Abstract
Phosphite (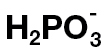 ) induces a range of physiological and developmental responses in plants by disturbing the homeostasis of the macronutrient phosphate. Because of its close structural resemblance to phosphate, phosphite impairs the sensing, membrane transport, and subcellular compartmentation of phosphate. In addition, phosphite induces plant defence responses by an as yet unknown mode of action. In this study, the acclimation of Arabidopsis thaliana plants to a sustained phosphite supply in the growth medium was investigated and compared with plants growing under varying phosphate supplies. Unlike phosphate, phosphite did not suppress the formation of lateral roots in several Arabidopsis accessions. In addition, the expression of well-documented phosphate-starvation-induced genes, such as miRNA399d and At4, was not repressed by phosphite accumulation, whilst the induction of PHT1;1 and PAP1 was accentuated. Thus, a mimicking of phosphate by phosphite was not observed for these classical phosphate-starvation responses. Metabolomic analysis of phosphite-treated plants showed changes in several metabolite pools, most prominently those of aspartate, asparagine, glutamate, and serine. These alterations in amino acid pools provide novel insights for the understanding of phosphite-induced pathogen resistance.
) induces a range of physiological and developmental responses in plants by disturbing the homeostasis of the macronutrient phosphate. Because of its close structural resemblance to phosphate, phosphite impairs the sensing, membrane transport, and subcellular compartmentation of phosphate. In addition, phosphite induces plant defence responses by an as yet unknown mode of action. In this study, the acclimation of Arabidopsis thaliana plants to a sustained phosphite supply in the growth medium was investigated and compared with plants growing under varying phosphate supplies. Unlike phosphate, phosphite did not suppress the formation of lateral roots in several Arabidopsis accessions. In addition, the expression of well-documented phosphate-starvation-induced genes, such as miRNA399d and At4, was not repressed by phosphite accumulation, whilst the induction of PHT1;1 and PAP1 was accentuated. Thus, a mimicking of phosphate by phosphite was not observed for these classical phosphate-starvation responses. Metabolomic analysis of phosphite-treated plants showed changes in several metabolite pools, most prominently those of aspartate, asparagine, glutamate, and serine. These alterations in amino acid pools provide novel insights for the understanding of phosphite-induced pathogen resistance.
Key words: Arabidopsis, phosphate, phosphate-starvation response, phosphite
Introduction
Phosphorus is an essential macronutrient for plants and is taken up and assimilated in the form of inorganic phosphate (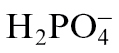 , Pi). The Pi concentrations in most soils are very low, and Pi availability is further decreased by immobilization, such as by the formation of insoluble complexes of Fe and Al oxides (Gerke, 1992). Plants have developed mechanisms to cope with Pi limitation. The inducible developmental and metabolic adaptations that enhance uptake and utilization are together referred to as the phosphate-starvation response (PSR) (Raghothama, 1999; Chiou and Lin, 2011). Uptake of Pi from the soil can be enhanced by secretion from the roots of organic acids, such as citrate or malate, and phosphatases to mobilize Pi from mineralized and organic forms (Duff et al., 1994; Ryan et al., 2001). Morphological alterations of root architecture, such as decreased primary root growth and increased lateral root and root-hair production, enhance the capacity to forage Pi from the richer topsoil layers (Lynch and Brown, 2001; Peret et al., 2011). Some plant species are able to generate specialized root structures composed of dense clusters of lateral roots or form associations with mycorrhizal fungi to form a symbiotic partnership to enhance Pi acquisition (Shane and Lambers, 2005; Bucher, 2007). The developmental and physiological processes controlled by Pi availability are under complex control through local and systemic signalling as well as external and internal Pi concentrations (Thibaud et al., 2010).
, Pi). The Pi concentrations in most soils are very low, and Pi availability is further decreased by immobilization, such as by the formation of insoluble complexes of Fe and Al oxides (Gerke, 1992). Plants have developed mechanisms to cope with Pi limitation. The inducible developmental and metabolic adaptations that enhance uptake and utilization are together referred to as the phosphate-starvation response (PSR) (Raghothama, 1999; Chiou and Lin, 2011). Uptake of Pi from the soil can be enhanced by secretion from the roots of organic acids, such as citrate or malate, and phosphatases to mobilize Pi from mineralized and organic forms (Duff et al., 1994; Ryan et al., 2001). Morphological alterations of root architecture, such as decreased primary root growth and increased lateral root and root-hair production, enhance the capacity to forage Pi from the richer topsoil layers (Lynch and Brown, 2001; Peret et al., 2011). Some plant species are able to generate specialized root structures composed of dense clusters of lateral roots or form associations with mycorrhizal fungi to form a symbiotic partnership to enhance Pi acquisition (Shane and Lambers, 2005; Bucher, 2007). The developmental and physiological processes controlled by Pi availability are under complex control through local and systemic signalling as well as external and internal Pi concentrations (Thibaud et al., 2010).
The PSR includes the transcriptional induction of genes such as high-affinity Pi transporters or phosphatases to increase Pi acquisition, remobilization from storage compounds such as phytate and phospholipids, and redistribution between tissues and/or subcellular compartments. At the post-transcriptional level, the miRNA399/PHO2/PHR1 regulon has been identified as an important mechanism to control Pi homeostasis. Under Pi deficiency, miRNA399 transcription is highly upregulated by the transcription factor PHR1 to direct the degradation of the mRNA for the E2 ubiquitin conjugase PHO2 (Bari et al., 2006; Chiou et al., 2006). The regulated degradation of PHO2 transcripts by miRNA399 is in turn attenuated by the regulatory RNAs IPS1 and At4 (Franco-Zorrilla et al., 2007). PHO2 controls Pi concentrations by regulating protein turnover through ubiquitination of a set of target proteins that include PHO1 and probably also Pi transporters (Liu et al., 2012). As a consequence, a pho2 mutant, as well as miRNA399 overexpressing Arabidopsis lines, hyperaccumulate Pi (Aung et al., 2006; Chiou et al., 2006). Furthermore, the phloem mobility of miRNA399 has been interpreted to indicate that miRNA399 is a signal in shoot-to-root communication (Pant et al., 2008). Transcriptional regulation has also been found for genes that are part of the acclimation to limited Pi supply, for example for SQD2 and NMT3 involved in the remodelling of membrane lipid composition (Essigmann et al., 1998; Hartel et al., 2000).
It is not clear how Pi levels are perceived in plants. In yeast, a phosphate transporter (PHO84) has been identified that signals Pi status through a conformational change induced by Pi binding that is independent of the transport of Pi across the membrane (Popova et al., 2010). A similar transceptor function has also been identified for the nitrate transporter NRT1.1/CHL1 in Arabidopsis, indicating that similar sensing mechanisms could exist for Pi in plants (Ho et al., 2009).
Post-translational regulation of the PSR involves metabolic adjustments to cope with the changed availability of Pi for enzymatic reactions and their regulation (Plaxton and Tran, 2011). Pi is a structural component of many organic components, such as DNA, RNA, phospholipids, and phosphorylated sugars. Therefore, Pi deficiency has profound effects on plant metabolism, such as leading to the accumulation of starch in the shoot (Ghosh and Preiss, 1966; Fliege et al., 1978). Another example is the remodelling of membrane lipid composition by replacing phospholipids with sulfo- and galactolipids to release Pi from this membrane component and conserve Pi (Essigmann et al., 1998; Hartel et al., 2000; Lambers et al., 2012). With Pi being a substrate in many more important pathways of primary metabolism, the metabolic pools of Pi in the various subcellular compartments need to be regulated, with the vacuole being the major storage compartment releasing Pi under a low external supply (Lee and Ratcliffe, 1993; Mimura et al., 1996; Danova-Alt et al., 2008; Pratt et al., 2009). Recent results from 31P-NMR studies gave estimates of subcellular Pi pools with concentrations in the range of 60–80 μM in the cytosol and 7mM in organellar mitochondrial and plastidal pools combined (Pratt et al., 2009). This same study provided evidence for considerable fluctuation of cytosolic but not of organellar concentrations under a Pi supply, which could be used to sense changes in cellular levels and trigger the signalling cascade of the PSR.
Phosphite (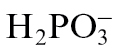 , Phi) is an analogue of phosphate in which a hydroxyl group is replaced by a hydrogen atom. It enters the cell via Pi transporters, therefore competing with Pi for uptake, and is mobile within the plant (Ouimette and Coffey, 1989; Carswell et al., 1996; Danova-Alt et al., 2008). On the other hand, Phi is used in the control of plant pathogens, especially oomycetes such as Phytophthora or Pythium species; it has been used for decades as the active agent in biostats such as Fosetyl-Al (Cohen and Coffey, 1986; Guest and Grant, 1991; Hardy et al., 2001). High concentrations of Phi directly inhibit growth of oomycete mycelia, and this has been attributed to interference of Phi with key reactions by competing with Pi for the enzyme catalytic sites, leading to altered abundances of metabolites such as ATP, NAD, polyphosphates, and pyrophosphate (Griffith et al., 1990; Barchietto et al., 1992; Niere et al., 1994). In addition to this direct effect of Phi on pathogens, it also activates plant defence responses (Molina et al., 1998; Friedrich et al., 2001; Eshraghi et al., 2011). This Phi-induced resistance depends on the signalling component NPR1 (non-expressor of PR1) and the accumulation of salicylic acid (Molina et al., 1998; Friedrich et al., 2001). Recently, it has been shown that Phi pre-treatment primes plants for a stronger defence response when subsequently challenged with Hyaloperonospora arabidopsidis and this priming involves the mitogen-activated protein kinase MPK4 (Massoud et al., 2012).
