Abstract
The Laboratory Without Walls is a modular field application of molecular biology that provides clinical laboratory support in resource-limited, remote locations. The current repertoire arose from early attempts to deliver clinical pathology and public health investigative services in remote parts of tropical Australia, to address the shortcomings of conventional methods when faced with emerging infectious diseases. Advances in equipment platforms and reagent chemistry have enabling rapid progress, but also ensure the Laboratory Without Walls is subject to continual improvement. Although new molecular biology methods may lead to more easily deployable clinical laboratory capability, logistic and technical governance issues continue to act as important constraints on wider implementation.
Introduction: the Origins of the Laboratory without Walls
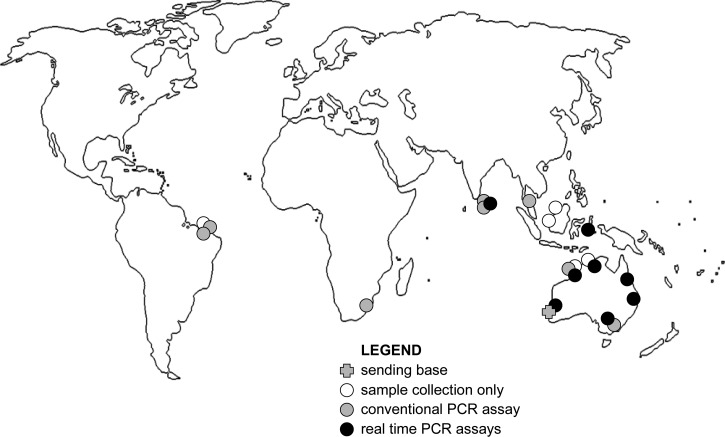
The current iteration of the Laboratory Without Walls (LWW) is far removed from its earliest version as a sample collection kit and light microscope flown into a remote Western Australian community during a melioidosis outbreak investigation over a decade ago1 (for chronology see Table 12–10). At that point the only rapid clinical laboratory method applicable to a bacterial emerging infectious disease was brightfield microscopy. Molecular confirmation had been described,11 but was not implemented in the State pathology service at that point. By the time a second melioidosis outbreak investigation was launched by the same laboratory group,2 a confirmatory molecular method was used in the state reference laboratory on a routine basis but was not easily transportable. It took repeated visits to insert the molecular method into a Brazilian public health laboratory (Figure 1 ). In the latter stages of that investigation it became clear that increasing restrictions on the movement of biosecurity sensitive biological agents would eventually place a barrier to international cooperation in the emerging infectious disease response, particularly shipment of live agents.12 Working closer to home and within national boundaries we recognized a corresponding need to take a highly targeted laboratory capability into a location of public health interest. We deployed our first molecular microbiology capability to a northern Australian mine site in 2006 and demonstrated Burkholderia pseudomallei, the bacterial cause of melioidosis, in environmental samples within hours of sample collection.3 The value of such a short turnaround time for potential health threat detection was immediately evident, because it was possible to conduct a second, more targeted sample acquisition while still on location.
Table 1.
Development chronology of the Laboratory Without Walls
| Year | Location | Diseases | Microscopy | PCR assays | Reference |
|---|---|---|---|---|---|
| 1997 | Northern WA | Melioidosis | Y | N | 1 |
| 1999 | Northern WA | Melioidosis | N | N | |
| 2000 | Northern WA | Melioidosis | N | N | |
| 2000 | Malaysia | Melioidosis | N | N | |
| 2003 | Malaysia | Melioidosis | N | N | |
| 2004 | NE Brazil | Melioidosis | N | N | 2 |
| 2005 | Northern WA | Melioidosis | N | N | 3 |
| 2006 | NE Brazil | Melioidosis | N | B. pseudomallei | |
| 2006 | Northern WA | Melioidosis | N | B. pseudomallei | 3 |
| 2008 | Malaysia | Melioidosis, Leptospirosis, Tuberculosis | Y | B. pseudomallei, Leptospira sp. | |
| 2009 | Sri Lanka | Melioidosis | N | B. pseudomallei | 4,5 |
| Leptospirosis | |||||
| 2009 | Northern WA | Influenza A | N | Influenza duplex | 6 |
| Urogenital infections, Staphylococcal infections | Urogenital duples | ||||
| MRSA assay | |||||
| 2009 | Central Queensland | Influenza A | N | Influenza duplex | 7 |
| Melioidosis | B. pseudomallei | ||||
| Leptospirosis | Leptospira sp. | ||||
| 2010 | Northern Queensland | Australian arboviruses | Y | Ross River, Murray Valley, Barmah Forest viruses, Leptospira sp., B. pseudomallei | 8 |
| Leptospirosis | |||||
