Abstract
Genetic relationships between plant height and root morphology were investigated in a diverse set of wheat germplasm [199 double-haploid progeny derived from a cross between Avalon and Cadenza (Triticum aestivum L.), Rht near-isogenic lines (NILs), and accessions from the Watkins Collection] to investigate whether Rht genes controlling shoot height also control seedling root growth. A germination paper screen was developed to measure seedling root length (distinguishing seminal axes from seminal lateral roots), surface area, volume, and dry weight, and these were compared with shoot dry weight and the root to shoot ratio. Field experiments were conducted to measure mature plant height (PH) and grain characteristics for the mapping population. Forty-three quantitative trait loci (QTLs) for PH, root and seed traits were identified. Some QTLs for roots and either height or seed characteristics were coincident: chromosome 2D had co-locating root and PH QTLs; chromosomes 4D had co-locating root, PH, and seed QTLs; chromosome 5A and 6A had co-locating root and seed QTLs; and other non-co-locating root and PH QTLs were found on chromosomes 3A and 3B. Rht NILs illustrated that some known dwarfing genes reduce both PH and root proliferation. However, analysis of 25 short and 23 tall lines from the Watkins wheat germplasm collection indicated that PH and root proliferation are not simply related.
Key words: Doubled-haploid population, plant height, QTLs, Rht, roots, wheat.
Introduction
Root architectural traits are important for the selection of wheat and cultivars that are efficient in the acquisition of nutrients and water from soil (Gregory, 2006), phenotypes which are critical for future sustainable agriculture. Furthermore, adequate water and soil nutrient supply underpin yield and quality characteristics in crop plants. Root traits are believed to be complex and controlled by many genes, each with a small genetic effect; genetic loci controlling such traits are called quantitative trait loci (QTLs) (Sharma et al., 2011). Studying root traits in mature field-grown crops is extremely challenging and there is a need for robust, high-throughput laboratory screens which may be a proxy for field performance. Therefore, researchers have adopted various hydroponic culture, agarose-filled gel chamber, and other rhizotron methods, principally for root characterization of seedlings (e.g. Zhu et al., 2005b ; Laperche et al., 2006; Ren et al., 2011; Liu et al., 2013). However, the growth system can substantially affect root architecture traits, with differences between wheat seedlings grown in gel chambers, in soil-filled columns, and in the field (Wojciechowski et al., 2009).
Plant height (PH) in wheat is controlled mainly by reduced height (Rht) genes, with contributions from many other minor genes (Ahmed et al., 2000). Wheat grain yields have increased following the widespread incorporation of Rht alleles to produce semi-dwarf varieties (Chapman et al., 2007), which have the advantages of favourable harvest indexes and decreased susceptibility to lodging, especially under high N inputs. Lodging is positively correlated with PH, and short plants are less susceptible to lodging (Navabi et al., 2006). As a consequence, PH reduction has been a key target for wheat breeding. However, the relationships between PH and root architecture and the effects of genes which control PH on root architecture are not clear. Miralles et al. (1997) found that dwarfing genes significantly reduced PH and above-ground biomass at anthesis, but total root length and root dry weight per unit area were increased with decreased PH of spring wheat in field experiments. McCaig and Morgan (1993) reported that stem and shoot (total above-ground tissue) dry matter per plant increased with PH and that there was no relationship between height and root dry matter. Siddique et al. (1990) reported that tall and dwarf isogenic lines had similar developmental and patterns of root:shoot dry matter accumulation.
In this study, a QTL approach is utilized to examine the genetic relationships between PH, seed size, and root characteristics in a wheat mapping population. The chosen doubled-haploid (DH) mapping population was generated from the cultivars Avalon and Cadenza as a focus for studies on wheat genetic improvement. Heading time and height QTLs for the Avalon×Cadenza population and populations derived from the crosses Savannah×Rialto, Spark×Rialto, and Charger×Badger have been studied by Griffiths et al. (2009, 2012) reporting 16 meta-QTLs (QTLs consistent in all crosses) for height on chromosomes 1A, 1B, 1D, 2A (two meta-QTLs), 2B, 2D, 3A, 3B, 4B, 4D, 5A, 5B, 6A, 6B, and 6D. The objective of this study was to identify QTLs responsible for seedling root traits using a rolled germination paper culture system and to analyse the relationship between QTLs of seedling root traits and QTLs of PH determined in the field, and additionally seed size using the DH population derived from Avalon and Cadenza.
Materials and methods
Plant materials
The population of DH individuals (199 lines) derived from F1 progeny of a cross between cultivars Avalon and Cadenza was developed by Clare Ellerbrook, Liz Sayers, and the late Tony Worland (John Innes Centre, Norwich, Norfolk, UK), as part of a Defra-funded project led by ADAS. The parents were originally chosen (to contrast for canopy architecture traits) by Steve Parker (CSL), Tony Worland and Darren Lovell (Rothamsted Research). The female parent, Avalon, is a short winter wheat, and the male parent, Cadenza, is a tall spring wheat.
Six Rht near-isogenic lines (NILs) in a cv. Mercia background and control Mercia (rht) were provided by the John Innes Centre. Mercia was introduced commercially in 1983, and was the last widely used non-semi-dwarf winter wheat cultivar suited for bread making in the UK. The Mercia NILs comprised: the parent line (rht, tall); gibberellin-insensitive semi-dwarf (Rht1-B1b, Rht1-D1b from ‘Norin 10’) and dwarf (Rht-B1c from ‘Tom thumb’; Rht-D1c from ‘Ai-Bian’) lines; and gibberellin-sensitive semi-dwarf (Rht8c from ‘Mara’) and dwarf (Rht12 from ‘Karcagi 522’) lines (Addisu et al., 2009).
