Abstract
Rats are considered the principal maintenance hosts of Leptospira. The objectives of this study were isolation and identification of Leptospira serovars circulating among urban rat populations in Kuala Lumpur. Three hundred urban rats (73% Rattus rattus and 27% R. norvegicus) from three different sites were trapped. Twenty cultures were positive for Leptospira using dark-field microscopy. R. rattus was the dominant carrier (70%). Polymerase chain reaction (PCR) confirmed that all isolates were pathogenic Leptospira species. Two Leptospira serogroups, Javanica and Bataviae, were identified using microscopic agglutination test (MAT). Pulsed-field gel electrophoresis (PFGE) identified two serovars in the urban rat populations: L. borgpetersenii serovar Javanica (85%) and L. interrogans serovar Bataviae (15%). We conclude that these two serovars are the major serovars circulating among the urban rat populations in Kuala Lumpur. Despite the low infection rate reported, the high pathogenicity of these serovars raises concern of public health risks caused by rodent transmission of leptospirosis.
Introduction
Leptospirosis, an infectious disease that affects both humans and animals, is recognized as one of the most widespread zoonoses worldwide.1 Annually, an estimated one-half million cases of severe leptospirosis are reported globally.2 However, this number is probably underestimated because of the lack of reported cases and the misdiagnosis of this disease in many countries.3
Leptospirosis is caused by the pathogenic strains of the bacterium Leptospira. Currently, there are nearly 300 known serovars,4 and most of them have their primary reservoirs in wild and domestic animals, of which rodents and rats are the most common source worldwide.5 Infected rats shed Leptospira spp. in their urine over an extended period of time,6 and humans and animals get infected through direct or indirect contact with urine, water, or soil contaminated by Leptospira spp.7 Approximately one-half of the pathogenic serovars belong to L. interrogans or L. borgpetersenii.8
Classically, the diagnosis of leptospirosis is based on serological tests, such as the microscopic agglutination test (MAT). In this test, reaction takes place between a leptospiral isolate and reference hyperimmune rabbit antisera.9 However, this method is laborious and time-consuming, and it requires extensive collection of reference strains and their corresponding rabbit antisera.10 Various molecular approaches have been developed, such as polymerase chain reaction (PCR)-based methods, to improve the diagnosis of leptospirosis. PCR has been successfully applied as a rapid, sensitive, and specific tool for the detection of several microorganisms, including Leptospira,11 in a variety of specimens from different hosts.12–14 The rapidity and reproducibility of pulsed-field gel electrophoresis (PFGE) makes it a very useful technique for typing Leptospira strains.15 PFGE is able to discriminate between closely related serovars of the Leptospira spp., which may aid in the identification of the different strains at serovar level,16 and this identification is important in understanding the epidemiology of leptospirosis in both humans and animals in any geographic region.17
Presently, leptospirosis is recognized as a globally re-emerging disease, with a marked increase in the number of cases in Latin America and Southeast Asia, including Malaysia.18 According to the Ministry of Health Malaysia, the numbers of reported cases of leptospirosis have increased from 263 in 2004 to 1,418 in 2009.19 Previous studies on leptospirosis in Malaysia focused mainly on humans and domestic animals in the vicinity of outbreak sites.20,21 However, there is very little information about this disease in Malaysian urban rat populations. Therefore, the purpose of this study was to isolate and detect the Leptospira spp. in the urban rats of Kuala Lumpur using MAT and PCR. In addition, the serovar identification of these isolates was determined by PFGE.
Materials and Methods
Choice of the study sites.
Kuala Lumpur, the capital city of Malaysia, is characterized by a tropical climate of high temperature and high humidity year round, with temperatures ranging between 30°C and 36°C; rainfall is fairly even throughout most of the year but typically heavier between October and February during the monsoon season. The choice of the study sites was based on the suitability of the habitat for rodents to forage and breed and the possibility of spreading the diseases. The first site, Chow Kit market (03°09′53.75″ N, 101°41′56.84″ E), is the largest fresh food wet market, commonly found in Asian countries, in Kuala Lumpur. Here, water is used extensively to wash the floors, keep the fruits and vegetables fresh, and keep fish and shellfish alive. This market provides an extensive range of raw foods, including fruits, vegetables, seafood, and meat to the public. In turn, tons of rubbish are deposited into several steel containers. Excess garbage falls to the ground, forming temporary grounds for rats to forage. In contrast, Bangsar (3°6′49″ N 101°39′45″ E) is an affluent residential suburb on the outskirts of Kuala Lumpur with mixed residential sites. The hawker centers, restaurants, and roadside stalls sell cooked food, and rodents found here thrive on leftovers. The third site, Setapak (3°11′32″ N 101°43′1″ E), is a highly dense residential site for the lower income bracket. Nearby, a small wet open market provides fresh produce to the community. Similarly, because of poor garbage management, this site also attracts rodents, stray dogs, and cats to breed and source for food.
Trapping and host identification.
