Abstract
In a previous study conducted in Cyprus, various spotted fever group Rickettsia species were detected and identified in ticks by molecular analysis. Among them, a partially characterized Rickettsia species was detected in Hyalomma anatolicum excavatum and Rhipicephalus turanicus ticks. We report characterization of this rickettsial strain by using polymerase chain reaction sequencing analysis of partial citrate synthase A, outer membrane protein A, outer membrane protein B, and 17-kD protein genes. We propose a provisional name Rickettsia sp. strain Tselenti for this strain until it is isolated and further characterized.
Introduction
Rickettsia spp. are obligate intracellular arthropod-borne bacteria that cause mild to severe diseases in humans and other vertebrate animals worldwide.1–3 Several controversial theories have been suggested for the taxonomic classification of rickettsiae. A widely accepted taxonomic scheme for classification of rickettsiae at the genus and species levels is based on molecular methods; sequencing of genes for 16S ribosomal RNA (rrs), citrate synthase A (gltA), outer membrane protein A (ompA), ompB, surface cell antigen 4 (sca4), and the 17-kD common antigen.4–6 However, other methods have been proposed.
One aspect of taxonomic classification was reported by Gillespie and others in examining rickettsial genes belonging to the core genome of rickettsial genomes.7,8 They proposed the transitional group of rickettsiae as an intermediate evolutionary stage between the ancestral group and the typhus group and the spotted fever group (SFG). Perlman and others9 and Weinert and others10 reported the influence of hosts, types of transmission, and habitat transitional events in the rickettsial evolutionary pathway, and created a robust phylogenetic tree of the genus.
Except for well-known rickettsial diseases (epidemic typhus caused by R. prowazekii, Rocky Mountain spotted fever caused by R. rickettsii, Mediterranean spotted fever caused by R. conorii, and murine typhus caused by R. typhi), at least 10 other rickettsial diseases have been described and recognized in the past 20 years, many of which have been attributed to Rickettsia species within the SFG.1,3 The number of pathogenic rickettsiae within the SFG has increased in recent decades and they are recognized as emerging pathogens worldwide. Seventeen distinct species within this group are currently recognized as human pathogens, causing the so-called tick-borne rickettsioses, which show clinical features such as fever, headache, rash, lymphadenitis, and occasional eschar formation at the site of the tick bite.3 Some species have been isolated from ticks, a large number of species have been recognized or are partially characterized as Rickettsia spp., and most species have been detected in arthropods, but not yet associated with disease.1–3
In Cyprus, three rickettsial species have been detected: R. conorii, R. typhi, and R. felis.11–15 In addition, SFG Rickettsia spp. have been detected by molecular methods in ticks and their animal hosts.16,17 In a recent study, five rickettsial species (R. aeschlimannii, R. massiliae, R. sibirica mongolotimonae, R. hoogstraalii, and “Candidatus R. barbariae”) were detected and genetically characterized by using polymerase chain reaction (PCR) sequencing analysis in naturally infected hard ticks (n = 3,950) collected from domestic and wild animals.18
Some Rickettsia species PCR-amplified DNA sequences have been compared with those of published Rickettsia spp.18 We detected a Rickettsia species in Rhiphicephalus ticks (11 of 805, infection rate = 1.3%), collected from goats, hares and, sheep and in Hyalomma anatolicum excavatum ticks (5 of 301, infection rate = 1.6%) collected from sheep.18 In the current study, we report characterization of this rickettsial strain by PCR sequencing analysis of portions of the gltA, ompA, ompB, and 17-kD genes.
Tick DNA extracts that were obtained during our previous study were stored at −20°C for further processing.18 The Rr19070p-Rr190602n, targeting a 532-basepair portion of the ompA gene; BG1-21/BG2-20, targeting a 650-basepair portion of the ompB gene; and 17kdf/17kdr, targeting a 434-basepair portion of the 17kDa protein gene.19–22 The four sets of primers were used in each of the 16 samples of the unrecognized Rickettsia sp. (total = 64 amplicons). Amplicons were purified using by the QIAquick PCR Purification Spin Kit (QIAGEN, Hilden, Germany).