Forty-eight lines from the Watkins Collection (Miller et al., 2001) were provided by the John Innes Centre. These comprised 25 short and 23 tall lines (Simon Griffiths, personal communication).
Field trials
Four field trials were conducted at Rothamsted, Harpenden, UK (latitude 51.8°N, longitude 0.4°W) from 2008 to 2011. In each trial, each line was grown in three replicate plots, 10.0 m×1.8 m, in a randomized block design, and grown according to standard agronomic practice including the use of chemical plant growth regulators at a dose to prevent lodging whilst not interfering with height segregation between the lines (Chlormequat at 2.25 l ha–1 except in 2008 where it was 1.0 l ha–1). Nitrogen treatments were 100kg N ha–1 in 2008 and 2009, and 200kg N ha–1 in 2010 and 2011. The PH of each line was measured: the mean measurement of 10 randomly plants was taken for each plot, excluding border plants. Thousand grain weight (TGW) was determined on 500 grains (pre-dried at 80 °C), with two technical replicates, using a seed counter (Elmor model C1, Switzerland), and drying the seed at 105 °C for 16h prior to weighing. The seeds for TGW QTL determination were from same set of seeds (from 2010) as used in the root experiments. The means of mature PHs from 2008 to 2011 were used for the QTL analysis; QTL analyses of the PHs for each independent year were performed (data not shown).
Paper culture system
A screening system based on the ‘cigar roll’ system was utilized (Zhu et al., 2005b ). Seeds were surface-sterilized with 0.5% calcium hypochlorite solution for 30min and then rinsed three times with sterilized water and left in the cold room (4 °C) overnight. Subsequently the seeds were germinated in Petri dishes on tissue with sterilized water. Four uniformly germinating seeds were placed on one germination paper (25×38cm, Anchor Paper Company, St. Paul, MN, USA), which was then rolled to final dimensions of 2cm in diameter and 38cm in height, supported vertically by a metal mesh with 2 cm×2cm holes. The experimental design was an unbalanced block design with three replicate rolls per line (in total 12 plants per line). The bases of the rolls were placed in a tray with nutrient solution, initially containing half-strength medium. After 3 d, nutrient solution was changed to full-strength medium and thereafter changed every day. The nutrient solution comprised: 1.5mM Ca(NO3)2, 5mM KNO3, 2mM NaNO3, 1mM MgSO4, 1mM KH2PO4, 25 μM FeEDTA, 160nM CuCl2, 9.2 μM H3BO3, 3.6 μM MnCl2, 16nM Na2MoO4, 5 μM KCl, and 770nM ZnCl2, at pH 5.8. Plants were grown in a controlled-environment growth room with 12/12h light/dark cycle, light intensity of 500 μmol m–2 s–1, 70% relative humidity during the day and 80% during the night, and temperature of 20 °C during the day and 16 °C during the night. After 11 d (at the two-leaf stage, one unfurled), plants were taken off the germination paper and kept in 30% ethanol prior to imaging. The intact root systems were scanned as a digital image with a scanner (STD-1600, Regent Instruments, QC, Canada). The root length, surface area, and volume were determined from the root images using the root image analysis software WinRHIZO Pro (Regent Instruments), differentiating the seminal axes and the seminal laterals with a distinguishing diameter of 0.25mm (verified in each experiment). Following scanning, the plants were oven-dried at 80 °C to determine root and shoot dry weights. To increase the reliability of QTL measurements, the mapping population experiment was repeated three times.
QTL identification and statistical analysis
The genetic map described by Griffiths et al. (2009, 2012) and available from the WGIN website (http://www.wgin.org.uk/) was utilized. The mean of three experiments (36 plants in total of each line in a DH population) was used to detect root QTLs by the software package QTL Cartographer v2.5 (Basten et al., 1994). Detection of QTLs was conducted by the composite interval mapping (CIM) method proposed by Zeng (1994). Forward regression was analysed using a window size of 10 cM, a walk speed of 2 cM, and five control markers. The threshold for QTL detection was determined with 1000 permutations at α=0.05 (Churchill and Doerge, 1994) which gave LOD values of 3.45 for total root length (TRL), 4.01 for seminal laterals length (SLL), 3.99 for seminal axes length (SAL), 3.76 for total root surface area (TRSA), 3.91 for seminal laterals surface area (SLSA), 3.59 for seminal axes surface area (SASA), 3.27 for total root volume (TRVol), 3.51 for seminal laterals volume (SLVol), 3.86 for seminal axes volume (SAVol), 4.16 for root dry weight (RDW), 3.70 for shoot dry weight (SDW), 3.86 for root to shoot ratio (R/S), 3.37 for PH, and 4.03 for TGW. These LOD scores were rescaled to a likelihood ratio (LR) test statistic [divide by log10 (e)/2=0.2171) (Churchill and Doerge, 1994). Naming of QTLs followed a standard formula (Yang et al., 2010): ‘q’ (abbreviation of QTL), followed by an abbreviation of the trait, then the linkage group. Correlation analysis was performed using SPSS 16.0 for Windows.