Trapping was conducted over a period of 6 days and 5 nights for each session, with a total of 13 trapping sessions conducted over a 6-month period (October of 2011 to March of 2012).
A total of 30 steel wire traps (18 × 12 × 28 cm) were placed in each site with baits such as dry fish, breads, and peanut butter. The traps were placed in the evening and collected in the early morning before the market was opened to the public. The captured rats were placed in black plastic bags to reduce stress and transported to the laboratory for examination. The rats were then euthanized with chloroform, and morphometric measurements were recorded.
Age and species identifications were carried out according to Medway22 based on the phenotypic characteristics, such as fur color (ventral and dorsal), body weight, hind foot, and head–body length.
Isolation of Leptospira and dark-field microscopy examination.
Euthanized rats were immediately killed, and selected organs, such as blood, livers, and kidneys, were collected. Skin, liver, and kidney lesions were recorded to investigate the relationship of these lesions with the transmissibility of Leptospira spp. Two drops of blood collected by cardiac puncture were inoculated into the modified semisolid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium. The urine was collected by direct puncture of the bladder and then cultured into the medium. Samples of liver and kidney tissues were removed using a sterile blade, and a small piece of tissue was placed in a sterile syringe without a needle and squeezed into the EMJH medium. All inoculated media were incubated aerobically at 30°C and examined under a dark-field microscope for the presence of Leptospira at 10-day intervals for a period of 3 months. Samples that failed to show any evidence of growth after 3 months were considered negative.
Two media types were used in this study: liquid and semisolid, in which 0.13% Bacto agar was added. Both media were enriched with 1.0% rabbit serum and bovine serum albumin and supplemented with 5-fluorouracil (Merck, Darmstadt, Germany) at a concentration of 400 μg/mL to minimize bacterial contamination.
Detection and determination of the pathogenic status of the leptospiral isolates by PCR.
DNA template preparation.
Genomic DNA was extracted from 7-day-old culture media using Wizard Genomic DNA Purification Kit (Promega, Madison, WI) using the manufacturer's instructions. The quantity of DNA was measured by Biophotometer (Eppendorf, Hamburg, Germany).
PCR detection and confirmation of Leptospira isolates.
PCR primers LA/LB ([5′-GGC GGC GCG TCT TAA ACA TG-3′] and [5′-TTC CCC CCA TTG AGC AAG ATT-3′]), which target the 16S rDNA gene, were used to confirm the genus Leptospira.23 The cycling conditions consisted of an initial denaturation at 94°C for 3 minutes, 35 cycles each of 94°C for 1 minute, 57°C for 1 minute, and 72°C for 2 minute, and additional extension at 72°C for 10 minutes. To determine the pathogenic status of the isolates, G1/G2 primers ([5′-CTG AAT CGC TGT ATA AAA GT-3′] and [5′-GGA AAA CAA ATG GTC GGA AG-3′]), which target the secY gene among the isolates except for L. kirschneri, were used.24 The cycling conditions consisted of an initial denaturation at 94°C for 10 minutes, 35 cycles each of 94°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute, and additional extension at 72°C for 5 minutes. In both PCRs, the reactions were done in a final volume of 25 μL containing 1× PCR buffer, 1.5 mM MgCl2, 200 μM each deoxynucleotide triphosphates (dNTPs), 0.3 μM each primer, 1 U Taq DNA polymerase (Intron Biotechnology, Sungnam, Kyungki-Do, South Korea), and 100 ng DNA template. The PCR products were analyzed by electrophoresis through a 1% TBE agarose gel (Promega, Madison, WI).
DNA sequencing.
Amplified DNA products from representative isolates were verified by DNA sequencing. The amplicons were purified using a DNA purification kit (Qiagen, Hilden, Germany) and sent to a commercial facility for sequencing (First BASE, Pte. Ltd., Singapore). The resulting DNA sequence data were compared with the GenBank database using the Basic Local Alignment Search Tool (BLAST) algorithm available at the website (http://www.ncbi.nih.gov).
MAT.
MAT was performed as described by the World Health Organization.25 A set of 25 reference hyperimmune antisera representing the major Leptospira serovars in Malaysia was provided by the Institute of Medical Research (IMR), Malaysia and used to determine the serogroup level of the isolates. The leptospiral isolates were cultured into liquid medium with additional 1.0% rabbit serum to increase bacterial density. A positive MAT was scored when there was 50% agglutination, leaving 50% free cells compared with negative control (culture diluted 1:2 in phosphate buffer saline only). Four known positive reference leptospiral cultures (Canicola, Pomona, Bataviae, and Javanica) were included to test the reactivity of the antisera. The antisera used in this study were raised against serovars Patoc (Patoc I), Ballum (Mus 127), Sejroe (M84), Javanica (Veldrat Batavia 46), Tarassovi (Perepelicin), Bratislava (Jez Bratislava), Canicola (Hond Ultrecht IV), Hebdomadis (Hebdomadis), Pomona (Pomona), Hardjo (Hardjoprajitno), Australis (Ballico), Bataviae (Swart), Pyrogenes (Salinem), Icterohaemorrhagiae (RGA), Paidjan (Paidjan), Gurungi (Gurung), Djasiman (Djasiman), Bangkinang (Bangkinang I), Autumnalis (Akiyami A), Samaranga (Veldrat Sem 173), Proechimys (1161 U), Grippotyphosa (Mandemakers), Grippotyphosa (Moskva V), Cynopteri (3522 C), and Celledoni (Celledoni).