Sequencing was performed twice by using the described primers in a CEQ 8000 Beckman Coulter Sequencer (Bioanalytica Genotype, Athens, Greece). The BLAST algorithm (National Center for Biotechnology Information, Bethesda, MD) (http://www.ncbi.nlm.nih.gov/BLAST) was used to test homology against known Rickettsia species. Product sequences were aligned by using ClustalW (http://www.clustal.org/) IUB DNA weight matrix with default pairwise and multiple parameters. Accession numbers were obtained from GenBank. Phylogenetic analysis was performed using the DAMBE version software package for data analysis in molecular biology and evolution.3 Tree-building parameters were analyzed by using the neighbor-joining together method and a bootstrap test of 1,000 replicates.23
All gltA amplicons (358 basepairs) showed 99.72% (357 of 358 basepairs) sequence homology with Rickettsia species RR01 (GU056205), R. rhipicephali strain HJ5 (DQ865206), “Candidatus R. Kulagini” strain Kertch (DQ365806), and R. rhipicephali 3-7-6 (U59721). The gltA amplicon was altered by a single amino acid (H282Y–DQ365806 numbering) when compared with Rickettsia species RR01 (GU056205). All partial ompA amplified sequences (510b basepairs) showed 99.21% (506 of 510 basepairs) homology with Rickettsia species ZJ43/2007 (EU258735). In this case, there was a nucleotide alteration, 14C > T, and a triplet insertion, 240^241insGAT, giving an amino acid alteration, S5F, and insertion D80^G81insD, respectively (EU258735 numbering). The ompB gene PCR products (616 basepairs) showed 99.83% (615 of 616 basepairs) sequence homology with Rickettsia species TwKM01 (EF219464).
The difference between the two sequences was made only on the basis of nucleotides because the single base mutation (1248 G > A) is a silent mutation (A416A) (EF219464 numbering). The 399-basepair nucleotide sequence of the 17-kD gene showed 99.24% (396 of 399 basepair) similarity with R. massiliae strain MTU5 (CP000683) and 99.24% (395 of 398 basepair) similarity with R. rhipicephali strain HJ5 (DQ865207). Nucleotide sequences compared with those of R. massiliae strain MTU5 differed at three nucleotide positions (225 A > G, 256 G > A, and 458 G > A), resulting in three amino acid mutations (I75M, A86T, and G153E, respectively).
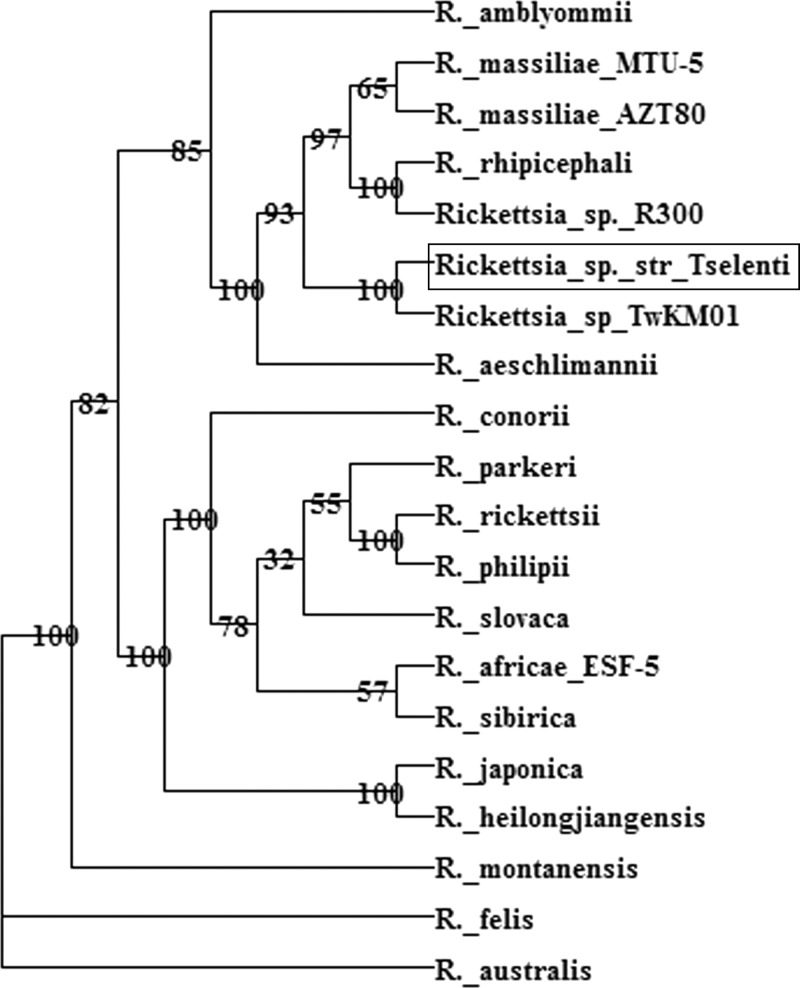
Phylogenetic analysis placed each gene differently in clusters of related phylogenetic trees, making it difficult to determine accurately the species of Rickettsia detected. On the basis of the gltA and ompB amplicon sequences, the new species was placed within the branch of the R. rhipicephali group. Conversely, on the basis of the ompA and 17-kD amplicons, this Rickettsia species seems to be closely related the R. rhipicephali–R. massiliae group, but it forms a separate evolutionary branch. Thus, for a more accurate classification of the organism, the four gene sequences were concatenated for the part of the sequences corresponding to the amplicons length. Phylogenetic analysis placed the new strain in a branch next to Rickettsia species TwKM01 and was supported by a strong bootstrap value (Figure 1).