Results
Phenotypic variation
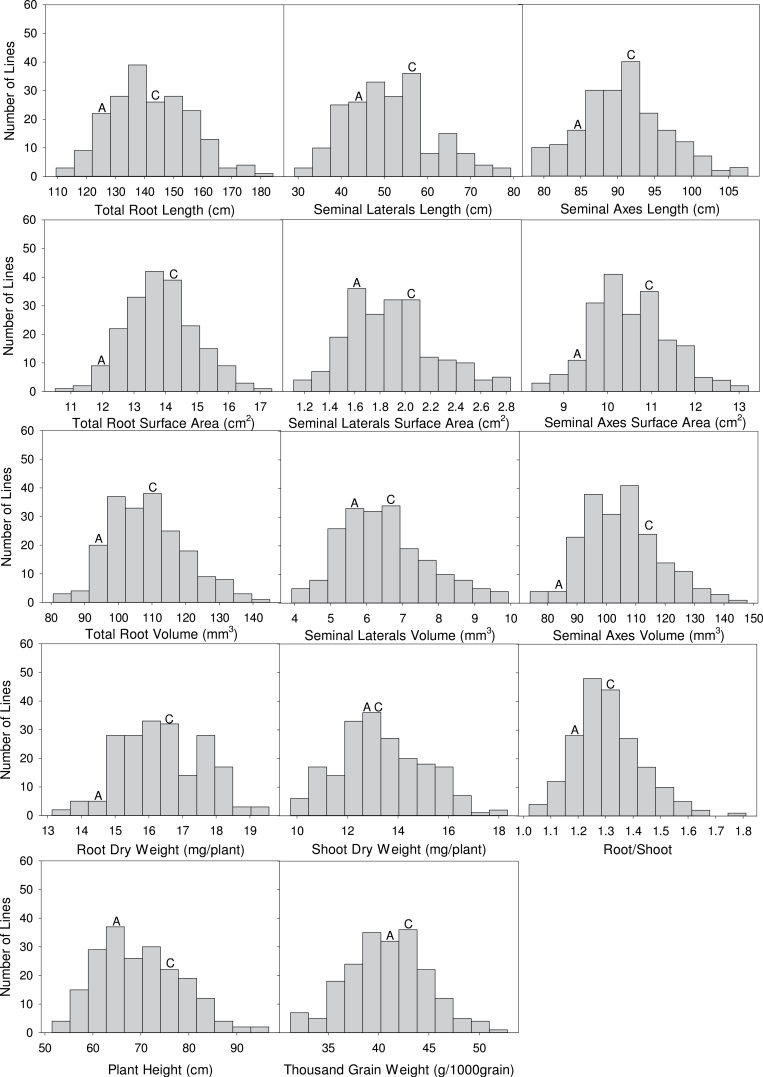
The phenotypic values of the DH population and parent varieties, Avalon and Cadenza, are presented in Table 1. The phenotypic mean of the population for each trait was between the parental values, except for SDW. The population exhibited significant transgressive segregation—phenotypic values beyond the values of parents—for all traits. PH and root system attributes were different between the two parents; the taller male parent, Cadenza, developed a larger root system (longer root length, larger root surface area and volume, and higher RDW) and larger TGW than the female parent, Avalon. The frequency distributions of the measured traits showed continuous variation with approximately normal distributions (Fig.1).
Table 1.
Phenotypes of parents and the DH population.
| Traits | Parents | DH population | |||
|---|---|---|---|---|---|
| Avalon | Cadenza | Mean ±SD | Minimum | Maximum | |
| TRL (cm) | 128 | 147 | 142±13.6 | 109 | 184 |
| TRSA (cm2) | 12.1 | 14.4 | 13.8±1.13 | 10.5 | 17.4 |
| TRVol (mm3) | 91.7 | 113 | 108±11.3 | 80.6 | 145 |
| SLL (cm) | 43.4 | 55.7 | 51.1±9.76 | 29.2 | 79.4 |
| SAL (cm) | 84.4 | 91.1 | 90.6±5.88 | 78.3 | 107.7 |
| SLSA (cm2) | 1.66 | 2.014 | 1.88±0.34 | 1.12 | 2.84 |
| SASA (cm2) | 9.19 | 10.9 | 10.5±0.89 | 8.28 | 13.2 |
| SLVol (mm3) | 5.91 | 6.87 | 6.51±1.22 | 3.91 | 10.0 |
| SAVol (mm3) | 85.6 | 111 | 105±12.8 | 74.1 | 148 |
| RDW (mg plant–1) | 14.5 | 16.6 | 16.4±1.23 | 13.1 | 19.6 |
| SDW (mg plant–1) | 12.7 | 13.3 | 13.3±1.70 | 9.7 | 18.3 |
| R/S | 1.20 | 1.32 | 1.31±0.125 | 1.02 | 1.81 |
| PH (cm) | 65.6 | 75.1 | 69.7±8.85 | 51.5 | 96.7 |
| TGW (g) | 40.1 | 42.6 | 40.8±3.97 | 31.3 | 52.8 |
In the DH population, data are means of 36 plants for root traits, SDW, and R/S, of 12 independent measurements of PH (4 years, three replicate plots per year), and three samples (single year, three replicate plots) for TGW. For the parent data, these sample numbers were doubled. In the DH population, the mean and standard deviation of the mean are of the 199 lines.
DH, doubled-haploid; SD, standard deviation; TRL, total root length; TRSA, total root surface area; TRVol, total root volume; SLL, seminal laterals length; SAL, seminal axes length; SLSA, seminal laterals surface area; SASA, seminal axes surface area; SLVol, seminal laterals volume; SAVol, seminal axes volume; RDW, root dry weight; SDW, shoot dry weight; R/S, root to shoot ratio; PH, plant height; TGW, thousand grain weight.
Fig. 1.
Histograms of frequency distribution of root traits, SDW, R/S, PH, and TGW for the Avalon×Cadenza doubled-haploid population used in this study. Data are means of 36 plants for root traits, SDW, and R/S, of 12 independent measurements of PH (4 years, three replicate plots per year), and three samples (single year, three replicate plots) for TGW.