, Phi) is an analogue of phosphate in which a hydroxyl group is replaced by a hydrogen atom. It enters the cell via Pi transporters, therefore competing with Pi for uptake, and is mobile within the plant (Ouimette and Coffey, 1989; Carswell et al., 1996; Danova-Alt et al., 2008). On the other hand, Phi is used in the control of plant pathogens, especially oomycetes such as Phytophthora or Pythium species; it has been used for decades as the active agent in biostats such as Fosetyl-Al (Cohen and Coffey, 1986; Guest and Grant, 1991; Hardy et al., 2001). High concentrations of Phi directly inhibit growth of oomycete mycelia, and this has been attributed to interference of Phi with key reactions by competing with Pi for the enzyme catalytic sites, leading to altered abundances of metabolites such as ATP, NAD, polyphosphates, and pyrophosphate (Griffith et al., 1990; Barchietto et al., 1992; Niere et al., 1994). In addition to this direct effect of Phi on pathogens, it also activates plant defence responses (Molina et al., 1998; Friedrich et al., 2001; Eshraghi et al., 2011). This Phi-induced resistance depends on the signalling component NPR1 (non-expressor of PR1) and the accumulation of salicylic acid (Molina et al., 1998; Friedrich et al., 2001). Recently, it has been shown that Phi pre-treatment primes plants for a stronger defence response when subsequently challenged with Hyaloperonospora arabidopsidis and this priming involves the mitogen-activated protein kinase MPK4 (Massoud et al., 2012).
The chemical resemblance of Phi to Pi has made it an attractive tool to investigate Pi sensing. Phi is metabolically inert in plants, does not enter the organic P pool and thus accumulates in the tissues (Carswell et al., 1996, 1997; Danova-Alt et al., 2008; Berkowitz et al., 2011). The treatment of severely Pi-starved plants with Phi suppresses the activation of many PSR genes and it is therefore assumed that Phi is recognized by the same sensing mechanism as Pi. The Phi-dependent downregulation of the PSR then leads to an accentuation of Pi deficiency, for example by decreasing Pi uptake capacity. This interference with the Pi regulatory network has been suggested to cause the plant growth-inhibiting effect of Phi (Carswell et al., 1996, 1997; Ticconi et al., 2001; Varadarajan et al., 2002). 31P-NMR studies using tobacco BY2 cells have shown that Phi accumulates in the cytoplasm of P-deprived cells and that resupply of Pi leads to efflux of Phi from the cell. In Pi-sufficient cells, however, Phi accumulates in the vacuole, illustrating the complex interactions of Pi and Phi with respect to cell compartmentation, sensing, and physiology (Danova-Alt et al., 2008).
To understand the specific effects of Phi on plant growth and metabolism, it is necessary to compare directly the effects of Phi-treated plants with plants under Pi limitation. In this study, Arabidopsis plants grown on medium containing Phi and Pi in varying ratios but at a constant combined concentration were investigated. Growth parameters, root development, and gene expression patterns of Phi-treated plant were compared with those of plants grown with various Pi supplies. In addition, Phi-induced metabolic adjustments with potential relevance for plant defence mechanisms were identified.
Materials and methods
Plant material
Seeds of accessions of Arabidopsis thaliana (L.) Heynh. used in this study were obtained from the European Arabidopsis stock centre (Col-0, N1093) and the Versailles Resource Centre (Col-4, AV187; Jea, AV25; Nd-1, AV220; Sha, AV236; Tsu-1, AV239). Arabidopsis plants were grown on a modified MS medium (Murashige and Skoog, 1962) in a vertical plate system for 10 d. The medium contained 2mM NH4NO3, 1.5mM CaCl2, 0.5mM MgSO4, 3mM KNO3, 3mM MES (pH 5.8), 0.5% (w/v) sucrose and 0.8% (w/v) agar. The agar contributed ~10 μM Pi. Phosphate was added as KH2PO4 (pH 5.8). KCl was substituted for KH2PO4 to maintain a constant osmotic potential. A freshly prepared 0.1M potassium phosphite stock solution was prepared from phosphorous acid by adjusting to pH 5.8 with KOH and was added to the medium as required. Micronutrients were used according to Murashige & Skoog (1962) and obtained from Austratec (Bayswater, Australia). After sterilization [2min in 70% (v/v) ethanol, 5min in 5% (v/v) sodium hypochlorite], the seeds were resuspended in 0.1% (w/v) agarose and stratified for 2 d at 4 °C. The seeds were transferred to 10×10cm square plates, which were then sealed with microporous tape, and the plants were grown in a controlled-environment growth chamber with a 10h photoperiod at ~100 μmol m–2 s–1. For metabolomic analyses, plants were grown for 3 weeks in horizontal 90mm Petri dishes sealed with microporous tape under the same growth conditions as described above. For analysis of growth responses, all experiments were repeated in five independent replicates consisting of eight seedlings each.
Phosphate and phosphite quantification
Phosphate and phosphite were extracted from ~10mg of shoot or root tissue with 1 % (v/v) acetic acid at 1:10 (w/v) using a tissue lyser (Qiagen, Doncaster, Australia) for 1min at 25 Hz. After clearing the lysate by centrifugation at 14 000g and 4 °C for 15min, the supernatant was assayed for phosphate and phosphite as described previously (Ames, 1966; Berkowitz et al., 2011).
Root growth measurements
For measurements of root growth, the position of the root tip was marked every 24h for the 10-day growth period. Plates were then scanned for analysis of root length, growth rate, and lateral root number using ImageJ software with the NeuronJ plug-in (Meijering et al., 2004).
Gene expression analyses
For each treatment, RNA was extracted from five independent biological replicate samples and reverse transcribed using Plant RNA Mini and Tetro cDNA synthesis kits according to the manufacturer’s instructions (Bioline, Alexandria, Australia). Quantitative real-time PCR on a Rotor Gene 6000 platform (Qiagen) was performed with 2ng of cDNA and a SensiFast SYBR No-ROX kit (Bioline) as described previously (Jost et al., 2007). Relative expression levels were calculated using the comparative ΔΔC T method (Livak and Schmittgen, 2001) and normalized to the internal reference genes PDF2, UBC9 and UPL7 (Czechowski et al., 2005). For evaluation of reference genes and amplification efficiencies, the geNORM and LinRegPCR algorithms, respectively, were used (Vandesompele et al., 2002; Ramakers et al., 2003). Primers were designed using Primer Express software (Applied Biosystems, Mulgrave, Australia) and the QuantPrime tool (Arvidsson et al., 2008). Primer sequences and Arabidopsis Genome Initiative (AGI) gene identifiers are given in the supplementary data (Table S1 at JXB online).
Metabolomic analysis by gas chromatography–mass spectrometry (GC-MS)
Polar metabolites were extracted from shoot tissue harvested from eight biologically independent experiments 3h after the onset of light and immediately frozen in liquid nitrogen. After homogenization in a tissue lyser (Tissue Lyser II; Qiagen), 30mg of frozen tissue powder was extracted with 500 μl extraction solution (20:3 methanol:water including 10 μg ml–1 of ribitol as an internal standard) for 20min at 75 °C with shaking at 1200rpm in a thermomixer (Thermo mixer compact; Eppendorf, North Ryde, Australia). After centrifugation at 16 100g for 3min at room temperature, 60 μl of the supernatant was dried under vacuum and the polar metabolites redissolved by incubation in 20 μl of methoxyamine hydrochloride (20mg ml–1) in pyridine at 37 °C with shaking at 750rpm for 90min. For derivatization, 30 μl of N-methyl-N-(trimethylsilyl)trifluoroacetamide was added and the mixture was incubated at 37 °C with shaking at 750rpm for 30min. A retention time standard (10 μl) containing 140 nl ml–1 each of n-dodecane, n-pentadecane, n-nonadecane, n-docosane, n-octacosane, n-dotriacontane and n-hexatriacontane in pyridine was added to each sample. GC-MS analysis was performed by injecting 1 μl of derivatized sample using the splitless mode on an Agilent 7890GC/5975MSD system (Agilent, Mulgrave, Australia) equipped with a Varian Factor 4 column and operated in scan mode (range: 50–650 m/z). Metabolites were identified by comparing retention times and ionization patterns with standards and the National Institute of Standards and Technology mass spectral library. The integrated signal of the total ion current for each peak was compared between samples after normalization. Metabolites were normalized first using the internal standard and then the sample weight.
Statistical analyses
For statistical analyses, the SPSS Statistics Base software package (version 17; IBM, New York, USA) was used. Analysis of variance with a Tukey HSD test was performed for multiple comparisons.