Pulsed-field gel electrophoresis.
Chromosomal DNA was prepared according to protocol16 with minor modifications. XbaI-digested Salmonella Braenderup H9812 was used as the DNA size marker. Leptospira DNA plugs were digested with 10 U restriction enzyme NotI (Promega, Madison, WI) at 37°C. The restriction fragments obtained were separated by electrophoresis in 0.5× 10X Tris-borate EDTA (TBE) buffer for 24 h at 14°C in a CHEF Mapper system (Bio-Rad, Hercules, CA) using pulsed times of 2.2–35 s. PFGE data were analyzed using BioNumerics Version 6.0 (Applied Maths, Sint-Martens-Latem, Belgium) software. Clustering was based on the unweighted pair group average method (UPGMA) with the position tolerance of 1.0.
Statistical analysis.
All statistical analyses were performed using SPSS software, version 17 (SPSS Inc., Chicago, IL). The data were subjected to an independent-samples t test, and the P values were generated. The confidence level was set at 95%. A test with a P value lower than 0.05 was considered statistically significant.
Results
A total of 300 rodents were captured from all three sites and comprised of two rat species, namely Rattus rattus (219, 73%) and R. norvegicus (81, 27%). Most rodents were captured from the vicinity of Chow Kit (160 rats, 53.3%) followed by Setapak and Bangsar with 90 and 50 rats, respectively. Based on the age and sex of the rats captured, 208 (69.3%) rats were adults, whereas 92 (30.7%) rates were juveniles; there were more males (167, 55.7%) compared with females (133, 44.3%). The trapping results from the three locations are as indicated in Table 1.
Table 1.
Summary of the trapping results from the three location in Kuala Lumpur city
| Location | Rat species | Sex | Age | Lesions | |||||
|---|---|---|---|---|---|---|---|---|---|
| R. rattus | R. norvegicus | Male | Female | Adult | Juvenile | Liver | Kidney | Skin | |
| Chow Kit | 113 (51.6%) | 47 (58.0%) | 94 (56.3%) | 66 (49.6%) | 105 (50.5%) | 55 (59.8%) | 27 (58.7%) | 12 (38.7%) | 24 (61.5%) |
| Setapak | 68 (31.0%) | 22 (27.2%) | 45 (26.9%) | 45 (33.8%) | 69 (33.2%) | 21 (22.8%) | 14 (30.4%) | 15 (48.4%) | 8 (20.5%) |
| Bangsar | 38 (17.4%) | 12 (14.8%) | 28 (16.8%) | 22 (16.6%) | 34 (16.3%) | 16 (17.4%) | 5 (10.9%) | 4 (12.9%) | 7 (18.0%) |
A total of 6.7% (20/300) were positive with cultures that showed the typical morphology and characteristic motility of Leptospira genus by dark-field microscopy from 16 urine and 4 kidney samples. R. rattus (14/20, 70%) had higher infection rates compared with R. norvegicus (6/20, 30%), and statistically significantly more male rats (15/20) were infected compared with females (5/20, P = 0.045). According to host age, more adults were infected with Leptospira (14/20) than juveniles (6/20, P = 0.663); however, the difference was not significant.
According to site, 10 positive samples were obtained from the Chow Kit vicinity followed by Setapak and Bangsar with 6 and 4 positive samples, respectively. Rats with liver lesions recorded higher positives (15.3%) compared with skin (13%) and kidney (10.3%) lesions.