Pearson’s correlation coefficients between traits were calculated (Table 2). Significant positive correlations (P < 0.01) were found amongst the traits. TRL, SLL, SAL, SLSA, SLVol, RDW, SDW, and R/S were significantly correlated with PH, whilst no significant positive correlations between TRSA, TRVol, SASA, SAVol, and PH were observed. All root architecture traits and PH were significantly correlated with TGW.
Table 2.
Correlation coefficients among the trait measurements in the DH population.
| TRSA | TRVol | SLL | SAL | SLSA | SASA | SLVol | SAVol | RDW | SDW | R/S | PH | TGW (g) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TRL (cm) | 0.80** | 0.32** | 0.93** | 0.78** | 0.90** | 0.49** | 0.84** | 0.29** | 0.66** | 0.35** | 0.24** | 0.25** | 0.38** |
| TRSA (cm2) | 0.83** | 0.57** | 0.90** | 0.50** | 0.91** | 0.42** | 0.80** | 0.84** | 0.50** | 0.15* | 0.12 | 0.53** | |
| TRVol (mm3) | 0.029 | 0.69** | –0.044 | 0.97** | –0.13 | 0.99** | 0.69** | 0.46** | 0.018 | –0.054 | 0.48** | ||
| SLL(cm) | 0.48** | 0.98** | 0.19** | 0.94** | 0.019 | 0.43** | 0.14* | 0.30** | 0.20** | 0.18* | |||
| SAL (cm) | 0.44** | 0.83** | 0.39** | 0.63** | 0.82** | 0.56** | 0.051 | 0.25** | 0.58** | ||||
| SLSA (cm2) | 0.11 | 0.99** | –0.072 | 0.39** | 0.13 | 0.30** | 0.24** | 0.17* | |||||
| SASA (cm2) | 0.018 | 0.96** | 0.78** | 0.51** | 0.041 | 0.020 | 0.52** | ||||||
| SLVol (mm3) | –0.17* | 0.33** | 0.10 | 0.28** | 0.27** | 0.15* | |||||||
| SAVol (mm3) | 0.66** | 0.43** | 0.030 | –0.096 | 0.44** | ||||||||
| RDW (mg plant–1) | 0.69** | 0.035 | 0.24** | 0.62** | |||||||||
| SDW (mg plant–1) | –0.58** | 0.46** | 0.62** | ||||||||||
| R/S | –0.33** | –0.27** | |||||||||||
| PH (cm) | 0.28** |
Correlations were performed using the 199 lines of the population. Data used were the means of 36 plants for root traits, SDW, and R/S, of 12 independent measurements of PH (4 years, three replicate plots per year), and three samples (single year, three replicate plots) for TGW.
Pairwise correlation coefficients were significant at the *5% or **1% significance level.
DH, doubled-haploid; TRL, total root length; TRSA, total root surface area; TRVol, total root volume; SLL, seminal laterals length; SAL, seminal axes length; SLSA, seminal laterals surface area; SASA, seminal axes surface area; SLVol, seminal laterals volume; SAVol, seminal axes volume; RDW, root dry weight; SDW, shoot dry weight; R/S, root to shoot ratio; PH, plant height; TGW, thousand grain weight.
QTL detection
A total of 43 QTLs for root, shoot, and seed traits were identified using QTL Cartographer. These QTLs were distributed on eight linkage groups (Fig. 2; Supplementary Table S1 available at JXB online). The majority of the QTLs were clustered on 2D, 3A, 3B, 4D, 5A, and 6A (Fig. 2). Thirty-one QTLs for root traits were detected on linkage groups 2A, 2D, 3A, 3B, 4D, 5A, 5BS, and 6A; amongst them, nine QTLs were detected for root length, nine QTLs for root surface area, 11 QTLs for root volume, and two QTLs for RDW. Two QTLs were identified for SDW and one for R/S. The five PH QTLs were distributed at five loci on five linkage groups, all of which corresponded to PH QTLs previously reported (Griffiths et al., 2012). Four QTLs for TGW were detected on four linkage groups. Seventeen alleles were contributed by Avalon, and 26 by Cadenza (Supplementary Table S1).
Fig. 2.
QTL mapping for root (blue), plant height (red), shoot (SDW and R/S; black), and TGW (purple) in molecular linkage maps. QTLs for all traits are indicated by bars and caps, which are 1- and 2-LOD likelihood intervals. Data are means of 36 plants for root traits, SDW, and R/S, of 12 independent measurements of PH (4 years, three replicate plots per year), and three samples (single year, three replicate plots) for TGW.
The wheat roots were resolved into seminal axes and laterals branching off these axes. No nodal roots were present. For total root length (seminal axes+laterals), four TRL QTLs were detected and mapped on chromosomes 5BS, 3B, 6A, and 4D; three SLL QTLs were detected and mapped on chromosomes 5BS, 3B, and 3A; two SAL QTLs were detected and mapped on chromosomes 4D and 6A. For root surface area, eight QTLs for TRSA located on chromosomes 2D, 4D, and 2A; three SLSA QTLs located on chromosomes 5BS, 3B, and 3A; and three SASA QTLs located on chromosomes 2D, 5A, and 4D. Eleven QTLs were detected for root volume. Four QTLs for TRVol were mapped on 5A, 3B, 2D, and 4D; four QTLs for SLVol were mapped on 3B, 5BS, 3A, and 6A; and three QTLs for SAVol were mapped on 5A, 3B, and 2D. These loci explained 6.03–16.0% of the variation for the respective root traits.