Results
Growth responses to sustained Phi supply
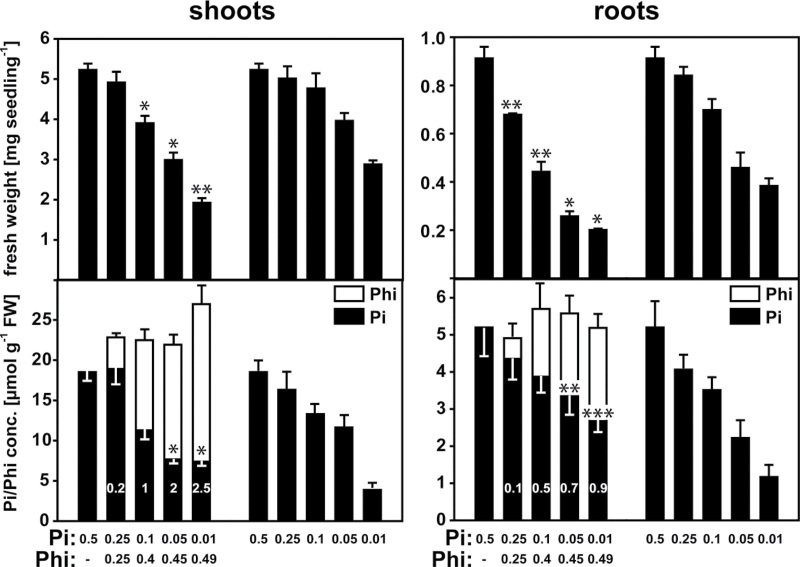
Because detailed reports on the acclimation of plants under sustained Phi supply that also take into account internal Pi and Phi concentrations are lacking, this study analysed the responses of Arabidopsis Col-0 seedlings to varying Pi and Phi concentrations in the growth medium at a constant combined total concentration of 0.5mM. Seeds were germinated directly on medium containing differing Phi:Pi ratios ranging from 1:1 to 50:1. Growth was compared with that of control seedlings grown on medium containing Pi only at concentrations over the same range (Fig. 1, Table 1). For the Phi-treated seedlings, the Pi and Phi concentrations of the shoots and roots varied according to the external supply. Interestingly, the total concentration of Pi+Phi in both tissues was remarkably constant, despite the substantial variation in the concentrations of the two anions in the growth medium (Fig. 1).
Fig. 1.
Fresh weights and Pi and Phi concentrations of Arabidopsis (Col-0) shoots and roots under varying Pi and Phi supplies. Plants were grown in a vertical plate system for 10 d before harvesting the shoot and root tissues. Numbers in bars represent the corresponding ratios of Phi and Pi concentrations in the tissue. For each sample, eight plants grown on one plate were pooled, and for each treatment five independent biological replicates were performed (n=5 pools of eight per treatment). Results are given as means ±standard error (SE). Significant differences between Phi treatments and their corresponding controls grown on the same Pi concentration are indicated (ANOVA Tukey: *P <0.05; ** P < 0.01; *** P < 0.001).
Table 1.
Comparison of fresh weights and Pi/Phi tissue concentrations of Phi-treated and non-treated plants of Arabidopsis thaliana (see also Fig. 1)Fresh weights (FW) and Pi tissue concentrations of Phi-treated plants are expressed relative to Pi-sufficient plants grown on 0.5mM Pi or control plants with the same Pi concentration in the medium but lacking Phi. Results are given as means of five independent biological replicates with eight seedlings each.
| Leaves | Roots | |||||||
|---|---|---|---|---|---|---|---|---|
| % of 0.5mM Pi | % of Pi control | % of 0.5mM Pi | % of Pi control | |||||
| Pw/Phi treatment | FW | [Pi] | FW | [Pi] | FW | [Pi] | FW | [Pi] |
| 0.25mM/0.25 mM | 93 | 88 | 98 | 115 | 74 | 78 | 80 | 107 |
| 0.1mM/0.4 mM | 74 | 71 | 81 | 85 | 48 | 67 | 63 | 110 |
| 0.05mM/0.45 mM | 57 | 62 | 75 | 66 | 28 | 42 | 56 | 151 |
| 0.01mM/0.49 mM | 36 | 20 | 66 | 190 | 21 | 22 | 52 | 229 |
Shoot fresh weights for seedlings grown on 0.25mM Pi+0.25mM Phi did not differ significantly from seedlings grown on 0.25mM Pi alone. Quantification of Pi and Phi tissue concentrations revealed that the seedlings supplied with Phi had the same shoot Pi concentration as plants supplied with Pi but not Phi. However, despite the equimolar supply of Pi and Phi, the Phi accumulation was only 20% of that of Pi (Fig. 1). Higher Phi supplies led to a decrease in shoot biomass that was accentuated compared with that of control seedlings grown at the same external Pi. Interestingly, the seedlings treated with 0.1mM Pi+0.4mM Phi and 0.05mM Pi had equal shoot Pi concentrations and similar shoot fresh weights. Thus, for this Phi treatment, the reduced biomass was explained mainly by a reduction in the Pi concentrations in the shoot tissue, and the accumulation of Phi seemed to have little direct effect on shoot biomass. However, the seedlings that contained the highest concentration of Phi in the shoots had an accentuated reduction in shoot biomass compared with seedlings grown on the lowest Pi supply. Surprisingly, in the two treatments containing 0.01mM Pi, the seedlings that were supplied with Phi showed a higher Pi concentration in the shoots.
Phosphite accumulation had a more pronounced effect on decreasing the fresh weight of the roots than on that of the shoots (Fig. 1, Table 1), which resulted in an increase in shoot:root ratio (Fig. S1 at JXB online). Across all treatments, seedlings grown on Phi-containing medium had a lower root biomass than seedlings that had a similar Pi tissue concentration in the absence of Phi. For example, the roots of seedlings exposed to the 0.05mM Pi+0.45mM Phi and the 0.1mM Pi treatments showed similar Pi concentrations, but the Phi-treated seedlings had a much lower root biomass. The Phi concentration in the roots was only comparable to the Pi concentration when Phi was in a ten- to 50-fold external excess (Fig. 1), and was only equimolar in the 0.01mM Pi+0.49mM Phi treatment. At this highest Phi:Pi ratio, the root fresh weight was reduced by about 80% compared with seedlings in the 0.5mM Pi treatment, whilst the shoot fresh weight was only reduced by ~25% (Table 1).
Across all Phi treatments tested, the Phi:Pi tissue ratio was consistently about twofold higher in the shoots than in the roots, indicating preferential transport and accumulation of Phi in the shoot. However, this did not lead to an accentuated impact on shoot biomass, as the relative decrease in biomass with increasing external Phi was higher in the roots. This discrepancy might indicate a higher Phi tolerance of the shoot tissue. A surprising observation was the consistently higher Pi concentration in roots of Phi-treated seedlings compared with seedlings grown at the same Pi supply. A similar effect was observed for shoot tissue but only at the highest external Phi:Pi ratios.
Phosphite does not suppress increases in lateral root density (LRD) under Pi limitation
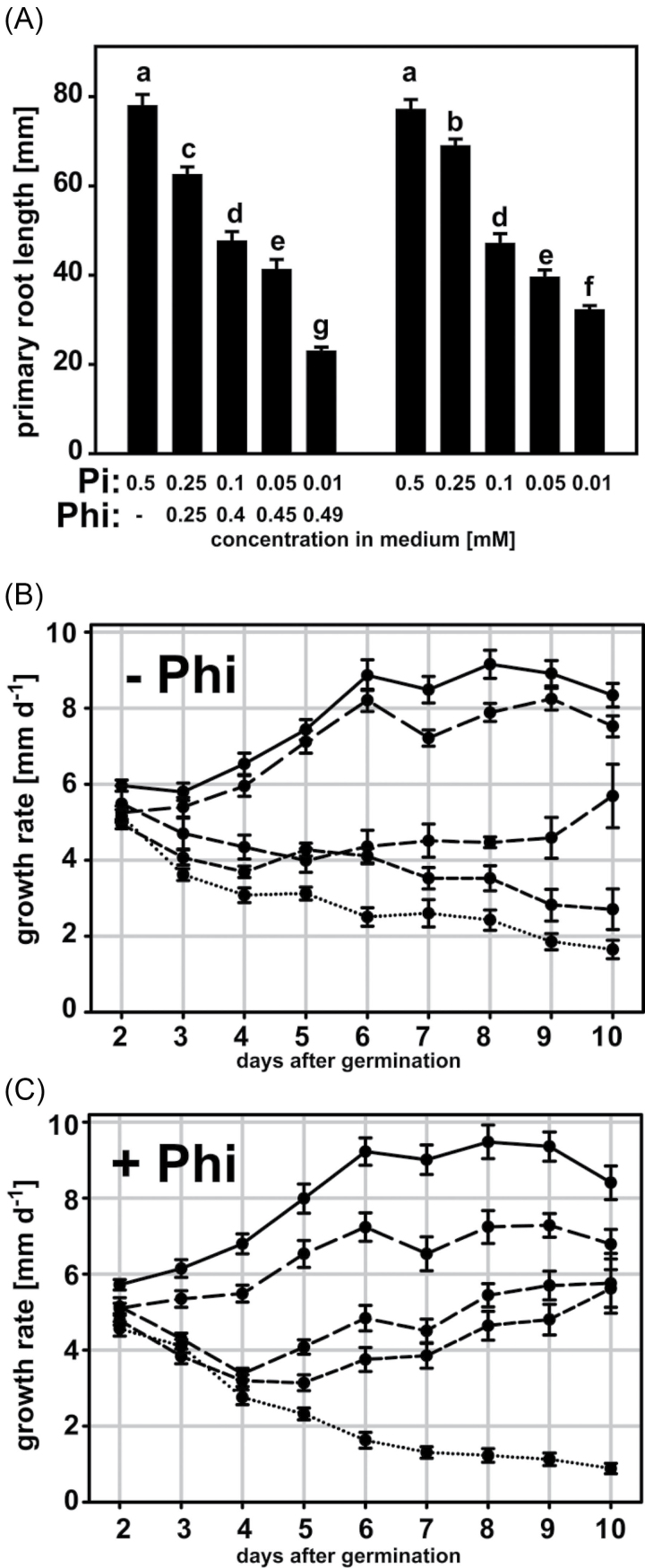
Next, the effect of Phi on root development under our growth conditions was examined. Plants grown at lower Pi supplies showed the characteristic reduction in primary root length (Fig. 2A), as reported previously (Peret et al., 2011). Phi only had an effect on primary root length at the highest concentration tested, with a 68% reduction in primary root length compared with seedlings grown at the same external Pi supply (Fig. 2A). The primary root growth rate in the 0.25mM Phi treatment was reduced by ~1mm d–1 from 4 d after germination compared with the control (Fig. 2B, C). Interestingly, primary roots exposed to either 0.1mM Pi+0.4mM Phi or 0.05mM Pi+0.45mM Phi initially had decreasing growth rates, which recovered from 4 d after germination and increased steadily for the rest of the experiment, leading to root lengths comparable to roots grown on the same Pi but without Phi in the medium. Only primary roots of seedlings growing at the lowest Pi supplies had a constant deceleration in growth that ultimately caused the shortest primary roots across treatments. This effect was stronger in the presence of Phi.