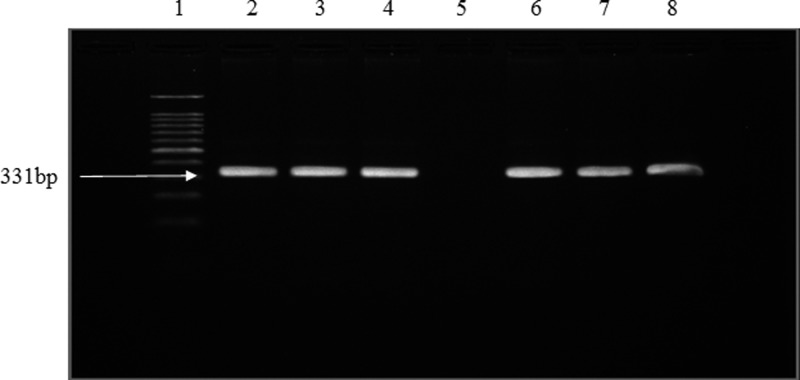
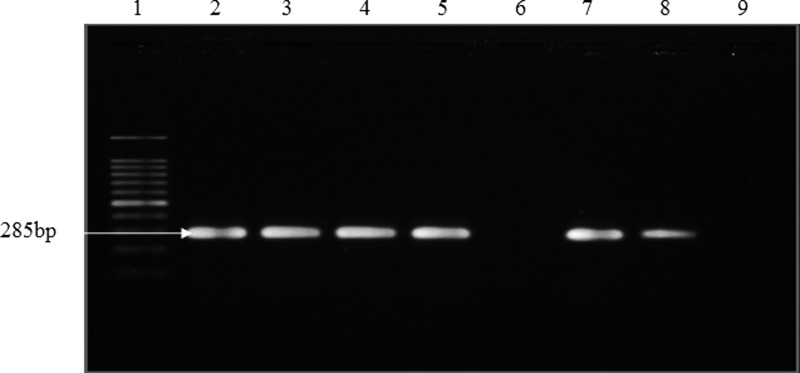
Leptospira genus confirmation by PCR showed that all 20 isolates gave the expected band size of 331 bp, indicating that all the positive isolates were Leptospira species (Figure 1). All isolates were pathogenic species as shown by the band size of 285 bp amplified by the primers G1/G2, which targets secY genes (Figure 2) (99% similarity; accession numbers EU358040 and DQ882852).
Figure 1.
Representative gel of PCR for detection of Leptospira genus using LA/LB primers. Lanes 1–8: DNA marker (100 bp); positive control; R2 (urine sample); R18 (urine sample); negative control; R71 (kidney sample); R77 (urine sample); R108 (kidney sample).
Figure 2.
Representative gel of PCR for confirmation of pathogenic Leptospira using G1/G2 primers. Lanes 1–8: DNA marker (100 bp); positive control; R77 (urine sample); R78 (urine sample); R84 (urine sample); negative control; R102 (urine sample); R108 (kidney sample).
The identification of the serogroup of the isolates was carried out using 25 different hyperimmune antisera. The results showed that 17 isolates had the agglutinating reaction against serovar Javanica antisera (titer > 1:400), whereas 3 isolates showed agglutination to serovar Bataviae (titer > 1:400).
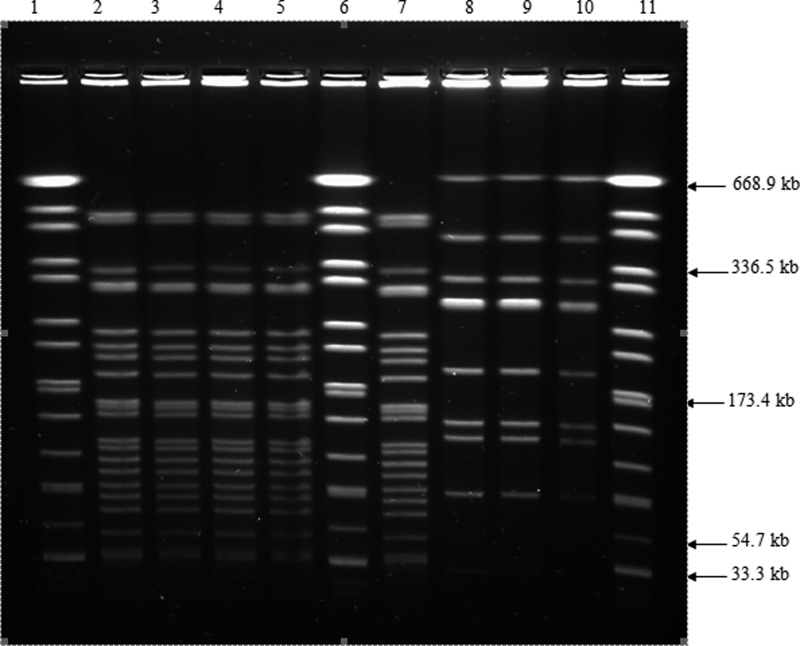
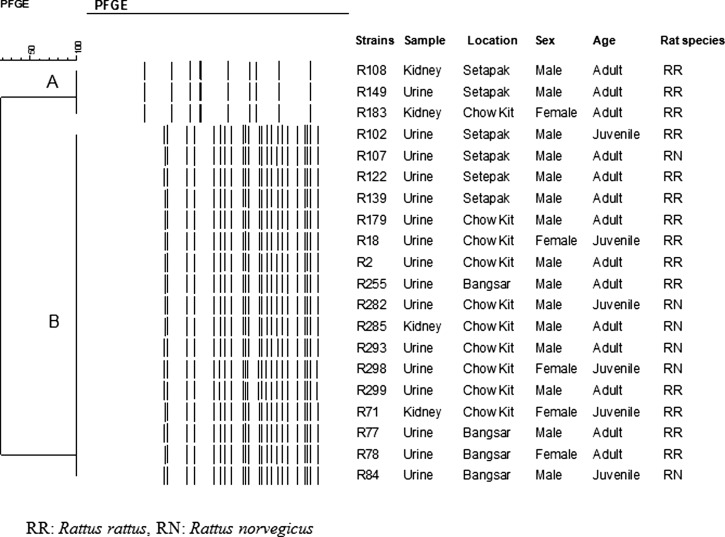
PFGE of NotI-digested chromosomal DNA subtyped the 20 isolates into two distinct PFGE profiles: LNot01 and LNot02. The number of DNA fragments generated ranged from 10 to 23, with sizes ranging from approximately 28 to 706 kb (Figure 3). Limited genetic diversity was found among the isolates. The dendrogram generated two major clusters: A and B (Figure 4). Cluster A consisted of three isolates isolated from R. rattus; two isolates were from Setapak, and one isolate was from Chow Kit market. Cluster B consisted of 17 isolates; 11 isolates were isolated from R. rattus, whereas 6 isolates were isolated from R. norvegicus. Of these isolates, nine isolates were from Chow Kit market, four isolates were from Setapak, and four isolates were from Bangsar.