The QTLs (SAVol and SASA) on chromosomes 2D and 5A were associated with seminal roots, whilst those on 3A and 5BS (SLVol, SLSA, and SLL) were associated with lateral roots; those QTLs on 3B, and possibly 6A, were associated with both.
For PH, five QTLs on chromosomes 2D, 4D, 3A, 3B, and 6A were detected and together explained 74.4% of the variation for PH. Two QTLs for SDW on chromosomes 4D and 5A (explaining 34.5% and 8.64% of the variation) and one QTL for R/S on chromosome 4D (explaining 21.1% of the variation) were detected. Four QTLs for TGW were detected on chromosomes 3A, 4D, 5A, and 6A, together explaining 82.5% of the TGW variation.
The major QTL for roots was located on chromosome 5BS and explained 16.0% of the variation for SLL. The main QTLs for PH and TGW were mapped on 4D and 6A, which explained 30.2% and 16.5% of the variation for PH and TGW, respectively.
Some but not all QTLs co-located on the genetic map (Fig. 2; Supplementary Table S2 at JXB online). The root traits PH, TGW, SDW, and R/S QTLs were found on chromosome 4D, and located between the same markers (Pt-0431.4B4D and wPt-5809.4B4D). The positive alleles were all from Cadenza, except for the R/S QTL, where the positive allele came from Avalon. Root traits, PH, and TGW QTLs were detected on 3A and 6A in different marker intervals. Cadenza alleles increased the SLL, SLSA, SLVol, and TGW, whereas Avalon alleles increased PH on chromosome 3A. On chromosome 6A, Avalon alleles increased TRL, SAL, SLVol, PH, and TGW. Both root traits and PH QTLs were detected on chromosome 2D in the same maker interval (gwm132 and wPt-9997.2DS) and on chromosome 3B in different marker intervals with positive contributions from both Avalon and Cadenza; both root traits and TGW QTLs were detected on chromosome 5A in the same marker intervals (gwm293b.5ASM–gwm156a.5ASM); only root traits were detected on chromosome 2A and 5BS, and all of the positive alleles came from Cadenza.
Effects of Rht genes on root growth
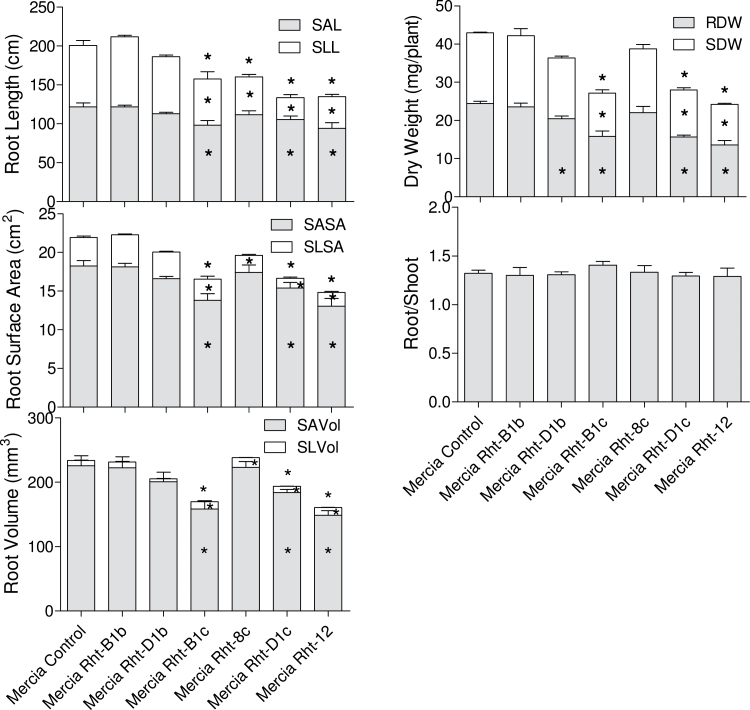
Root traits were determined for a series of Rht NILs and for control Mercia rht plants using the same cigar roll paper culture system. TRL, SLL, SAL, TRSA, SLSA, SASA, TRVol, SLVol, SAVol, RDW, SDW, and R/S were measured (Fig. 3). TRL, SLL, SLSA, and SLVol were significantly reduced in the dwarf and semi-dwarfing lines (Rht-B1c, Rht-D1c, Rht-8c, and Rht12) compared with the control (rht). The dwarf lines (Rht-B1c, Rht-D1c, and Rht12) had significantly shorter SAL, and smaller TRSA, TRVol, SASA, SAVol, and SDW compared with the control. The genotypes (Rht-D1b, Rht-B1c, Rht-D1c, and Rht12) showed a significantly decreased RDW. There was no significant difference between Rht lines and the control in R/S.
Fig. 3.
Effect of Rht alleles on seminal axes (shaded bar), seminal lateral roots (white bar), and total root traits (shaded + white). Data are the means of three replicates, each replicate consisting of four plants. Error bars are standard errors of these means. Significant differences from the Mercia control are indicated with an asterisk (*P < 0.05) in the appropriate bar or above the stacked bar for the total value.