Fig. 2.
Lengths and growth rates of Arabidopsis (Col-0) primary roots in response to external Phi and Pi supply. (A) Primary root lengths of plants grown in a vertical plate system for 10 d with varying Phi and Pi concentrations in the growth medium. Means with different letters are significantly different (ANOVA Tukey, P <0.05). (B, C) Root growth rates for plants grown with decreasing Pi (B) and variation of Phi:Pi ratios (C) in the medium. Data points are means ±SE of five biological replicates with eight seedlings each. Medium concentrations in (B) and (C): solid line, 0.5mM Pi; long-dashed line, 0.25mM Pi (+0.25mM Phi); medium-dashed line, 0.1mM Pi (+0.4mM Phi); short-dashed line, 0.05mM Pi (+0.45mM Phi); dotted line, 0.01mM Pi (+0.49mM Phi).
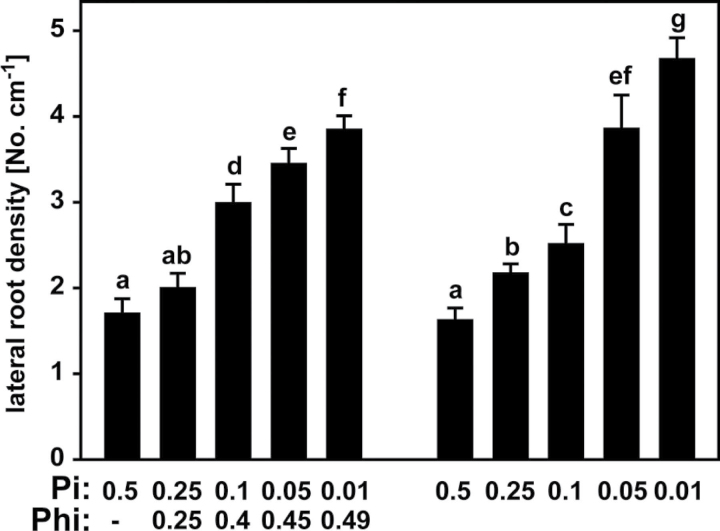
LRD (the number of lateral roots per cm of primary root) for seedlings grown in the absence of Phi increased with decreasing Pi supply, as expected (Fig. 3). Intriguingly, Phi treatment had no effect on LRD at the lowest Phi:Pi ratio tested, whilst plants growing on 0.1mM Phi+0.4mM Pi had a higher LRD than plants on 0.1mM Pi alone (Fig. 3). This was surprising, as the combined total concentration of Pi+Phi in the root and shoot tissues was unchanged, and also across all treatments the combined concentration of Pi and Phi in the growth medium was constant at 0.5mM (Fig. 1). Thus, Phi did not seem to have a suppressing effect on LRD similar to that of Pi. Only for the two highest Phi concentrations was the increase in LRD reduced compared with the corresponding control seedlings, but these Phi-treated seedlings also had a higher Pi concentration than the control seedlings grown on the same Pi concentration in the absence of Phi (Fig. 1). Therefore, even at these higher concentrations, Phi had very little impact on suppressing lateral root formation, and instead had rather an indirect effect leading to elevated Pi in the root tissue, which subsequently resulted in a decreased LRD when compared with the corresponding control plants.
Fig. 3.
LRD of Arabidopsis (Col-0) plants grown on medium with different Phi and Pi concentrations (mM). Primary root lengths and the numbers of lateral roots (>100 μm) were determined from five biological replicates with eight seedlings each. Results are given as means ±SE. Means with different letters are significantly different (ANOVA Tukey, P <0.05).
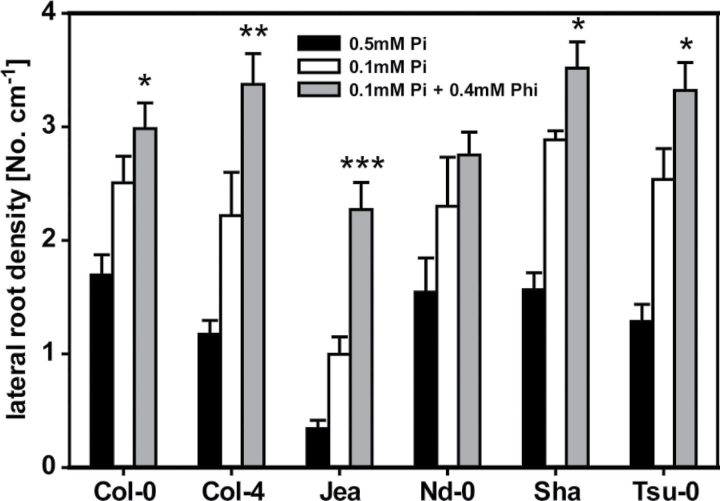
As a first step to analyse the natural genetic variation of Arabidopsis accessions in the response to Phi, the LRD of another five accessions in addition to the Col-0 accession used in our other experiments was analysed. All of these accessions showed an increase in LRD on 0.1mM Pi when compared with growth on 0.5mM Pi (Fig. 4). Interestingly, we observed an even higher LRD for plants grown on 0.1mM Pi+0.4mM Phi, especially for the accessions Col-4, Jea, Sha and Tsu-0 with a 1.5- to twofold increase in LRD, and a less pronounced increase for Col-0 and Nd-0. Thus, Phi did not suppress lateral roots in a similar fashion to Pi but instead led to an accentuated increase in LRD.
Fig. 4.
Comparison of LRD for six Arabidopsis accessions grown on growth medium containing 0.5mM Pi, 0.1mM Pi, or 0.1mM Pi+0.4mM Phi. For each accession, at least 16 seedlings per treatment were analysed (four independent replicates with four seedlings each). Results are given as means ±SE. Statistically significant differences between the 0.1mM Pi and the 0.1mM Pi+0.4mM Phi treatments are indicated (ANOVA Tukey, *P < 0.05; **P < 0.01; ***P < 0.001).
Gene expression analyses
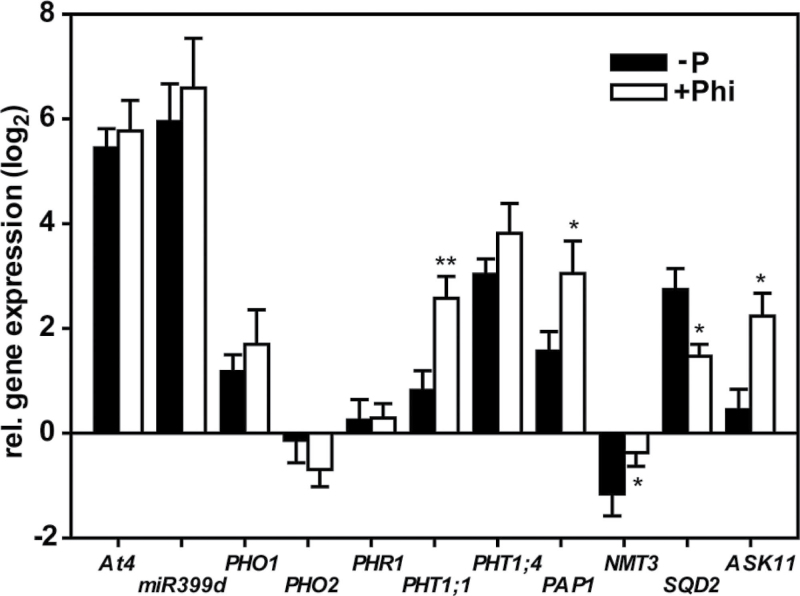
As Phi was not able to suppress an increase in LRD under Pi limitation, a known PSR, the effects of Phi treatment on the expression of genes involved in the PSR and others associated with Pi metabolism were examined next. For these experiments, shoots from seedlings exposed to 0.1mM Pi+0.4mM Phi and 0.05mM Pi were compared. These two shoot sets had comparable Pi concentrations, which were lower than those of seedlings supplied with 0.5mM Pi (Fig. 1). The shoots of these Phi-treated seedlings also accumulated similar concentrations of both Phi and Pi. The transcript levels for a set of genes involved in the primary Pi regulatory network (At4, miRNA399d, PHR1, PHO1, and PHO2) and downstream genes involved in the adaptation to low-Pi levels (PHT1;1, PHT1;4, PAP1, NMT3, SQD2, and ASK11) were quantified. As expected, transcripts for the well-documented Pi-starvation-induced genes At4 and miRNA399d showed strong induction (~60-fold) under Pi limitation (Fig. 5). Transcripts of PHT1;1, PHT1;4, SQD2, PAP1 and PHO1 showed a milder induction (between 2.5- and eightfold), whilst NMT3 expression was repressed (approx. twofold). These results confirmed that these seedlings were sensing and responding to the reduced availability and tissue accumulation of Pi (Bari et al., 2006; Chiou et al., 2006). Surprisingly, At4, miRNA399d, PHO1 and PHT1;4 transcripts had a very similar response in the Phi-treated seedlings (Fig. 5). The expression of these genes was not affected by the substantial accumulation of Phi in the leaf tissue. Thus, under these growth conditions, seedlings were still able to sense and respond to the depletion of Pi and this was not suppressed by Phi. However, Phi suppressed the induction and repression of genes important for the remodelling of membrane lipid composition, SQD2 and NMT3, respectively. By contrast, the observed induction in gene expression of PHT1;1, PAP1 and ASK11 under Pi limitation was accentuated in the presence of Phi. These results demonstrated a multifaceted impact of Phi on gene regulation beyond the direct mimicking of Pi in sensing mechanisms.