Figure 1.
Evolutionary relationships of four concatenated genes for citrate synthase A, outer membrane protein A, outer membrane protein B, and 17-kD protein of Rickettsia species strain Tselenti. Box indicates sequences of the new Rickettsia species. Evolutionary history was inferred by using the neighbor-joining method with bootstrap testing (1,000 replicates) with DAMBE version 5.2.68 software.23
According to genotypic criteria and Rickettsia species definition,5 the species analyzed in this study was difficult to assign to one of the valid Rickettsia species. Under these conditions and until isolates are obtained and clearly characterized, we suggest that the species detected in Cyprus be provisionally named Rickettsia species strain Tselenti.
This new Rickettsia strain seems to belong to the SFG, which constitute a phylogenetically distinct clade of rickettsiae composed of more than 30 recognized and validated rickettsial species. Seveteen sequences of Candidatus species are in the process of validation (National Center for Biotechnology Information). In addition, more than 150 non-characterized Rickettsia species, most of whose pathogenicity to humans or animals is unknown, have been deposited and await further identification and description.
Current and past studies18 had detected other SFG rickettsiae in Cyprus, in addition to those previously identified (R. conorii, R. typhi, and R. felis).12 We report a new strain of Rickettsia species that has been characterized and classified as belonging to the SFG. Further investigations are warranted to determine if this organism causes disease to humans. The disease ecology of SFG in Cyprus is complex and needs further research.
GenBank accession numbers of genes sequenced in this study are EU448155, EU448156, EU448157, EU448158, EU448159, EU448160, JF803898, JF803899, GU353188, and GU353185. Six additional (three for ompB and three for 17-kD) accession numbers are still pending.
Footnotes
Authors' addresses: Vassilios Sandalakis, Dimosthenis Chochlakis, and Anna Psaroulaki, Laboratory of Clinical Bacteriology, Parasitology, Zoonoses and Geographical Medicine, Medical School, University of Crete, Voutes, Heraklion, Crete, Greece, E-mails: vassand@gmail.com, surreydimos@hotmail.com, and annapsa@med.uoc.gr. Ioannis Ioannou, Veterinary Services, Nicosia, Cyprus, E-mail: gioannou@vs.moa.gov.cy.
References
- 1.Merhej V, Raoult D. Rickettsial evolution in the light of comparative genomics. Biol Rev Camb Philos Soc. 2011;86:379–405. doi: 10.1111/j.1469-185X.2010.00151.x. [DOI] [PubMed] [Google Scholar]
- 2.Parola P, Paddock CD, Raoult D. Tick-borne rickettsioses around the world: emerging diseases challenging old concepts. Clin Microbiol Rev. 2005;18:719–756. doi: 10.1128/CMR.18.4.719-756.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raoult D, Roux V. Rickettsioses as paradigms of new or emerging infectious diseases. Clin Microbiol Rev. 1997;10:694–719. doi: 10.1128/cmr.10.4.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fournier PE, Dumler JS, Greub G, Zhang J, Wu Y, Raoult D. Gene sequence-based criteria for identification of new Rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J Clin Microbiol. 2003;41:5456–5465. doi: 10.1128/JCM.41.12.5456-5465.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raoult D, Fournier PE, Eremeeva M, Graves S, Kelly PJ, Oteo JA, Sekeyova Z, Tamura A, Tarasevich I, Zhang L. Naming of rickettsiae and rickettsial diseases. Ann N Y Acad Sci. 2005;1063:1–12. doi: 10.1196/annals.1355.002. [DOI] [PubMed] [Google Scholar]
- 6.Eremeeva M, Dasch GA. The genus Rickettsia: microbiology, diversity, and taxonomy. Rev Tunis Infect 5 (Suppl 1) 2011:S7–S14. [Google Scholar]
