 273, 733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKintosh RW, Davies SP, Clarke PR, Weekes J, Gillespie JG, Gibb BJ, Hardie DG. 1992. Evidence for a protein kinase cascade in higher plants. European Journal of Biochemistry 209, 923–931 [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, et al. 2009. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptors. Nature 462, 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Giraudat J. 1997. Genetic analysis of abscisic acid signal transduction. Plant Physiology 114, 751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. 2001. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signaling pathway. The Plant Journal 25, 295–303 [DOI] [PubMed] [Google Scholar]
- Meskiene I, Bogre L, Glaser W, Balog J, Brandstotter M, Zwerger K, Ammerer G, Hirt H. 1998. MP2C, a plant protein phosphatase 2C, functions as a negative regulator of mitogen-activated protein kinase pathways in yeast and plants. Proceedings of the National Academy of Sciences, USA 95, 1938–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. 1994. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana . Science 264, 1452–1455 [DOI] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, et al. 2009. Structural basis of abscisic acid signalling. Nature 462, 609–614 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. 2009. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326, 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salentijn EMJ, Aharoni A, Schaart JG, Boone MJ, Krens FA. 2003. Differential gene expression analysis of strawberry cultivars that differ in fruit-firmness. Physiologia Plantarum 118, 571–578 [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Márquez JA. 2009. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462, 665–668 [DOI] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, et al. 2010. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of transcription repressors to relieve ABA-responsive genes of inhibition. The Plant Cell 22, 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. 1998. Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proceedings of the National Academy of Sciences, USA 95, 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RD. 1996. Plant protein phosphatases. Annual Review of Plant Physiology and Plant Molecular Biology 47, 101–125 [DOI] [PubMed] [Google Scholar]
- Sopory SK, Munshi M. 1998. Protein kinases and phosphatases and their role in cellular signaling in plants. Critical Reviews in Plant Science 17, 245–318 [Google Scholar]
- Sun L, Sun Y, Zhang M, et al. 2012. Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiology 158, 283–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wang YP, Chen P, Ren J, Ji K, Li Q, Li P, Dai SJ, Leng P. 2011. Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. Journal of Experimental Botany 62, 5659–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei YZ, Hu FC, Hu GB, Li XJ, Huang XM, Wang HC. 2011. Differential expression of anthocyanin biosynthetic genes in relation to anthocyanin accumulation in the pericarp of Litchi chinensis Sonn. PLoS One 6, e19455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T, Wang D, Zhang S, Ehlting J, Ni F, Jakab S, Zheng C, Zhong Y. 2008. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics 9, 550 [DOI] [PMC free article] [PubMed] [Google Scholar]