| 2010 | Sri Lanka | Melioidosis | Y | B. pseudomallei | 9 |
| Leptospirosis | Leptospira sp. | ||||
| Scrub typhus | Orienta sp. | ||||
| Tuberculosis | Mycobacteria | ||||
| M. tuberculosis genotyping | |||||
| 2010 | Timor Leste | Tuberculosis | Y | Mycobacterial panel | 10 |
| Malaria | Malaria panel | ||||
| Septicemia | Bacteremia panel | ||||
| Urogenital infections | Urogenital duplex | ||||
| Mycobacterial genotyping | |||||
| 2011 | Central Queensland | Influenza | Y | Influenza duplex | |
| Leptospirosis | Leptospirosis | ||||
| Australian arboviruses | Ross River, Murray Valley & Barmah Forest viruses | ||||
| 2012 | Northern WA | Australian arboviruses | Y | Ross River, Murray Valley & Barmah Forest viruses | |
| Septicaemia | Bacteremia panel | ||||
| 2012 | Rural New South Wales | Australian arbovirus infection | N | Ross River, Murray Valley & Barmah Forest viruses | |
| Epidemic gastroenteritis | Influenza duplex | ||||
| Influenza duplex | Norovirus 1, 2 | ||||
| 2012 | Central WA | Septicaemia panel | Y | Ross River, Murray Valley & Barmah Forest viruses | |
| Influenza | Bacteremia panel | ||||
| Australian arbovirus infection | Influenza duplex | ||||
| 2012 | Sri Lanka | Septicaemia, malaria, dengue, scrub typhus | Y | Bacteremia panel | |
| Melioidosis | Malaria panel | ||||
| Leptospirosis | Arbovirus panel | ||||
| Orienta assay | |||||
| B. pseudomallei | |||||
| Leptospira sp. |
Figure 1.
Field laboratory deployments leading to current Laboratory Without Walls real-time polymerase chain reaction (PCR) repertoire.
Emerging infectious disease response.
In 2008 the World Health Organization Laboratory Capability-Building Program approved a proposal to develop a laboratory response to a small group of locally emergent bacterial infections, including melioidosis, leptospirosis, and scrub typhus. Although the intent was to establish an indigenous stand-alone capability, the initial step was a methods demonstration with a deployable molecular laboratory. The LWW initially fielded a melioidosis confirmation capability in Central Province, Sri Lanka in early 2009 after a trial run in remote northern Malaysia.4 This was followed up by a return visit 2 years later using a wider range of methods including a field genotyping method for Mycobacterium tuberculosis.9 Sri Lankan colleagues were able to subsequently publish on molecular confirmation of Rickettsial infections and Leptospirosis without further input from the Western Australian capability-building group.13,14 The next step in this development path is to enable a truly stand-alone capability with a degree of laboratory diversification, stratification, and national surveillance networking.
Around that time the Australian Public Health Laboratory Network (PHLN) had to contend with a major disease response initiative at least every second year, with smaller scale challenges more frequently. The majority of these infections was of viral etiology and included severe acute respiratory syndrome, human influenza of avian origin, and periodic dengue virus incursions.15–17 The arrival of influenza A/H1N1/09 in May 2009 prompted a coordinated PHLN response, resulting in swift implementation of the laboratory component of the Australian Influenza Pandemic Plan. Within weeks a Western Australian-based group had demonstrated a deployable capability in northwestern Australia,6 and shortly afterward deployed its portable laboratory to support Commonwealth government agencies in central Queensland.7 Field deployable real-time influenza assay results were consistent with subsequent reference laboratory analysis of samples retained for confirmatory analysis, and were significantly more sensitive than an influenza rapid antigen test in use for preliminary screening. The effect was an immediate shortening of clinical laboratory turnaround time from days to hours, enabling more timely initiation of influenza-specific therapy and specific infection control measures.18
Tropical diseases in northern Australia and Timor Leste.