Fig. 5.
Gene expression analysis for leaf tissue of 10-d-old Arabidopsis (Col-0) seedlings. Shown are quantitative RT-PCR results for Pi-deficient plants grown on 0.05mM Pi (–P) and 0.1mM Pi+0.4mM Phi (+Phi). For both treatments, expression levels are given relative to control plants with a sufficient Pi supply (0.5mM Pi) and normalized to the reference genes PDF2, UBC9 and UPL7. Results are given as means ±SE (n=5). Statistically significant differences between the –P and +Phi treatments are indicated (ANOVA Tukey, *P < 0.05; **P < 0.01).
Metabolic changes induced by phosphite treatment
Current evidence suggests that plants do not assimilate Phi into organic compounds and that oxidation of Phi to Pi as occurs in microbes is unlikely (Carswell et al., 1997; Danova-Alt et al., 2008; Berkowitz et al., 2011). However, this does not rule out the possibility that Phi affects metabolism by mimicking Pi, for example by competing for catalytic sites or in the allosteric regulation of enzymatic reactions.
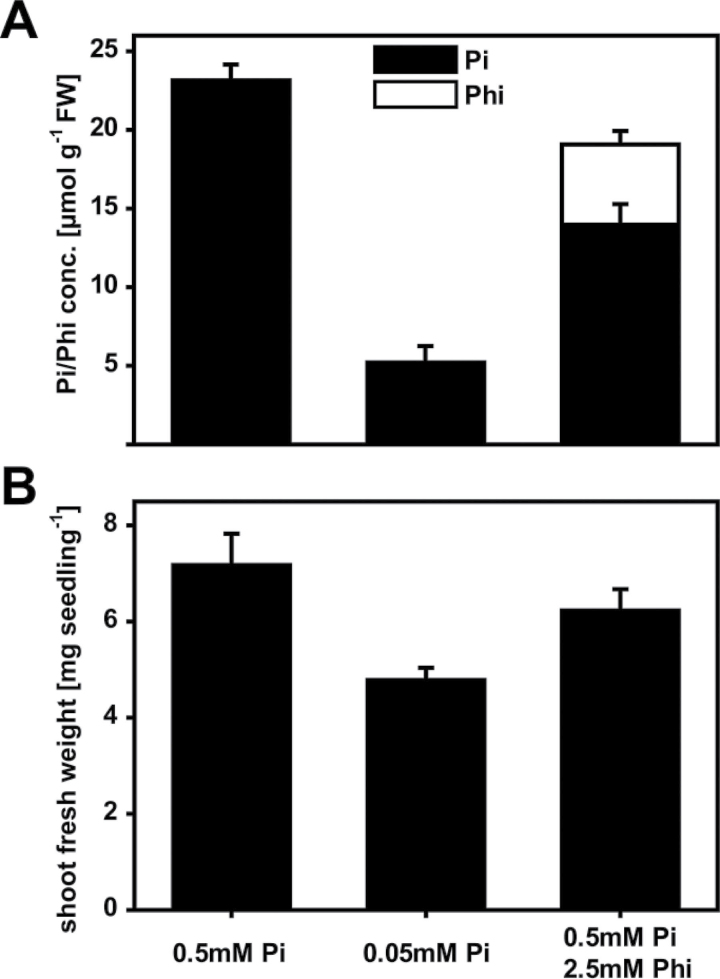
An untargeted approach to measure metabolite levels in Phi-treated and untreated control plants by GC-MS was carried out to probe for Phi-induced metabolomic changes. To differentiate between Pi-dependent and Phi-specific differences in metabolite pools, control plants were supplied with either high (0.5mM) or low (0.05mM) Pi in the growth medium. Accordingly, these growth conditions resulted in high and low Pi concentrations, respectively, in the harvested shoot tissues after 3 weeks of growth (Fig. 6A). In parallel, and on the basis of earlier experiments, the Phi treatment was exposure of plants to 0.5mM Pi+2.5mM Phi in the growth medium. This treatment yielded plants that had accumulated shoot Pi to concentrations that were intermediate compared with those obtained in the shoots of the control plants exposed to either high or low Pi supply. The Phi concentrations at the time of harvest were below the threshold where Phi indirectly affects shoot growth. The fresh weight of the Phi-treated plants was reduced by only 13% compared with the high-Pi-treated plants, whereas the fresh weight of plants supplied with low Pi was reduced by 36% (Fig. 6B). The plants exposed to low Pi, however, had not accumulated anthocyanins (data not shown) and were therefore not severely P limited. This conclusion was supported by the still substantial Pi concentration in the shoots. Therefore, after being exposed to either Phi or low Pi, these plants were not severely P limited, and any differences in metabolite abundance were expected to represent metabolic adjustments rather than a stress response brought on by severe Pi deficiency.
Fig. 6.
Metabolomic profiling. Arabidopsis plants (Col-0) were grown for 3 weeks on three different growth media to allow comparison of Phi-treated (0.5mM Pi+2.5mM Phi) and Pi-limited (0.05mM Pi) plants with Pi-sufficient (0.5mM Pi) control plants. (A) Shoot Pi and Phi concentrations. (B) Shoot fresh weights. Results are given as means ±SE from eight independent biological replicates.
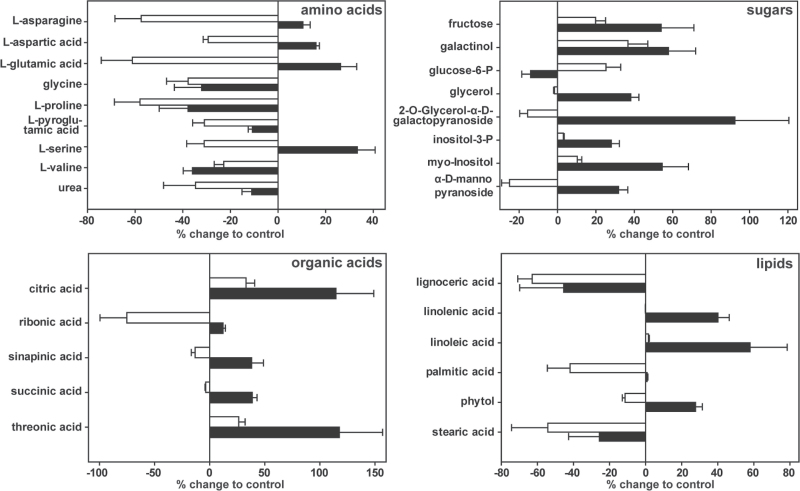
In total, we were able to robustly quantify 112 polar metabolites by GC-MS. Of these, 84 metabolites could be identified through peak comparison with mass spectra libraries. For quantitative comparison of the metabolites present in the shoots of plants exposed to the low-Pi and Phi treatments, we normalized the data to the metabolite levels obtained from shoots of plants supplied with high (0.5mM) Pi. A total of 28 metabolites showed a significant difference (ANOVA Tukey, P <0.05) in abundance of more than 20% in at least one comparison with the control (Fig. 7).
Fig. 7.
Effect of Phi accumulation and Pi limitation on levels of polar metabolites in 3-week-old Arabidopsis (Col-0) shoots. Changes in metabolite pools of Phi-treated (0.5mM Pi+2.5mM Phi, open bars) and Pi-limited (0.05mM Pi, filled bars) plants are given in as percentages of the results for the Pi-sufficient (0.5mM Pi) control plants. Shown are only metabolites identified by GC-MS analysis as having a greater than 20% change for at least one comparison are shown (ANOVA Tukey, P <0.05). Results are given as means ±SE of eight independent biological replicates per treatment.
The most prominent group of primary metabolites for which differences across treatments were observed were the amino acids asparagine, glutamic acid, aspartic acid, and serine, which showed contrasting changes in the two treatments. The abundance of each of these amino acids was 40–60% lower in the shoots of the Phi-treated plants, but slightly higher (10–20%) in the shoots of plants exposed to low Pi when compared with the control plants.