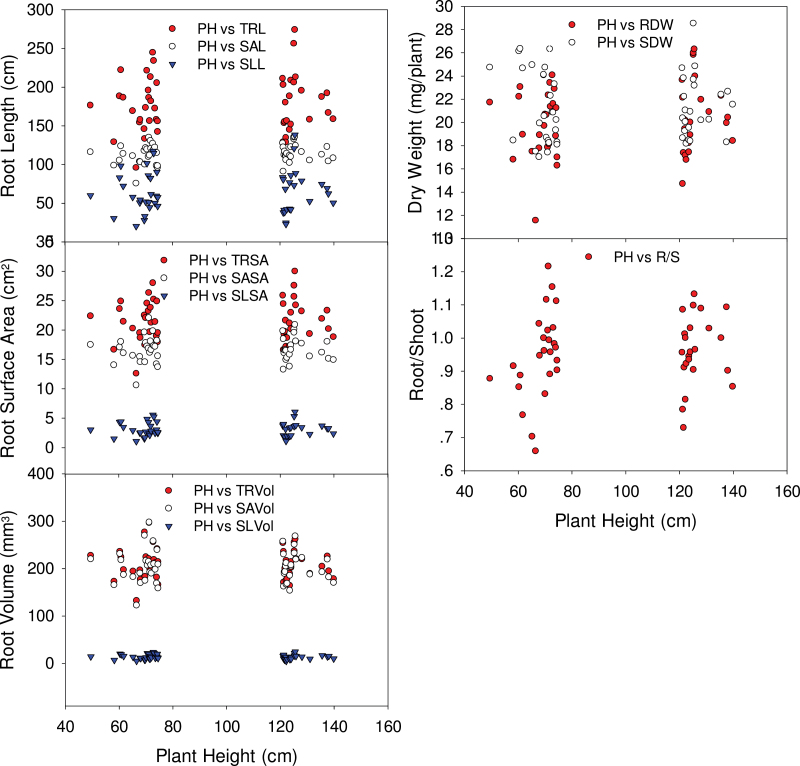
Correlations between PH and root traits, SDW, and R/S in a diverse set of wheat cultivars
Figure 4 shows the relationships of PH to root traits (TRL, SAL, SLL, TRSA, SASA, SLSA, TRVol, SAVol, SLVol, and RDW), SDW, and R/S for 25 short and 23 tall wheat varieties. In spite of there being a wide range of PH (~50–140cm), no correlations between PH and any of these traits was apparent, either within or across the two groups of varieties.
Fig. 4.
Plant height (PH) as a function of selected root traits for 25 short and 23 tall lines from the Watkins collection of wheat cultivars. Data points are the means of three replicates, each replicate consisting of four plants.
Discussion
Components of root architecture and PH are important quantitative traits of wheat. TRL, SLL, SAL, SLSA, SLVol, RDW, and SDW were significantly and positively correlated with PH in this work. Understanding the genetic control of root morphology and PH is crucial for wheat breeding for yield and sustainability. However, roots grow below the soil surface and it is difficult to determine the root traits (Nagel et al., 2009). Many laboratory-based root culture systems (e.g. soil, sand, hydroponic, and paper systems) have been developed to investigate root morphology, mainly at the seedling stage (Zhu et al., 2005a , b ; Yang et al., 2010). The paper roll culture system provides a high-throughput screen to investigate root morphology on a large scale, albeit limited to young seedlings. Clearly extrapolation to field performance for mature plants should be made with caution; for example, in the seedlings used here, there were no nodal roots, and, in addition, mature wheat plants produce roots which penetrate to a depth of in excess of 1 m (Thorup-Kristensen et al., 2009) which may involve quite different processes. In spite of these limitations, QTLs identified at the young seedling stage with this system corresponded to mature PH determined in the field and TGW QTLs. This indicates that there are common genes, or at least closely linked genes, contributing to both traits. Furthermore, the correlation of seedling rht QTLs with PH rht QTLs determined in the field was indicative of the validity of the seedling screen. There is an impact of seed size on early seedling root growth (see, for example, Bremner et al., 1963; Evans and Bhatt, 1977). Of the four QTLs for TGW, three appeared to correspond to root traits: two principally involved in seminal axes traits, and one with the seminal lateral traits. The impact of a large seed size on promoting seedling root growth is predictable, and these QTLs probably have no relevance for extrapolation to more mature roots. The other identified root trait QTLs which are independent of TGW are clearly expressed at the seedling stage and it would be interesting to know if these traits continue to be expressed as the plant matures.
In the present study, the paper culture system together with the imaging technique employed was able to distinguish between seminal axes and seminal lateral roots. Some QTLs for root and shoot traits were co-located and some were independent, and QTLs for seminal axes roots and seminal laterals were quite distinct. Seminal axes traits and PH were co-located (on chromosomes 2D, 4D, and 6A), but seminal lateral root QTLs were mostly independent of PH (with the possible exception of the QTL on 6A). Furthermore, it was not always the case for co-locating QTLs that positive alleles for root traits and PH came from the same background (the QTL on 2D): either there are separate but closely located genes, or the same gene has quite contrasting effects on shoots and roots. QTLs for seminal laterals which were probably completely independent of PH or seed size were found on 3A, 3B, and 5BS; determining the molecular basis of these QTLs is a next step. Chromosome 4D had QTLs for seminal axes traits (TRL, SAL, TRSA, SASA, TRVol, SAVol), RDW, SDW, R/S, PH, and TGW between RhtMrkD1.4D and wPt-0431.4B4D/wPt-5809.4B4D, with the only positive allele from Avalon being R/S, and all of the others having a positive allele originating from Cadenza.