Figure 3.
PFGE patterns of NotI digested chromosomal DNA of Leptospira isolates from urban rats of Kuala Lumpur. Lanes 1, 6, 11: XbaI digested chromosomal DNA of Salmonella H9812 marker strain; lanes: 2–5, 7–10: L. interrogans serovar Javanica R2; L. interrogans serovar Javanica R18; L. interrogans serovar Javanica R7; L. interrogans serovar Javanica R78; L. interrogans serovar Javanica R84; L. interrogans serovar Bataviae R108; L. interrogans serovar Bataviae R149; L. interrogans serovar Bataviae R183.
Figure 4.
Dendrogram based on cluster analysis of the PFGE profiles of NotI digested chromosomal Leptospira isolated from urban rats generated using BioNumerics Version 6.0 (Applied Maths, Belgium) software and unweighted pair group arithmetic means methods (UPGMA). Cluster A represents L. interrogans serovar Bataviae, cluster B represents L. borgpetersenii serovar Javanica.
The two PFGE profiles produced were compared with the NotI patterns from the database provided by the Centers for Disease Control and Prevention (CDC). The results showed that LNot01 has a similar pattern to L. interrogans serovar Bataviae, whereas LNot02 has the same pattern as L. borgpetersenii serovar Javanica.
Discussion
In Kuala Lumpur city, R. rattus and R. norvegicus were found to be the two most dominant commensal rat species living close to human habitation.26 Urbanization of cities and towns has created favorable conditions for commensal rats to live, because these rats often depend on direct contact with human food. The two species are closely related, and formerly, R. norvegicus was classed as a subspecies of R. rattus.27 Similar to the report by Gordon-Smith and others,28 more than twofold R. rattus were captured compared with R. norvegicus from the three sites, suggesting the population dominance of R. rattus in urban settings. Both species have successfully adapted and established themselves, despite a continuous and extensive rodent control program in the city. Therefore, it is essential that the rodent population be regularly monitored.
Rodents are known as the maintenance hosts of Leptospira in Malaysia. To date, 37 serovars of Leptospira from 13 serogroups have been identified in Malaysia, and more than one-half of these serovars are carried by rats.20 However, previous studies focused on rodent populations associated with outbreak areas, such as national training service centers, oil palm estates, and forest habitat.29–31 The present study serves to highlight this disease in the urban setting, where rodents are known to live closely with humans.
Chow Kit market is the largest market in Malaysia, with plentiful resources for rodents. Rubbish left in open garbage containers attracts high numbers of rats to forage, especially R. rattus. Similarly, in Setapak, a smaller market nearby also attracts this pest. In addition, open sewers and poor drainage serve as thriving grounds for rodents to breed. The more affluent residential area, Bangsar, had the smallest number of rats captured. This result could be explained by better sanitary conditions and garbage management practices in this affluent area.
R. rattus was found to be the more prevalent rat species infected with Leptospira compared with R. norvegicus. Gordon-Smith and others28 reported that the prevalence of the infected R. norvegicus was higher compared with R. rattus (2/259). Other rat species have also been reported as carriers of the leptospires, such as R. tiomanicus, R. exulans, and R. argentiventer.29,30 Faria and others32 showed that the higher population density of R. norvegicus was the main factor facilitating transmission of leptospirosis in Brazil, whereas R. tiomanicus was predominant in various habitats in Malaysia.30
In this study, the overall prevalence of rats being infected was low (6.7%). This finding is concordant to previous studies.28,30,33 Studies in Brazil and the Philippines reported a high prevalence of leptospirosis among urban rats.32,34
Of 300 captured rats, 38.7% had lesions in liver, kidney, or skin. The R. rattus species showed more lesions compared with R. norvegicus (P = 0.029); however, the frequency of the clinical lesions was not statistically significant (P > 0.05) between the rats species and the prevalence of the positive infected rats. A total of 16 urine samples was positive compared with only 4 kidney samples.