The largest differences in metabolite abundance in the shoots of plants exposed to low Pi were found for the organic acids citrate and threonate, which were about twofold more abundant than in control tissues. For the Phi treatments, both metabolites were about 25% more abundant. Organic acids such as citrate have frequently been found to be elevated under P deficiency, probably to increase pools to allow for excretion and improve Pi uptake from the soil (Koyama et al., 2000; Narang et al., 2000). An increase in myo-inositol and inositol-3-phosphate in the low-Pi samples and to a lesser degree for Phi treatments might indicate the mobilization of Pi from, or a reduced flux into, storage pools. Both these metabolites are intermediates in the biosynthesis and degradation of the phosphate-storage compound phytate (inositol hexakisphosphate) (Raboy, 2003). Similarly, replacement of phospholipids by galacto- and sulfolipids is an important means of releasing Pi from membrane lipids and remodelling plant lipid composition by providing precursors for lipid biosynthesis (Essigmann et al., 1998; Hartel et al., 2000; Li et al., 2006). Correspondingly, we were able to observe changes for the major fatty acids linolenic and linoleic acid. For both, levels decreased in the low-Pi but not in the Phi-treated samples. Palmitic acid on the other hand shows a decreased abundance in the Phi-treated shoots only, whilst stearic acid levels dropped in both treatments. A slight decrease in glucose-6-phosphate in the Pi-limited plants and an increase in the Phi treatments was also detected. Several metabolomic studies have reported decreases in levels of phosphorylated sugars under P deficiency (Rychter and Randall, 1994; Morcuende et al., 2007; Huang et al., 2008).
Overall, 11 and one metabolite(s) were specifically more abundant in the low-Pi and Phi treatments, respectively, whilst four metabolites increased in abundance in both treatments (Fig. S2A at JXB online). For the Phi-treated plants, nine metabolites, most notably the central amino acids asparagine, aspartic acid, serine, and glutamic acid and its degradation product pyroglutamic acid were less abundant and six metabolites differed in their abundance between both treatments (Fig. S2A).
Discussion
Impact of phosphite accumulation on plant growth
Understanding the consequences of Phi accumulation in plants is complicated by the different molecular processes that Phi may impact on and their possible interactions. Several studies have reported an inhibitory effect of Phi on the growth of plants (Carswell et al., 1996, 1997; Ticconi et al., 2001; Varadarajan et al., 2002; Thao et al., 2008a, b). However, knowledge on the cause of the inhibitory effect of Phi on plant growth is limited, and detailed studies on the interaction of Pi and Phi on root architecture are not available. Phi competes with Pi for uptake by the same membrane transport system and should therefore, at least at high external concentrations, reduce the amount of Pi entering the cells and lead to Pi depletion of the plant (Carswell et al., 1996; Danova-Alt et al., 2008). In addition, Phi influences subcellular compartmentation of Pi (Danova-Alt et al., 2008; Pratt et al., 2009) and both processes will impinge on Pi homeostasis with downstream effects on Pi-dependent metabolism. At the same time, the pseudo-pyramidal Phi closely resembles tetrahedral Pi in its geometry, which suggests a potential for Phi to mimic Pi and impair Pi-sensing mechanisms or Pi-dependent enzyme reactions and their allosteric regulation. In this study, a constant combined tissue concentration of Phi+Pi in Phi-treated plants across a wide range of external concentration ratios was observed, suggesting a mechanism integrating Pi and Phi concentrations and regulating the homeostasis in both shoot and root tissues. Intriguingly, a higher Pi concentration in roots of seedlings grown in the presence of Phi was detected when compared with those grown on the same medium lacking Phi. A correspondingly higher shoot Pi concentration with Phi treatment was apparent only for the highest Phi supply. As plants are unable to oxidize Phi to Pi and the shoot accumulated Phi to a higher extent than the roots, this increased root Pi concentration is probably caused by an increased uptake of Pi from the medium and/or retention in the root. As the expression of miRNA399d was not suppressed by Phi, the uptake of Pi by high-affinity transporters might not be downregulated under sustained Phi treatments. Inhibition of Pi assimilation into the organic P pool as a consequence of Phi accumulation could also lead to increased Pi concentrations in the roots. This could occur either through forcing more Pi into the vacuolar storage pool, thereby decreasing Pi availability in the cytosol and organelles, or by directly inhibiting metabolic processes. These results highlight the necessity to determine Phi and Pi concentrations in the tissues to be able to differentiate between direct effects of Phi, for example on the PSR or metabolism, and indirect effects on Pi homeostasis.
Suppression of the PSR by phosphite
The perception of Phi by the plant has the potential to suppress the PSR. In support of this hypothesis, several studies have shown the capacity of Phi to suppress the PSR in severely Pi-starved plants when transferred to medium containing high concentrations of Phi, for example suppression of Pi-transporter genes and the inhibition of root-hair growth or anthocyanin accumulation (Carswell et al., 1996; Ticconi et al., 2001; Varadarajan et al., 2002). Another characteristic developmental pattern influenced by Pi availability is the alteration of root architecture as part of the PSR. Plants respond to reduced Pi supply by slowing primary root growth and increasing LRD, which is regulated locally by the external Pi concentration (Thibaud et al., 2010). Surprisingly, under acclimation to Phi, seedlings showed an increase in LRD comparable to Pi-deficient plants in this study. This was the case even though the combined concentrations of both anions in the external medium were constant and overall tissue concentrations were also the same, i.e. there were no changes in external or internal [Pi+Phi] across all treatments. These results suggest that plants are able to differentiate between the two anions at least to some extent and sense the depletion of Pi even in the presence of Phi. This was not only the case for the accession Col-0 but also for several other Arabidopsis accessions we tested for changes in LRD, which responded even more strongly. Similar studies have also shown a varying sensitivity of Arabidopsis accessions to Pi deficiency (Chevalier et al., 2003; Reymond et al., 2006).
In addition, our gene expression studies also indicated that plants accumulating Phi can still sense Pi deficiency. Pi-starvation-induced (PSI) genes of the primary regulatory network (At4, miRNA399d, PHO1, and PHT1;4) remained upregulated, although Phi accumulation resulted in a combined Pi+Phi tissue concentration that was comparable to plants on a high Pi supply. By contrast, several other studies have reported repression of several PSI genes (e.g. At4, PAP1, PHT1;1 and IPS1) by Phi treatment in severely Pi-starved plants (Ticconi et al., 2001; Varadarajan et al., 2002; Ribot et al., 2008), but Ribot et al. (2008) also found no suppression of PHO1 by Phi. A possible explanation for these differences in response might be a distinct subcellular localization of Phi and/or Pi under different experimental conditions. In Pi-sufficient plants, Phi preferentially accumulates in the vacuole, whereas in Pi-starved plants, Phi accumulates in the cytoplasm (Danova-Alt et al., 2008). Such differential compartmentation might explain differences in the ability of Phi to interfere with Pi-sensing mechanisms and subsequently the regulation of gene expression. In our experiments, the induction and repression of SQD2 and NMT3, respectively, was attenuated by Phi, whilst the induction of PHT1;1, PAP1 and ASK11 was accentuated. At the same time, the induction of miRNA399d, At4 and PHT1;4 was not affected by Phi. These results indicate alternative sensing mechanisms of the Pi (and Phi) status within the plant. A possible mechanism allowing the plant to differentiate between Pi and Phi could be the additional sensing of a P-containing or a derived component as a measure of flux of free Pi into the organic P pool. Members of the NMT gene family have already been implicated in a stress signalling pathway involving the phospholipid-derived second messenger phosphatidic acid (Jost et al., 2009). Such regulation would be reminiscent of other two major macronutrients nitrate and sulfate, for which downstream assimilation products, i.e. glutamate/glutamine and glutathione, respectively, are part of the nutrient-sensing network (Lappartient et al., 1999; Gutierrez et al., 2008).
Metabolomic response to phosphite accumulation
Several enzymes, such as glyceraldyde-3-phosphate dehydrogenase and phosphorylases such as starch phosphorylase, use Pi as a substrate and many enzymes of primary metabolism are regulated by Pi, such as phosphofructokinase (PFK), sucrose phosphate synthase, ADP-glucose pyrophosphorylase, and glutathione synthetase (Sowokinos and Preiss, 1982; Doehlert and Huber, 1984; Hausler et al., 1989; Jez and Cahoon, 2004). Because of its steric similarity to Pi, Phi probably mimics and competes with Pi for binding in catalytic or regulatory sites of enzymes, causing competitive or allosteric inhibition. Indeed, for ATP-dependent PFK, but not PPi-dependent PFK, of Brassica nigra, Phi inhibits in vitro enzyme activity (Carswell et al., 1996). Similarly, several glycolytic enzymes from Phytophthora palmivora and yeast are also inhibited by Phi (Stehmann and Grant, 2000), indicating that the regulation of enzyme activity by Phi may be of wider significance.
Metabolomic analyses identified alterations in a number of metabolite pools. These results showed the ability of Phi to induce specific shifts, which for some metabolites contrasted with those of Pi-limited plants. We found that Phi specifically decreased levels of the central amino acids asparagine, aspartate, glutamate, and serine (Figs 7 and S2). Alterations in amino acid levels have also been observed for severely P-limited Arabidopsis and barley plants (Morcuende et al., 2007; Huang et al., 2008).