During the 2009 influenza epidemic response, a fully field portable laboratory module was taken overland to a distant location and operated with portable generator power, as had already been done in northern Malaysia. The portable module comprised a self-contained molecular microbiology laboratory bench capable of operation from the back of a vehicle powered by a portable generator. The modular concept extended to the main field laboratory unit consisting of another module capable of independent operation, whereas the smaller module was in use at a distant location. The epidemic influenza deployment led to use of both modules in another laboratory deployment to remote northern Queensland, supporting an army environmental health threat assessment.8 Later that year, we established the Laboratory Without Walls as a not-for-profit agency specifically to develop, deploy, demonstrate, and otherwise contribute to clinical laboratory capability-building in remote, resource-limited locations. The first formal LWW deployment was a preliminary feasibility study that involved sending a laboratory team to Dili in Timor Leste to pave the way for insertion of a molecular tuberculosis laboratory service.10 This LWW deployment was the first successful operation of molecular mycobacteriology and parasitology in a clinical laboratory in Timor Leste and resulted in the establishment of a permanent molecular mycobacteriology capability funded by the Australian Commonwealth under its AusAid development program.
Equipment and reagents.
At first, LWW relied on a conventional thermocycler (ABI 2720, Applied Biosystems, Foster City, CA) and a microfluidic chip (Expert 2100, operating DNA 1000 chip, Agilent, Little Falls, DE) to resolve amplicons in a field setting.3,4 The bioanalyzer proved to be highly robust under extreme conditions such as the dust of central Queensland and the humidity of a Malaysian rainforest, but the thermocycler was more vulnerable to fluctuations in environmental conditions (Table 2). Variable mains power supply in remote settings makes molecular equipment susceptible to surges and power interruptions, necessitating the use of uninterrupted power supply devices (UPS). Even with the use of UPS, some power fluctuations have been more than these devices can cope with, therefore the Timor Leste clinic needed to install a reliable generator before a molecular mycobacteriology service could be sustained. The steady move toward real-time polymerase chain reaction (PCR) assays has taken the conventional thermocycler out of the limelight, though it still has a niche use for functions such as genotyping. The weight of the early real-time thermocyclers made them less suitable for LWW deployment. The first real-time thermocycler to demonstrate a combination of robust construction, suitability for extreme environmental conditions, and incorporation of a shipping lock on delicate components was a 48-well, three-channel model (StepOne, ABI). This saw service during the influenza pandemic of 2009,7 the arbovirus surveillance activity in May 2010, and the Timor Leste LWW deployment in December 2012.8,10 During these activities the real-time thermocycler was paired with a magnetic bead DNA purification robot (MagMax-24, ABI) for sample preparation. The extraction robot improved reproducibility of nucleic acid extraction and reduced reliance on repetitive fluid handling, which proved difficult to sustain in a warm climate. More recently smaller, easily portable real-time thermocyclers have come into use. The result is a reduction in the size and weight of molecular equipment, raising the potential for a backpack-portable laboratory.
Table 2.
Practical application of molecular microbiology methods by stage of deployment
| Lessons learned | Assays | Locations |
|---|---|---|
| 1. Preparation | ||
| Pre-departure assay checks | Burkholderia assay, 2005, 2008 | Brazil, Ceará; South Africa |
| Pre-departure equipment checks include small mission-essential items | Influenza, Burkholderia, Leptospira assays, 2009; Arbovirus assays, 2010 | Australia, Queensland |
| 2. Insertion | ||
| Equipment arrival delay | Norovirus, influenza, 2012 | Australia, New South Wales |
| Equipment and assay checks on arrival | Influenza assay, 2009 | Australia, Queensland |
| Reagent cold chain during prolonged insertion phase | Full assay range, 2011 | Australia, Queensland |
| 3. Operation | ||
| Power interruption during PCR assay | Mycobacteria, Plasmodium, Burkholderia, Leptospira, Sepsis panel assays, 2009–12 | Central Sri Lanka, Timor Leste, rural Western Australia |
| Thermocycler failure caused by humidity | Burkholderia assay, 2008 | Malaysia, Perak |
| Dust contamination during sample preparation | Sepsis panel, 2012 | rural Western Australia |
| Dust contamination during amplicons analysis | Leptospira, Burkholderia remote field assays, 2009 | Australia, Queensland |
| Reagent deterioration during prolonged operation | Influenza epidemic, 2009 | Australia, Queensland |
| 4. Withdrawal | ||
| Confirmatory assay sample deterioration during withdrawal | Arbovirus assays, 2012 | Australia, Queensland |
| Delayed equipment return during withdrawal | Influenza, 2009; Norovirus and influenza, 2012 | Australia, Queensland; rural Western Australia |
The switch from conventional to real-time PCR assays has simplified some aspects of the methods used by the LWW. It is now possible to deploy a molecular laboratory for some applications without the need for microfluidic laboratory chips, their reagents requiring different cold chain conditions. However, complex fluid handling requirements for making up master mix are not suited to field work. Mixes therefore require pre-dispensing, limiting their shelf life, and forcing careful calculation of expected assay demand. The improved performance of newer real-time thermocyclers and molecular probes reduces cross talk between detection channels. However, methods originally designed for fixed laboratories results in a need to dispense individual reagent aliquots in the field. This step is potentially vulnerable to contamination and has led us to use a comprehensive set of positive, non-template and extraction controls with each assay.7
The logistics of deployable PCR assays.