According to host age, adult rats (15/208) were infected more frequently compared with juvenile rats (5/92). This finding is in agreement with other reports that showed that adult rats can carry more Leptospira than immature rats.35,36 Carter and Cordes37 reported that mature urban R. rattus were four times more likely to carry Leptospira compared with immature R. rattus. Aggression, fighting, and biting among the adult rats can also facilitate the transmission of Leptospira between the adults.30 Other factors include the dynamic movement of adults compared with the young and length of exposure to the environment contaminated with Leptospira from rat urine. Transmission by sexual contact in adults can also take place for this age class only. Moreover, young rats are generally confined to the nesting burrows; hence, they experience less exposure to the pathogen.
According to host sex, more male carrier rats were observed compared with females (P = 0.045). This result could be explained by the dynamic movement and aggressiveness of male rats, which is a sign of maturity.30 However, this observation differed from previous studies. Among them, a study on urban rats of Baltimore, Maryland by Easterbrook and others38 reported more females carrying Leptospira compared with male rats. Faria and others,32 however, found similar carriage between male and female rats. In contrast, Agudelo-Florez and others39 reported no correlation between sex, age, and health condition of the host and prevalence of the infection.
Many studies reported the low sensitivity of the culture technique in detection of Leptospira spp. compared with other techniques, such as PCR, immunofluorescence, and nucleic acid hybridization.40,41 The success of the isolation step is influenced by various factors, such as the number of organisms per inoculation, the type of media used, and the type of specimen. The difficulty in the isolation of Leptospira and the slow growth of this fastidious organism made the culture technique time consuming with low sensitivity. In the present study, PCR was able to successfully determine the genus and pathogenic status of the isolates. With its specificity and rapidity, PCR has become a promising tool for the early detection of Leptospira in various specimens.12,13,33
However, MAT was able to identify successfully 20 positive Leptospira isolates at the serogroup level using 25 different sera. Two serogroups were identified; serogroup Javanica was found to be the predominant serogroup infecting 17 (85%) positive rats, whereas serogroup Bataviae was found only in 3 rats. In a recent study, the prevalence of leptospiral antibody among municipal workers in Kota Bharu, Kelantan, Malaysia showed that antibodies against serogroups of Bataviae and Javanica were predominant.42
The rat species R. rattus was found to be the main carrier for both serogroups. Interestingly, R. norvegicus was the main carrier of serogroup Javanica rather than serogroup Icterohaemorrhagiae, which was recognized as the maintenance host of these leptospires. This finding indicates that carriers of this pathogen are not dependent on host species but the distribution of the respective rat species in the various habitats. Gordon-Smith and others28 reported that, among the 42 identified strains from Javanica serogroup, 37 (88%) strains were isolated from the rice field rat, R. argentiventer. L. borgpetersenii serovar Javanica was also one of the predominant strains isolated from rice field R. norvegicus trapped in rural areas of Tamil Nadu, India.43 The existence of this serovar among the urban rats could be explained by the migration of infected rats from the rural area to the urban cities by transportation of goods, such as rice, and animals between the two areas. This reason probably explained the high number of positive rats infected with serogroup Javanica from Chow Kit market, where raw foods were brought from rural areas to be sold in the city.
PFGE results were in concordance with MAT results. Two PFGE profiles were generated using the NotI enzyme. With reference to PFGE profiles available in the leptospiral database at CDC, we were able to identify L. interrogans serovar Bataviae in 3 isolates and L. borgpetersenii serovar Javanica in 17 isolates.
PFGE showed a limited genetic diversity within the serovars of Javanica and Bataviae. All 17 Javanica isolates had the same PFGE profiles, although these isolates were collected from different locations, different rat species, and different sampling times. Similarly, all the Bataviae that originated from different geographical locations were indistinguishable. This finding was in agreement with the observation of Galloway and Levett15 that PFGE was unable to discriminate strains that are closely related, such as serovars Icterohaemorragiae and Copenhageni of L. interrogans species. However, PFGE showed a high discriminatory power to differentiate different serovars of L. borgpetersenii and L. interrogans. Hence, PFGE could be applied as a complementary tool to the serological technique to differentiate leptospiral serovars.15,44
In conclusion, this study provides important information regarding the infection levels and identification of the pathogenic serovars circulating among the urban rat populations of Kuala Lumpur, namely Leptospira serovars Javanica and Bataviae. Despite the low infection rate present in the population, because of its high pathogenicity, long-term monitoring is necessary to keep the safety of the public in check.
ACKNOWLEDGMENTS
The authors thank the Director and staff of Kuala Lumpur City Hall (DBKL) for their kind cooperation and the Institute for Medical Research, Ministry of Health, Malaysia (IMR) for providing the reference hyperimmune antisera for this study. We are grateful to Professor Dr. Ben Adler for his advice in this research project. The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
Disclaimer: No competing interests exist.