Coincidence of QTLs as described here is suggestive of a genetic relationship between Rht genes and the genes controlling root traits. Rht-B1b and Rht-D1b have been mapped to chromosomes 4B and 4D, respectively (Ellis et al., 2002), Rht-B1c and Rht-D1c on chromosome 4BS (Börner et al., 1997), and Rhtl2 and Rht-8c on chromosome 5A (Worland et al., 1994) and chromosome 2D (Korzun et al., 1998), respectively. Rht NILs were investigated using the rolled germination paper screening system and the results were broadly similar to those reported for soil columns and field assessments (Wojciechowski et al., 2009). In this study, there were no significant differences between control and NILs with Rht-B1b and Rht-D1b for TRL, SAL, and SLL. Wojciechowski et al. (2009) found that neither total root length nor any of the components constituting the total length for Rht-B1b, Rht-D1b, and Rht8c NILs were significantly different compared with the control plants in gel chambers, in soil-filled columns, and in the field. Similarly the TRL of isogenic semi-dwarfing lines did not decrease in the study of Miralles et al. (1997). The NIL with Rht-8c was found to have significantly shorter TRL and SLL, but not SAL. This is in contrast to the data of Wojciechowski et al. (2009), and may be because the methods used were different in these two studies, or because the imaging procedure used here is able to distinguish primary and lateral roots. Four dwarfing genes in this study (Rht-B1c, Rht-D1c, Rht-8c, and Rht12) reduced root lengths significantly; this is coincident with the results in the field experiment (with the exception of Rht-8c) of Wojciechowski et al. (2009). Furthermore, in the present study, Rht-B1c, Rht-D1c, and Rht-12 significantly reduced TRSA, TRVol, SASA, SAVol, SLSA, SLVol, RDW, and SDW; Rht-8c reduced SLSA and SLVol significantly; and Rht-D1b only reduced RDW significantly. However, Wojciechowski et al. (2009) found that Rht-D1b and Rht-12 increased the root dry masses in the field and gel chamber experiments, but the other NILs showed no significant differences to the rht control. Only Rht-D1c reduced shoot dry weight (there was no significant difference with the other genes) in the gel chamber experiment and there were no significant differences in the soil-filled column and field experiments; no significant differences between the root/shoot dry mass ratios of Rht NILs were observed in the soil-filled column experiment. These contrasting findings may be due to the effect of either different genetic backgrounds or different growing environments, or both. Rht genes did not significantly influence the R/S in this study, because RDW and SDW of NILs varied similarly.
In the present study, short and tall lines from the Watkins Collection were investigated using the paper roll culture system. Root traits did not correlate with mature PH in field trials. The older taller varieties had no propensity for longer seedling seminal roots, at least at this seedling stage in the screening system employed. In addition to the complexity of multiple genes contributing to PH (Ahmed et al., 2000), the Watkins Collection varies for many traits which will confound a simple correlation.
In this study, QTLs for root traits, seed size, and PH were not always co-located, indicating an opportunity for independent selection. The results indicated that there is a close relationship between root traits and PH, as some genes, or closely linked genes, control both root traits and PH; however, other genes only influence root traits or PH. Furthermore, separate QTLs for seminal axes and seminal lateral roots were distinguished, offering further possibilities for selective breeding.
Supplementary data
Supplementary data are available at JXB online.
Table S1. QTLs for traits at sufficient nutrient supply using composite interval mapping, listed by trait.
Table S2. Summary of QTL loci by chromosome.
Acknowledgements
We thank Simon Griffiths and Simon Orford (John Innes Centre) for providing the Watkins Collection lines with associated height data, and Peter Buchner, Saroj Parmar, Professor Yingang Hu, and Dr Jing Wang for help and advice on the paper roll technique. Rothamsted Research receives support from the BBSRC of the UK. This research forms part of the 20:20 Wheat® programme. The field trials were conducted as part of the Defra-funded WGIN project. CB was supported by a CSC Chinese Joint PhD scholarship.
Glossary
Abbreviations:
- DH
doubled-haploid
- NIL
near isogenic line
- PH
plant height
- QTL
quantitative trait locus
- RDW
root dry weight
- R/S
root to shoot ratio
- SAL
seminal axes length
- SASA
seminal axes surface area
- SAVol
seminal axes volume
- SDW
shoot dry weight
- SLL
seminal laterals length
- SLSA
seminal laterals surface area
- SLVol
seminal laterals volume
- TGW
thousand grain weight
- TRL
total root length
- TRSA
total root surface area
- TRVol
total root volume.