Although these results do not allow the underlying specific targets of Phi causing these changes to be identified, they reveal that even moderate accumulation of Phi influences plant metabolic processes. This may provide clues to increase our understanding of the mechanisms of Phi-induced resistance of plants to oomycete pathogens such as Phytophthora and Pythium species (Guest and Grant, 1991). The molecular mode of action of Phi in inducing resistance has not yet been identified, but studies in Arabidopsis have implicated salicylic acid and its major signalling component NPR1, as well as a MAP kinase regulatory network (Molina et al., 1998; Friedrich et al., 2001; Eshraghi et al., 2011; Massoud et al., 2012). Several recent studies have identified a role of primary metabolites, especially amino acids, in the establishment of resistance against plant pathogens (Song et al., 2004; Chanda et al., 2008; van Damme et al., 2009; Liu et al., 2010; Park et al., 2010; Brauc et al., 2011, 2012; Chanda et al., 2011; Hwang et al., 2011; Stuttmann et al., 2011; Voll et al., 2012). These studies show that the alteration of specific metabolite levels by feeding or genetic manipulation of plant metabolic pathways can lead to resistance against pathogens. Interestingly, alterations in the abundance of amino acids derived from the aspartate pathway have been identified to be important for resistance against the oomycete pathogen H. arabidopsidis and the bacterial pathogen Pseudomonas syringae (van Damme et al., 2009; Stuttmann et al., 2011; Navarova et al., 2012). As we found Phi-induced changes in levels of aspartate and the related amino acids asparagine and glutamate (Fig. S2B), we surmise that these are similarly important for Phi-induced resistance against oomycete pathogens. Currently, it is not clear how these metabolic changes induce resistance. It has been suggested that, due to the evolutionary relationship between plants and oomycetes, metabolic pathways are conserved and metabolic perturbations in the host decrease the colonization ability of the pathogen (Stuttmann et al., 2011). These results indicate the possibility for a mode of Phi action by inducing alterations in primary metabolite pools leading to pathogen resistance.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Shoot:root ratios of plant under +Phi and –Pi treatments as in shown in Fig. 1.
Supplementary Fig. S2. (A) Representation of metabolites with overlapping and contrasting changes in Phi-treated and Pi-limited Arabidopsis plants. (B) Simplified metabolic pathways showing interconnections of metabolites with altered abundance (in bold) under Phi treatment or Pi limitation.
Supplementary Table S1. Sequences of the primers used in gene expression experiments.
Acknowledgements
We thank Laurent Nussaume and Wolf-Rüdiger Scheible for helpful discussions. This work was funded by the Australian Research Council Linkage Project (LP0776252) and its partners, the Minerals and Energy Research Institute of Western Australia, the Department of Environment and Conservation (Western Australia), Alcoa, BHP Billiton, Tronox, and Western Power.
Glossary
Abbreviations:
- GC-MS
gas chromatography–mass spectrometry
- LRD
lateral root density
- PFK
phosphofructokinase
- Phi
phosphite; inorganic phosphate
- PSR
phosphate-starvation response
- SE
standard error.
References
- Ames BN. 1966. Assay of inorganic phosphate, total phosphate and phosphatases. In: Neufeld G, Ginsburg V, eds. Methods in enzymology: complex carbohydrates , Vol. 8, New York: Academic Press; 115–118 [Google Scholar]
- Arvidsson S, Kwasniewski M, Riano-Pachon DM, Mueller-Roeber B. 2008. QuantPrime—a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9, 465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ. 2006. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiology 141, 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchietto T, Saindrenan P, Bompeix G. 1992. Physiological responses of Phytophthora citrophthora to a subinhibitory concentration of phosphonate. Pesticide Biochemistry and Physiology 42, 151–166 [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. 2006. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiology 141, 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz O, Jost R, Pearse SJ, Lambers H, Finnegan PM, Hardy GE, O’Brien PA. 2011. An enzymatic fluorescent assay for the quantification of phosphite in a microtiter plate format. Analytical Biochemistry 412, 74–78 [DOI] [PubMed] [Google Scholar]
- Brauc S, De Vooght E, Claeys M, Geuns JMC, Höfte M, Angenon G. 2012. Overexpression of arginase in Arabidopsis thaliana influences defence responses against Botrytis cinerea. Plant Biology 14, 39–45 [DOI] [PubMed] [Google Scholar]
- Brauc S, De Vooght E, Claeys M, Höfte M, Angenon G. 2011. Influence of over-expression of cytosolic aspartate aminotransferase on amino acid metabolism and defence responses against Botrytis cinerea infection in Arabidopsis thaliana . Journal of Plant Physiology 168, 1813–1819 [DOI] [PubMed] [Google Scholar]
- Bucher M. 2007. Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytologist 173, 11–26 [DOI] [PubMed] [Google Scholar]
- Carswell C, Grant BR, Theodorou ME, Harris L, Niere JO, Plaxton WC. 1996. The fungicide phosphonate disrupts the phosphate-starvation response in Brassica nigra seedlings. Plant Physiology 110, 105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carswell MC, Grant BR, Plaxton WC. 1997. Disruption of the phosphate-starvation response of oilseed rape suspension cells by the fungicide phosphonate. Planta 203, 67–74 [DOI] [PubMed] [Google Scholar]
- Chanda B, Venugopal SC, Kulshrestha S, Navarre DA, Downie B, Vaillancourt L, Kachroo A, Kachroo P. 2008. Glycerol-3-phosphate levels are associated with basal resistance to the hemibiotrophic fungus Colletotrichum higginsianum in Arabidopsis . Plant Physiology 147, 2017–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda B, Xia Y, Mandal MK, Yu K, Sekine KT, Gao QM, Selote D, Hu Y, Stromberg A, Navarre D, Kachroo A, Kachroo P. 2011. Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nature Genetics 43, 421–427 [DOI] [PubMed] [Google Scholar]
- Chevalier F, Pata M, Nacry P, Doumas P, Rossignol M. 2003. Effects of phosphate availability on the root system architecture: large-scale analysis of the natural variation between Arabidopsis accessions. Plant, Cell and Environment 26, 1839–1850 [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL. 2006. Regulation of phosphate homeostasis by microRNA in Arabidopsis . Plant Cell 18, 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI. 2011. Signaling network in sensing phosphate availability in plants. Annual Reviews of Plant Biology 62, 185–206 [DOI] [PubMed] [Google Scholar]
- Cohen Y, Coffey MD. 1986. Systemic fungicides and the control of oomycetes. Annual Review of Phytopathology 24, 311–338 [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis . Plant Physiology 139, 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danova-Alt R, Dijkema C, De Waard P, Kock M. 2008. Transport and compartmentation of phosphite in higher plant cells—kinetic and P nuclear magnetic resonance studies. Plant, Cell and Environment 31, 1510–1521 [DOI] [PubMed] [Google Scholar]
- Doehlert DC, Huber SC. 1984. Phosphate inhibition of spinach leaf sucrose phosphate synthase as affected by glucose-6-phosphate and phosphoglucoisomerase. Plant Physiology 76, 250–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff SMG, Sarath G, Plaxton WC. 1994. The role of acid phosphatases in plant phosphorus metabolism. Physiologia Plantarum 90, 791–800 [Google Scholar]
- Eshraghi L, Anderson J, Aryamanesh N, Shearer B, McComb J, Hardy GES, O’Brien PA. 2011. Phosphite primed defence responses and enhanced expression of defence genes in Arabidopsis thaliana infected with Phytophthora cinnamomi . Plant Pathology 60, 1086–1095 [Google Scholar]
- Essigmann B, Guler S, Narang RA, Linke D, Benning C. 1998. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 95, 1950–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliege R, Flugge UI, Werdan K, Heldt HW. 1978. Specific transport of inorganic phosphate, 3-phosphoglycerate and triosephosphates across the inner membrane of the envelope in spinach chloroplasts. Biochimica et Biophysica Acta 502, 232–247 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics 39, 1033–1037 [DOI] [PubMed] [Google Scholar]
- Friedrich L, Lawton K, Dietrich R, Willits M, Cade R, Ryals J. 2001. NIM1 overexpression in Arabidopsis potentiates plant disease resistance and results in enhanced effectiveness of fungicides. Molecular Plant–Microbe Interactions 14, 1114–1124 [DOI] [PubMed] [Google Scholar]
- Gerke J. 1992. Phosphate, aluminum and iron in the soil solution of three different soils in relation to varying concentrations of citric acid. Zeitschrift für Pflanzenernährung und Bodenkunde 155, 339–343 [Google Scholar]
- Ghosh HP, Preiss J. 1966. Adenosine diphosphate glucose pyrophosphorylase. A regulatory enzyme in the biosynthesis of starch in spinach leaf chloroplasts. Journal of Biological Chemistry 241, 4491–4504 [PubMed] [Google Scholar]
- Griffith JM, Smillie RH, Grant BR. 1990. Alterations in nucleotide and pyrophosphate levels in Phytophthora palmivora following exposure to the antifungal agent potassium phosphonate (phosphite). Journal of General Microbiology 136, 1285–1291 [Google Scholar]
- Guest D, Grant B. 1991. The complex action of phosphonates as antifungal agents. Biological Reviews of the Cambridge Philosophical Society 66, 159–187 [Google Scholar]
- Gutierrez RA, Stokes TL, Thum K, et al. 2008. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1 . Proceedings of the National Academy of Sciences, USA 105, 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy GES, Barrett S, Shearer BL. 2001. The future of phosphite as a fungicide to control the soilborne plant pathogen Phytophthora cinnamomi in natural ecosystems. Australasian Plant Pathology 30, 133–139 [Google Scholar]
- Hartel H, Dormann P, Benning C. 2000. DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis . Proceedings of the National Academy of Sciences, USA 97, 10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausler RE, Holtum JA, Latzko E. 1989. Cytosolic phosphofructokinase from spinach leaves: I. Purification, characteristics, and regulation. Plant Physiology 90, 1498–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. 2009. CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194 [DOI] [PubMed] [Google Scholar]