Logistics has been described as the detailed organization and implementation of a complex operation, and specifically in a military context the activity of organizing the movement, equipment, and accommodation of troops.19 Supply chain logistics is specifically concerned with the demand, distance, and duration of supply while operating a particular service. Supply logistics has been one of the two main challenges to LWW deployment; both within Australia and to overseas destinations.5 Considerations that rarely occur to the staff of a large public health reference laboratory or clinical pathology service are important obstacles to safe and effective operation of a molecular biology laboratory in the field. These logistic issues can be divided into four time-delimited phases: preparation, insertion, operations, and withdrawal (Table 2). The overriding concern in all of these stages is the need to remain self-sufficient throughout the expedition, in terms of staff, equipment, dry consumables, reagents, and maintenance. Nevertheless, a field laboratory does not function well in a professional vacuum. It needs technical governance and reachback to a definitive or confirmatory laboratory. This demands reliable communication for professional networking, which is often patchy in remote locations.
Technical governance.
Contemporary clinical laboratory standards demand a quality management system. This in turn requires quality assurance, diligent record keeping, sample tracking, and staff credentialing. Though the LWW is a work in progress, its operators are not exempt from the obligations of quality control. The current debate on point-of-care tests is instructive and may provide LWW with guidance on how a remote quality system could operate.20 In the interim, the following approach has been taken: assays have been developed in conjunction with and under the guidance of an established molecular laboratory quality management system. Sample handling number, worksheet, and reporting conventions have been followed. Positive, non-template, and extraction controls have been used to determine reliability of results. Complementary test systems have been used for clinical tests, e.g., influenza direct antigen test,7 and positive samples have been shipped to the reference laboratory for confirmatory work. An additional measure found to improve reliability has been the running of trial assays and systems checks immediately before LWW deployments.
Opportunities and challenges to come.
The increasing number of graduates with molecular biology expertise should make operation of LWW procedures more feasible in the future. Miniaturization of equipment platforms and the use of solid-state technology will make the future field deployable laboratory even more portable. However, the replacement of culture-based microbiology with solely molecular methods is still some way off. Falling costs will enable some remote locations to run selected molecular assays, given the initial confidence-building support of LWW or similar initiative. The other key ingredient of these developments; field-compatible microscopy, has been overshadowed by recent advances in molecular biology. The speed of microscopy and its low cost suit it to guiding the selection of appropriate molecular tests, as is beginning to happen in better resourced clinical laboratories investigating the cause of septicemia.21 The chemistry currently used for LWW molecular assays has moved from conventional to real-time PCR. Given the pace of development, this approach is unlikely to be used indefinitely. The low technology demands of loop-mediated PCR (LAMP) have seen this promising chemistry developed for remote applications.22–24 Once issues such as low sensitivity and a technically demanding LAMP development pathway have been overcome, LAMP may have wider application in the LWW. Perhaps the greatest challenge to the field application of molecular biology is the emerging debate between centralized and devolved pathology services. It is apparent that the greatest benefits of molecular biology for the greatest number of patients will be found when rapid clinical pathology assays are placed closest to the patient. The issue is therefore not whether to concentrate clinical molecular biology services in large centralized laboratories or alternatively in small peripheral hospital pathology units. It is more a question of how best to establish effective operational links between the field and the center of excellence, and whether this can be done with simple-to-operate/inflexible laboratory platforms or more complex but flexible molecular systems that require more highly skilled operators. These questions will only be answered by more field-led operational research.