Footnotes
Financial support: This work was supported by University of Malaya Research Grant RG053/11BIO.
Authors' addresses: Douadi Benacer and Kwai Lin Thong, Institute of Biological Sciences, Faculty of Science and Laboratory of Biomedical Science and Molecular Microbiology, Institute of Graduate Studies, University of Malaya, Kuala Lumpur, Malaysia, E-mails: benacer_dvt@hotmail.com and thongkl@um.edu.my. Siti Nursheena Mohd Zain, Institute of Biological Sciences, Faculty of Science, University of Malaya, Kuala Lumpur, Malaysia, E-mail: nsheena@um.edu.my. Fairuz Amran, Bacteriology Unit, Institute for Medical Research, Ministry of Health, Kuala Lumpur, Malaysia, E-mail: fairuz@imr.gov.my. Renee L. Galloway, Bacterial Zoonoses Special Pathogens Branch, Centers for Disease Control and Prevention, Atlanta, GA, E-mail: zul0@cdc.gov.
References
- 1.Palaniappan RU, Ramanujam S, Chang YF. Leptospirosis: pathogenesis, immunity, and diagnosis. Curr Opin Infect Dis. 2007;20:284–292. doi: 10.1097/QCO.0b013e32814a5729. [DOI] [PubMed] [Google Scholar]
- 2.Hartskeerl RA, Collares-Pereira M, Ellis WA. Emergence, control and re-emerging leptospirosis: dynamics of infection in the changing world. Clin Microbiol Infect. 2011;17:494–501. doi: 10.1111/j.1469-0691.2011.03474.x. [DOI] [PubMed] [Google Scholar]
- 3.Socolovschi C, Angelakis E, Renvoisé A, Fournier PE, Marie JL, Davoust B, Stein A, Raoult D. Strikes, flooding, rats, and leptospirosis in Marseille, France. Int J Infect Dis. 2011;15:710–715. doi: 10.1016/j.ijid.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 4.Valverde ML, Ramirez JM, Montes de Oca LG, Goris MG, Ahmed N, Hartskeerl RA. Arenal, a new Leptospira serovar of serogroup Javanica, isolated from a patient in Costa Rica. Infect Genet Evol. 2008;8:529–533. doi: 10.1016/j.meegid.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Green-Mckenzie J, Shoff WH. Leptospirosis in Emergency Medicine. 2012. http://emedicine.medscape.com/article/788751-overview E Medicine (WebMD) Available at. Accessed August 15, 2012.
- 6.Turk N, Milas Z, Margaletic J, Staresina V, Slavica A, Riquelme-Sertour N, Bellenger E, Baranton G, Postic D. Molecular characterisation of Leptospira spp. strains isolated from small rodents in Croatia. Epidemiol Infect. 2003;130:159–166. doi: 10.1017/s0950268802008026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, Vinetz JM. Peru-United States Leptospirosis Consortium Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3:757–771. doi: 10.1016/s1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- 8.Victoriano AF, Smythe LD, Gloriani-Barzaga N, Cavinta LL, Kasai T, Limpakarnjanarat K, Ong BL, Gongal G, Hall J, Coulombe CA, Yanagihara Y, Yoshida S, Adler B. Leptospirosis in the Asia Pacific region. BMC Infect Dis. 2009;9:147. doi: 10.1186/1471-2334-9-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dikken H, Kmety E. Serological typing methods of leptospires. In: Bergan T, Norris JR, editors. Methods in Microbiology. London, UK: Academic Press; 1978. pp. 259–307. [Google Scholar]
- 10.Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bal AE, Gravekamp C, Hartskeerl RA, De Meza-Brewster J, Korver H, Terpstra WJ. Detection of leptospires in urine by PCR for early diagnosis of leptospirosis. J Clin Microbiol. 1994;32:1894–1898. doi: 10.1128/jcm.32.8.1894-1898.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fonseca Cde A, Teixeira MM, Romero EC, Tengan FM, Silva MV, Shikanai-Yasuda MA. Leptospira DNA detection for the diagnosis of human leptospirosis. J Infect. 2006;52:15–22. doi: 10.1016/j.jinf.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 13.Bomfim MR, Barbosa-Stancioli EF, Koury MC. Detection of pathogenic leptospires in urine from naturally infected cattle by nested PCR. Vet J. 2008;178:251–256. doi: 10.1016/j.tvjl.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 14.Vital-Brazil JM, Balassiano IT, Oliveira FS, Costa AD, Hillen L, Pereira MM. Multiplex PCR-based detection of Leptospira in environmental water samples obtained from a slum settlement. Mem Inst Oswaldo Cruz. 2010;105:353–355. doi: 10.1590/s0074-02762010000300020. [DOI] [PubMed] [Google Scholar]