- Huang CY, Roessner U, Eickmeier I, Genc Y, Callahan DL, Shirley N, Langridge P, Bacic A. 2008. Metabolite profiling reveals distinct changes in carbon and nitrogen metabolism in phosphate-deficient barley plants (Hordeum vulgare L.). Plant and Cell Physiology 49, 691–703 [DOI] [PubMed] [Google Scholar]
- Hwang IS, An SH, Hwang BK. 2011. Pepper asparagine synthetase 1 (CaAS1) is required for plant nitrogen assimilation and defense responses to microbial pathogens. The Plant Journal 67, 749–762 [DOI] [PubMed] [Google Scholar]
- Jez JM, Cahoon RE. 2004. Kinetic mechanism of glutathione synthetase from Arabidopsis thaliana . Journal of Biological Chemistry 279, 42726–42731 [DOI] [PubMed] [Google Scholar]
- Jost R, Berkowitz O, Masle J. 2007. Magnetic quantitative reverse transcription PCR: a high-throughput method for mRNA extraction and quantitative reverse transcription PCR. BioTechniques 43, 206–211 [DOI] [PubMed] [Google Scholar]
- Jost R, Berkowitz O, Shaw J, Masle J. 2009. Biochemical characterization of two wheat phosphoethanolamine N-methyltransferase isoforms with different sensitivities to inhibition by phosphatidic acid. Journal of Biological Chemistry 284, 31962–31971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama H, Kawamura A, Kihara T, Hara T, Takita E, Shibata D. 2000. Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus-limited soil. Plant and Cell Physiology 41, 1030–1037 [DOI] [PubMed] [Google Scholar]
- Lambers H, Cawthray G, Giavalisco P, Kuo J, Laliberté E, Pearse SJ, Scheible WR, Stitt M, Teste F, Turner BL. 2012. Proteaceae from severely phosphorus-impoverished soils extensively replace phospholipids by galactolipids and sulfolipids during leaf development to achieve a high photosynthetic phosphorus-use efficiency. New Phytologist 196, 1098–1108 [DOI] [PubMed] [Google Scholar]
- Lappartient AG, Vidmar JJ, Leustek T, Glass AD, Touraine B. 1999. Inter-organ signaling in plants: regulation of ATP sulfurylase and sulfate transporter genes expression in roots mediated by phloem-translocated compound. The Plant Journal 18, 89–95 [DOI] [PubMed] [Google Scholar]
- Lee RB, Ratcliffe RG. 1993. Subcellular distribution of inorganic phosphate, and levels of nucleoside triphosphate, in mature maize roots at low external phosphate concentrations—measurements with 31P-NMR. Journal of Experimental Botany 44, 587–598 [Google Scholar]
- Li M, Welti R, Wang X. 2006. Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases Dζ1 and Dζ2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiology 142, 750–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Ji Y, Bhuiyan NH, Pilot G, Selvaraj G, Zou J, Wei Y. 2010. Amino acid homeostasis modulates salicylic acid-associated redox status and defense responses in Arabidopsis . Plant Cell 22, 3845–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ. 2012. PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis . Plant Cell 24, 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔC T method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Lynch JP, Brown KM. 2001. Topsoil foraging—an architectural adaptation of plants to low phosphorus availability. Plant and Soil 237, 225–237 [Google Scholar]
- Massoud K, Barchietto T, Le Rudulier T, Pallandre L, Didierlaurent L, Garmier M, Ambard-Bretteville F, Seng JM, Saindrenan P. 2012. Dissecting phosphite-induced priming in Arabidopsis infected with Hyaloperonospora arabidopsidis . Plant Physiology 159, 286–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JCF, Steiner P, Hirling H, Unser M. 2004. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry Part A 58A, 167–176 [DOI] [PubMed] [Google Scholar]
- Mimura T, Sakano K, Shimmen T. 1996. Studies on the distribution, re-translocation and homeostasis of inorganic phosphate in barley leaves. Plant, Cell and Environment 19, 311–320 [Google Scholar]
- Molina A, Hunt MD, Ryals JA. 1998. Impaired fungicide activity in plants blocked in disease resistance signal transduction. Plant Cell 10, 1903–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, et al. 2007. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant, Cell and Environment 30, 85–112 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497 [Google Scholar]
- Narang RA, Bruene A, Altmann T. 2000. Analysis of phosphate acquisition efficiency in different Arabidopsis accessions. Plant Physiology 124, 1786–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarova H, Bernsdorff F, Doring AC, Zeier J. 2012. Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell[Epub ahead of print] doi:10.1105/tpc.112.103564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niere JO, Deangelis G, Grant BR. 1994. The effect of phosphonate on the acid-soluble phosphorus components in the genus Phytophthora . Microbiology 140, 1661–1670 [Google Scholar]
- Ouimette DG, Coffey MD. 1989. Phosphonate levels in avocado (Persea americana) seedlings and soil following treatment with fosetyl-Al or potassium phosphonate. Plant Disease 73, 212–215 [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR. 2008. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. The Plant Journal 53, 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Mirabella R, Bronstein PA, Preston GM, Haring MA, Lim CK, Collmer A, Schuurink RC. 2010. Mutations in γ-aminobutyric acid (GABA) transaminase genes in plants or Pseudomonas syringae reduce bacterial virulence. The Plant Journal 64, 318–330 [DOI] [PubMed] [Google Scholar]
- Peret B, Clement M, Nussaume L, Desnos T. 2011. Root developmental adaptation to phosphate starvation: better safe than sorry. Trends in Plant Science 16, 442–450 [DOI] [PubMed] [Google Scholar]
- Plaxton WC, Tran HT. 2011. Metabolic adaptations of phosphate-starved plants. Plant Physiology 156, 1006–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova Y, Thayumanavan P, Lonati E, Agrochao M, Thevelein JM. 2010. Transport and signaling through the phosphate-binding site of the yeast Pho84 phosphate transceptor. Proceedings of the National Academy of Sciences, USA 107, 2890–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt J, Boisson AM, Gout E, Bligny R, Douce R, Aubert S. 2009. Phosphate (Pi) starvation effect on the cytosolic Pi concentration and Pi exchanges across the tonoplast in plant cells: an in vivo 31P nuclear magnetic resonance study using methylphosphonate as a Pi analog. Plant Physiology 151, 1646–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboy V. 2003. myo-Inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 64, 1033–1043 [DOI] [PubMed] [Google Scholar]
- Raghothama KG. 1999. Phosphate acquisition. Annual Reviews of Plant Physiology and Plant Molecular Biology 50, 665–693 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339, 62–66 [DOI] [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T. 2006. Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana . Plant, Cell and Environment 29, 115–125 [DOI] [PubMed] [Google Scholar]
- Ribot C, Wang Y, Poirier Y. 2008. Expression analyses of three members of the AtPHO1 family reveal differential interactions between signaling pathways involved in phosphate deficiency and the responses to auxin, cytokinin, and abscisic acid. Planta 227, 1025–1036 [DOI] [PubMed] [Google Scholar]
- Ryan P, Delhaize E, Jones D. 2001. Function and mechanism of organic anion exudation from plant roots. Annual Reviews of Plant Physiology and Plant Molecular Biology 52, 527–560 [DOI] [PubMed] [Google Scholar]
- Rychter AM, Randall DD. 1994. The effect of phosphate deficiency on carbohydrate metabolism in bean roots. Physiologia Plantarum 91, 383–388 [Google Scholar]
- Shane MW, Lambers H. 2005. Cluster roots: a curiosity in context. Plant and Soil 274, 101–125 [Google Scholar]
- Song JT, Lu H, Greenberg JT. 2004. Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, aberrant growth and death2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. Plant Cell 16, 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowokinos JR, Preiss J. 1982. Pyrophosphorylases in Solanum tuberosum: III. Purification, physical, and catalytic properties of ADP-glucose pyrophosphorylase in potatoes. Plant Physiology 69, 1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehmann C, Grant BR. 2000. Inhibition of enzymes of the glycolytic pathway and hexose monophosphate bypass by phosphonate. Pesticide Biochemistry and Physiology 67, 13–24 [Google Scholar]
- Stuttmann J, Hubberten HM, Rietz S, Kaur J, Muskett P, Guerois R, Bednarek P, Hoefgen R, Parker JE. 2011. Perturbation of Arabidopsis amino acid metabolism causes incompatibility with the adapted biotrophic pathogen Hyaloperonospora arabidopsidis . Plant Cell 23, 2788–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao HTB, Yamakawa T, Myint AK, Sarr PS. 2008a. Effects of phosphite, a reduced form of phosphate, on the growth and phosphorus nutrition of spinach (Spinacia oleracea L.). Soil Science and Plant Nutrition 54, 761–768 [Google Scholar]
- Thao HTB, Yamakawa T, Shibata K, Sarr PS, Myint AK. 2008b. Growth response of komatsuna (Brassica rapa var. peruviridis) to root and foliar applications of phosphite. Plant and Soil 308, 1–10 [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. 2010. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. The Plant Journal 64, 775–789 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Abel S. 2001. Attenuation of phosphate starvation responses by phosphite in Arabidopsis . Plant Physiology 127, 963–972 [PMC free article] [PubMed] [Google Scholar]
- van Damme M, Zeilmaker T, Elberse J, Andel A, de Sain-van der Velden M, van den Ackerveken G. 2009. Downy mildew resistance in Arabidopsis by mutation of HOMOSERINE KINASE. Plant Cell 21, 2179–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3, RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan DK, Karthikeyan AS, Matilda PD, Raghothama KG. 2002. Phosphite, an analog of phosphate, suppresses the coordinated expression of genes under phosphate starvation. Plant Physiology 129, 1232–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voll LM, Zell MB, Engelsdorf T, Saur A, Wheeler MG, Drincovich MF, Weber AP, Maurino VG. 2012. Loss of cytosolic NADP-malic enzyme 2 in Arabidopsis thaliana is associated with enhanced susceptibility to Colletotrichum higginsianum . New Phytologist 195, 189–202 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.