Conclusions
The Laboratory Without Walls is an approach to clinical molecular microbiology that seeks to place emerging technology wherever emerging infectious diseases occur. Those who have joined the LWW on a deployment recognize that this is a work in progress, however have not been dissuaded by the apparent scale of the capability gap. Real-time PCR and other molecular technologies can be operated reliably in infectious diseases hot spots, with appropriate controls and adjustment to environmental conditions. Logistic and technical governance challenges can best be identified by field experience, and can be addressed with the help of industry, service providers, and regulatory bodies. Deployment of a laboratory capability outside the clinical laboratory walls is the necessary response to diseases that readily cross international borders. The Laboratory Without Walls is our answer to diseases without borders.
ACKNOWLEDGMENTS
The author thanks the following for their respective support in the development of the Lab Without Walls: PathWest staff – Adam Merritt, Avram Levy, Gerry Harnett, Glenys Chidlow, David Speers, David Smith; Health Department of Western Australia – Mike Lindsay, Peter Neville, Stuart Garrow; University of Western Australia – Cheryl Johanson, Jay Nicholson, Wendy Erber, Barbara Chang; Lab Without Walls – Barry Mendelawitz, John Kevan, Heather Inglis; 17 Combat Services Support Brigade – Georgeina Whelan; Richard Mallet, Lachlan Sinclair, Ian Marsh, Stan Papastomatis. and Geoff Matthews; Michael Houston, Nathan Flindt, and Natalie Wigg; Richard Bradbury, Adam Scholler, Patricia Veitheer, Jane Currie, and Brady McPherson; Agilent Technologies Australia – Russell McInnes and Rod Minett; Applied Biosystems – Michael Tavaria and Jim Frames; BioRad Australia – Richard Harrison, Thao Nguyen, and Eli Mrkusich.
Footnotes
Financial support: The author gratefully acknowledges funding support from the Government of Western Australia (MERIWA), the World Health Organization, Lab Without Walls, and the Rotary Clubs of Western Australia.
Author's address: Timothy J. J. Inglis, Pathology and Laboratory Medicine, The University of Western Australia, Crawley WA 6009, Australia, E-mail: tim.inglis@uwa.edu.au.
References
- 1.Inglis TJ, Garrow SC, Adams C, Henderson M, Mayo M. Dry-season outbreak of melioidosis in Western Australia. Lancet. 1998;352:1600. doi: 10.1016/S0140-6736(05)61047-1. [DOI] [PubMed] [Google Scholar]
- 2.Rolim DB, Vilar DC, Sousa AQ, Miralles IS, de Oliveira DC, Harnett G, O'Reilly L, Howard K, Sampson I, Inglis TJ. Melioidosis, northeastern Brazil. Emerg Infect Dis. 2005;11:1458–1460. doi: 10.3201/eid1109.050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inglis TJ, Levy A, Merritt AJ, Hodge M, McDonald R, Woods DE. Melioidosis risk in a tropical industrial environment. Am J Trop Med Hyg. 2009;80:78–84. [PubMed] [Google Scholar]
- 4.Inglis TJ, Merritt A, Montgomery J, Jayasinghe I, Thevanesam V, McInnes R. Deployable laboratory response to emergence of melioidosis in central Sri Lanka. J Clin Microbiol. 2008;46:3479–3481. doi: 10.1128/JCM.01254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inglis TJ, Merritt A, Jayasinghe I, Montgomery J, Thevanesam V. Logistic aspects of a deployable molecular microbiology laboratory. J Mil Vet Health. 2008;17:6–10. [Google Scholar]
- 6.Inglis TJ, Murray RJ, Watson M. Trouble in paradise – Tropical, Emergency and Disaster Medicine Conference and Tropical Medicine Summit, Broome, Western Australia, 22–24 May 2009. Med J Aust. 2009;191:597–598. doi: 10.5694/j.1326-5377.2009.tb03343.x. [DOI] [PubMed] [Google Scholar]
- 7.Inglis TJ, Merritt AJ, Levy A, Vietheer P, Bradbury R, Scholler A, Chidlow G, Smith DW. Deployable laboratory response to influenza pandemic: PCR assay field trials and comparison with reference methods. PLoS ONE. 2011;6:e25526. doi: 10.1371/journal.pone.0025526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brumpton BM, McPherson BA, Frances SP, Inglis TJ, McCall B. Townsville Field training area health assessment. ADF Health Journal. 2011;12:45–50. [Google Scholar]
- 9.Merritt AJ, Keehner T, O'Reilly LC, McInnes RL, Inglis TJ. Multiplex-amplified nominal tandem repeat analysis (MANTRA), a rapid method for genotyping Mycobacterium tuberculosis by use of multiplex PCR and a microfluidic laboratory chip. J Clin Microbiol. 2010;48:3758–3761. doi: 10.1128/JCM.00471-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lab Without Walls expedition reports on the worldwide web. 2012. http://labwithoutwalls.org/projects Available at. Accessed January 2013.