- 7.Gillespie JJ, Beier MS, Rahman MS, Ammerman NC, Shallom JM, Purkayastha A, Sobral BS, Azad AF. Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS ONE. 2007;2:e266. doi: 10.1371/journal.pone.0000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillespie JJ, Williams K, Shukla M, Snyder EE, Nordberg EK, Ceraul SM, Dharmanolla C, Rainey D, Soneja J, Shallom JM, Vishnubhat ND, Wattam R, Purkayastha A, Czar M, Crasta O, Setubal JC, Azad AF, Sobral BS. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS ONE. 2008;3:e2018. doi: 10.1371/journal.pone.0002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perlman SJ, Hunter MS, Zchori-Fein E. The emerging diversity of Rickettsia. Proc Biol Sci. 2006;273:2097–2106. doi: 10.1098/rspb.2006.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinert LA, Werren JH, Aebi A, Stone GN, Jiggins FM. Evolution and diversity of Rickettsia bacteria. BMC Biol. 2009;7:6. doi: 10.1186/1741-7007-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Psaroulaki A, Loukaidis F, Hadjichristodoulou C, Tselentis Y. Detection and identification of the aetiological agent of Mediterranean spotted fever (MSF) in two genera of ticks in Cyprus. Trans R Soc Trop Med Hyg. 1999;93:597–598. doi: 10.1016/s0035-9203(99)90061-5. [DOI] [PubMed] [Google Scholar]
- 12.Psaroulaki A, Antoniou M, Papaeustathiou A, Toumazos P, Loukaides F, Tselentis Y. First detection of Rickettsia felis in Ctenocephalides felis fleas parasitizing rats in Cyprus. Am J Trop Med Hyg. 2006;74:120–122. [PubMed] [Google Scholar]
- 13.Koliou M, Psaroulaki A, Georgiou C, Ioannou I, Tselentis Y, Gikas A. Murine typhus in Cyprus: 21 paediatric cases. Eur J Clin Microbiol Infect Dis. 2007;26:491–493. doi: 10.1007/s10096-007-0310-8. [DOI] [PubMed] [Google Scholar]
- 14.Christou C, Psaroulaki A, Antoniou M, Toumazos P, Ioannou I, Mazeris A, Chochlakis D, Tselentis Y. Rickettsia typhi and Rickettsia felis in Xenopsylla cheopis and Leptopsylla segnis parasitizing rats in Cyprus. Am J Trop Med Hyg. 2010;83:1301–1304. doi: 10.4269/ajtmh.2010.10-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Psaroulaki A, Antoniou M, Toumazos P, Mazeris A, Ioannou I, Chochlakis D, Christophi N, Loukaides P, Patsias A, Moschandrea I, Tselentis Y. Rats as indicators of the presence and dispersal of six zoonotic microbial agents in Cyprus, an island ecosystem: a seroepidemiological study. Trans R Soc Trop Med Hyg. 2010;104:733–739. doi: 10.1016/j.trstmh.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Ioannou I, Chochlakis D, Kasinis N, Anayiotos P, Lyssandrou A, Papadopoulos B, Tselentis Y, Psaroulaki A. Carriage of Rickettsia spp., Coxiella burnetii and Anaplasma spp. by endemic and migratory wild birds and their ectoparasites in Cyprus. Clin Microbiol Infect. 2009;15((Suppl 2)):158–160. doi: 10.1111/j.1469-0691.2008.02207.x. [DOI] [PubMed] [Google Scholar]
- 17.Ioannou I, Sandalakis V, Kassinis N, Chochlakis D, Papadopoulos B, Loukaides F, Tselentis Y, Psaroulaki A. Tick-borne bacteria in mouflons and their ectoparasites in Cyprus. J Wildl Dis. 2011;47:300–306. doi: 10.7589/0090-3558-47.2.300. [DOI] [PubMed] [Google Scholar]
- 18.Chochlakis C, Ioannou I, Sandalakis V, Dimitriou T, Kassinis K, Papadopoulos B, Psaroulaki A. Spotted fever group rickettsiae in ticks, in Cyprus. Microb Ecol. 2011 doi: 10.1007/s00248-011-9926-4. [DOI] [PubMed] [Google Scholar]
- 19.Tselentis Y, Psaroulaki A, Maniatis J, Spyridaki I, Babalis T. Genotypic identification of murine typhus Rickettsia in rats and their fleas in an endemic area of Greece by the polymerase chain reaction and restriction fragment length polymorphism. Am J Trop Med Hyg. 1996;54:413–417. doi: 10.4269/ajtmh.1996.54.413. [DOI] [PubMed] [Google Scholar]
- 20.Fournier PE, Roux V, Raoult D. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int J Syst Bacteriol. 1998;48:839–849. doi: 10.1099/00207713-48-3-839. [DOI] [PubMed] [Google Scholar]
- 21.Roux V, Raoult D. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB) Int J Syst Evol Microbiol. 2000;50:1449–1455. doi: 10.1099/00207713-50-4-1449. [DOI] [PubMed] [Google Scholar]
- 22.Roux V, Rydkina E, Eremeeva M, Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int J Syst Bacteriol. 1997;47:252–261. doi: 10.1099/00207713-47-2-252. [DOI] [PubMed] [Google Scholar]
- 23.Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]