References
- Addisu M, Snape JW, Simmonds JR, Gooding MJ. 2009. Reduced height (Rht) and photoperiod insensitivity (Ppd) allele associations with establishment and early growth of wheat in contrasting production systems. Euphytica 166, 249–267 [Google Scholar]
- Ahmed TA, Tsujimoto H, Sasakuma T. 2000. QTLs associated with plant height and related characters in haploid wheat. Breeding Science 50, 267–273 [Google Scholar]
- Basten C, Weir BS, Zeng ZB. 1994. Zmap—a QTL cartographer. In: Smith C, Gavora JS, Benkel B, Chesnais J, Fairfull W, Gibson JP, Kennedy BW, Burnside EB, eds. Proceedings of the 5th World Congress on Genetics Applied to Livestock Production: Computing Strategies and Software, Guelph, Organizing Committee; 22, 85–66 [Google Scholar]
- Börner A, Röder M, Korzun V. 1997. Comparative molecular mapping of GA insensitive Rht loci on chromosomes 4B and 4D of common wheat (Triticum aestivum L.). Theoretical and Applied Genetics 95, 1133–1137 [Google Scholar]
- Bremner PM, Eckersall RN, Scott RK. 1963. The relative importance of embryo size and endosperm size in causing the effects associated with seed size in wheat. Journal of Agricultural Science 61, 139–145 [Google Scholar]
- Chapman SC, Mathews KL, Trethowan RM, Singh RP. 2007. Relationships between height and yield in near-isogenic spring wheats that contrast for major reduced height genes. Euphytica 157, 391–397 [Google Scholar]
- Churchill G, Doerge R. 1994. Empirical threshold values for quantitative trait mapping. Genetics 138, 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MH, Spielmeyer W, Gale KR, Rebetzke GJ, Richards RA. 2002. ‘Perfect’ markers for the Rht B1b and Rht D1b dwarfing genes in wheat. Theoretical and Applied Genetics 105, 1038–1042 [DOI] [PubMed] [Google Scholar]
- Evans LE, Bhatt GM. 1977. Influence of seed size, protein content and cultivar on early seedling vigor in wheat. Canadian Journal of Plant Science 57, 929–935 [Google Scholar]
- Gregory P. 2006. Roots and the architecture of root systems. In: Plant roots: growth, activity and interactions with soils. Oxford, Blackwell Publishing, 18–44 [Google Scholar]
- Griffiths S, Simmonds J, Leverington M, et al. 2009. Meta-QTL analysis of the genetic control of ear emergence in elite European winter wheat germplasm. Theoretical and Applied Genetics 119, 383–395 [DOI] [PubMed] [Google Scholar]
- Griffiths S, Simmonds J, Leverington M, Wang Y, Fish L, Sayers L, Alibert L, Orford S, Wingen L, Snape J. 2012. Meta-QTL analysis of the genetic control of crop height in elite European winter wheat germplasm. Molecular Breeding 29, 159–171 [DOI] [PubMed] [Google Scholar]
- Korzun V, Röder MS, Ganal MW, Worland AJ, Law CN. 1998. Genetic analysis of the dwarfing gene (Rht8) in wheat. Part I. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat (Triticum aestivum L.). Theoretical and Applied Genetics 96, 1104–1109 [Google Scholar]
- Laperche A, Devienne-Barret F, Maury O, Gouis JL, Ney B. 2006. A simplified conceptual model of carbon/nitrogen functioning for QTL analysis of winter wheat adaptation to nitrogen deficiency. Theoretical and Applied Genetics 113, 1131–1146 [DOI] [PubMed] [Google Scholar]
- Liu X, Li R, Chang X, Jing R. 2013. Mapping QTLs for seedling root traits in a doubled haploid wheat population under different water regimes. Euphytica 189, 51–66 [Google Scholar]
- McCaig TN, Morgan JA. 1993. Root and shoot dry matter partitioning in near-isogenic wheat lines differing in height. Canadian Journal of Plant Science 73, 679–689 [Google Scholar]
- Miller TE, Ambrose MJ, Reader SM. 2001. The Watkins Collection of landrace-derived wheats. In: Caligari PDS, Brandham PE, eds. Wheat taxonomy: the legacy of John Percival. The Linnean Society of London, Special Issue No. 3. An edited volume arising from papers presented at The Percival Symposium: Wheat—Yesterday, Today and Tomorrow, The University of Reading, UK, 12–13 July 1999 London: Academic Press, 113–120 [Google Scholar]
- Miralles DJ, Slafer GA, Lynch V. 1997. Rooting patterns in near-isogenic lines of spring wheat for dwarfism. Plant and Soil 197, 79–86 [Google Scholar]
- Nagel KA, Kastenholz B, Jahnke S, et al. 2009. Temperature responses of roots: impact on growth, root system architecture and implications for phenotyping. Functional Plant Biology 36, 947–959 [DOI] [PubMed] [Google Scholar]
- Navabi A, Iqbal M, Strenzke K, Spaner D. 2006. The relationship between lodging and plant height in a diverse wheat population. Canadian Journal of Plant Science 86, 723–726 [Google Scholar]
- Ren Y, He X, Liu D, Li J, Zhao X, Li B, Tong Y, Zhang A, Li Z. 2011. Major quantitative trait loci for seminal root morphology of wheat seedlings. Molecular Breeding 30, 139–148 [Google Scholar]
- Sharma S, Xu S, Ehdaie B, Hoops A, Close TJ, Lukaszewski AJ, Waines JG. 2011. Dissection of QTL effects for root traits using a chromosome arm-specific mapping population in bread wheat. Theoretical and Applied Genetics 122, 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique KHM, Belford RK, Tennant D. 1990. Root:shoot ratios of old and modern, tall and semi-dwarf wheats in a mediterranean environment. Plant and Soil 121, 89–98 [Google Scholar]
- Thorup-Kristensen K, Cortasa MS, Loges R. 2009. Winter wheat roots grow twice as deep as spring wheat roots, is this important for N uptake and N leaching losses? Plant and Soil 322, 101–114 [Google Scholar]
- Wojciechowski T, Gooding MJ, Ramsay L, Gregory PJ. 2009. The effects of dwarfing genes on seedling root growth of wheat. Journal of Experimental Botany 60, 2565–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worland AJ, Sayers EJ, Börner A. 1994. The genetics and breeding potential of Rhtl2, a dominant dwarfing gene in wheat. Plant Breeding 113, 187–196 [Google Scholar]
- Yang M, Ding G, Shi L, Feng J, Xu F, Meng J. 2010. Quantitative trait loci for root morphology in response to low phosphorus stress in Brassica napus . Theoretical and Applied Genetics 121, 181–193 [DOI] [PubMed] [Google Scholar]
- Zeng Z. 1994. Precision mapping of quantitative trait loci. Genetics 136, 1457–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Kaeppler SM, Lynch JP. 2005a Mapping of QTL controlling root hair length in maize (Zea mays L.) under phosphorus deficiency. Plant and Soil 270, 299–310 [Google Scholar]
- Zhu J, Kaeppler SM, Lynch JP. 2005b Mapping of QTLs for lateral root branching and length in maize (Zea mays L.) under differential phosphorus supply. Theoretical and Applied Genetics 111, 688–695 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.