- 11.Kunakorn M, Markham RB. Clinically practical seminested PCR for Burkholderia pseudomallei quantitated by enzyme immunoassay with and without solution hybridization. J Clin Microbiol. 1995;33:2131–2135. doi: 10.1128/jcm.33.8.2131-2135.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearson JE, Edwards S. Transportation of reagents, reference materials and samples: the international perspective. Dev Biol (Basel) 2006;126:61–70. [PubMed] [Google Scholar]
- 13.Agampodi SB, Peacock SJ, Thevanesam V, Nugegoda DB, Smythe L, Thaipadungpanit J, Craig SB, Burns MA, Dohnt M, Boonsilp S, Senaratne T, Kumara A, Palihawadana P, Perera S, Vinetz JM. Leptospirosis outbreak in Sri Lanka in 2008: lessons for assessing the global burden of disease. Am J Trop Med Hyg. 2011;85:471–478. doi: 10.4269/ajtmh.2011.11-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Silva N, Wijesundara S, Liyanapathirana V, Thevanesam V, Stenos J. Scrub typhus among pediatric patients in Dambadeniya: a base hospital in Sri Lanka. Am J Trop Med Hyg. 2012;87:342–344. doi: 10.4269/ajtmh.2012.12-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cameron PA, Rainer TH, de Villiers Smit P. The SARS epidemic: lessons for Australia. Med J Aust. 2003;178:478–479. doi: 10.5694/j.1326-5377.2003.tb05318.x. [DOI] [PubMed] [Google Scholar]
- 16.Isaacs D, Dwyer DE, Hampson AW. Avian influenza and planning for pandemics. Med J Aust. 2004;181:62–63. doi: 10.5694/j.1326-5377.2004.tb06172.x. [DOI] [PubMed] [Google Scholar]
- 17.Hanna JN, Ritchie SA, Richards AR, Humphreys JL, Montgomery BL, Ehlers GJ, Pyke AT, Taylor CT. Dengue in north Queensland, 2005–2008. Commun Dis Intell. 2009;33:198–203. [PubMed] [Google Scholar]
- 18.Currie AJ, Heslop DJ, Winter SM. H1N1 in the field: the impact on Australian Defence Force Field exercise Talisman Sabre 09. Australas Emerg Nurs J. 2011;14:103–107. [Google Scholar]
- 19.Oxford Dictionary online definition of logistics. 2012. http://oxforddictionaries.com/definition/logistics?q=logistics Available at. Accessed December 2012.
- 20.Park S, Zhang Y, Lin S, Wang TH, Yang S. Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol Adv. 2011;29:830–839. doi: 10.1016/j.biotechadv.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallet F, Nseir S, Bauman L, Herwegh S, Sendid B, Boulo M, Roussel-Delvallez M, Durocher AV, Courcol RJ. Preliminary clinical study using a multiplex real-time PCR test for the detection of bacterial and fungal DNA directly in blood. Clin Microbiol Infect. 2010;16:774–779. doi: 10.1111/j.1469-0691.2009.02940.x. [DOI] [PubMed] [Google Scholar]
- 22.Parida M, Horioke K, Ishida H, Dash PK, Saxena P, Jana AM, Islam MA, Inoue S, Hosaka N, Morita K. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription loop-mediated isothermal amplification assay. J Clin Microbiol. 2005;43:2895–2903. doi: 10.1128/JCM.43.6.2895-2903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poschl B, Waneesorn J, Thekisoe O, Chitipongvivate S, Karanis P. Comparative diagnosis of malaria infections by microscopy, nested PCR, and LAMP I northern Thailand. Am J Trop Med Hyg. 2010;83:56–60. doi: 10.4269/ajtmh.2010.09-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagdev KV, Kashyap RS, Parida MM, Kapgate RC, Purohit HJ, Taori GM, Daginawala HF. Loop-mediated isothermal amplification for rapid and reliable diagnosis of tuberculosis meningitis. J Clin Microbiol. 2011;49:1861–1865. doi: 10.1128/JCM.00824-10. [DOI] [PMC free article] [PubMed] [Google Scholar]