- 15.Galloway RL, Levett PN. Evaluation of a modified pulsed-field gel electrophoresis approach for the identification of Leptospira serovars. Am J Trop Med Hyg. 2008;78:628–632. [PubMed] [Google Scholar]
- 16.Galloway RL, Levett PN. Application and validation of PFGE for serovar identification of Leptospira clinical isolates. PLoS Negl Trop Dis. 2010;4:e824. doi: 10.1371/journal.pntd.0000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levett PN. Sequence-based typing of Leptospira: epidemiology in the genomic era. PLoS Negl Trop Dis. 2007;1:e120. doi: 10.1371/journal.pntd.0000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartskeerl RA. Leptospirosis: current status and future trends. Indian J Med Microbiol. 2006;24:309. doi: 10.4103/0255-0857.29404. [DOI] [PubMed] [Google Scholar]
- 19.Lokman HS. Guidelines for the Diagnosis, Management, Prevention and Control of Leptospirosis in Malaysia. 1st Ed. Disease Control Division, Department of Public Health, Ministry of Public; Health; Malaysia: 2011. [Google Scholar]
- 20.Bahaman AR, Ibrahim AL. A review of leptospirosis in Malaysia. Vet Res Commun. 1988;12:179–189. doi: 10.1007/BF00362799. [DOI] [PubMed] [Google Scholar]
- 21.Koay TK, Nirmal S, Noitie L, Tan E. An epidemiological investigation of an outbreak of leptospirosis associated with swimming, Beaufort, Sabah. Med J Malaysia. 2004;59:455–459. [PubMed] [Google Scholar]
- 22.Medway L. The Wild Mammals of Malaya (Peninsula Malaysia) and Singapore. 2nd Ed. Oxford, UK: Oxford University Press; 1983. p. 131. [Google Scholar]
- 23.Merien F, Amouriaux P, Perolat P, Baranton G, Saint Girons I. Polymerase chain reaction for detection of Leptospira spp. in clinical samples. J Clin Microbiol. 1992;30:2219–2224. doi: 10.1128/jcm.30.9.2219-2224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gravekamp C, Van de Kemp H, Franzen M, Carrington D, Schoone GJ, Van Eys GJ, Everard CO, Hartskeerl RA, Terpstra WJ. Detection of seven species of pathogenic leptospires by PCR using two sets of primers. J Gen Microbiol. 1993;139:1691–1700. doi: 10.1099/00221287-139-8-1691. [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization . Human Leptospirosis: Guidance for Diagnosis, Surveillance and Control. Geneva: World Health Organization and International Leptospirosis Society; 2003. [Google Scholar]
- 26.Mohd Zain SN, Behnke JM, Lewis JW. Helminth communities from two urban rat populations in Kuala Lumpur, Malaysia. Parasit Vectors. 2012;5:47. doi: 10.1186/1756-3305-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corbet GB, Harris S. The Handbook of British Mammals. Oxford Blackwell; UK: 1991. p. 588. [Google Scholar]
- 28.Gordon-Smith CE, Turner LH, Harrison JL, Broom JC. Animal leptospirosis in Malaya: 1. methods, zoogeographical background, and broad analysis of results. Bull World Health Organ. 1961;24:5–21. [PMC free article] [PubMed] [Google Scholar]
- 29.Mohamed-Hassan SN, Bahaman AR, Mutalib AR, Khairani-Bejo S. Serological prevalence of leptospiral infection in wild rats at the National Service Training Centres in Kelantan and Terenggaanu. Trop Biomed. 2010;27:30–32. [PubMed] [Google Scholar]
- 30.Mohamed-Hassan SN, Bahaman AR, Mutalib AR, Khairani-Bejo S. Prevalence of pathogenic leptospires in rats from selected locations in peninsular Malaysia. Res J Anim Sci. 2012;6:12–25. [Google Scholar]
- 31.Ridzlan FR, Bahaman AR, Khairani-Bejo S, Mutalib AR. Detection of pathogenic Leptospira from selected environment in Kelantan and Terengganu, Malaysia. Trop Biomed. 2010;27:632–638. [PubMed] [Google Scholar]
- 32.Faria MT, Calderwood MS, Athanazio DA, McBride AJA. Carriage of Leptospira interrogans among domestic rats from an urban setting highly endemic for leptospirosis in Brazil. Acta Trop. 2008;108:1–5. doi: 10.1016/j.actatropica.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khairani-Bejo S, Oii SS, Bahaman AR. Rats: leptospirosis reservoir in Serdang Selangor residential area. J Anim Vet Adv. 2004;3:66–69. [Google Scholar]
- 34.Villanueva SY, Ezoe H, Baterna RA, Yanagihara Y, Muto M, Koizumi N, Fukui T, Okamoto Y, Masuzawa T, Cavinta LL, Gloriani NG, Yoshida S. Serologic and molecular studies of Leptospira and leptospirosis among rats in the Philippines. Am J Trop Med Hyg. 2010;82:889–898. doi: 10.4269/ajtmh.2010.09-0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krøjgaard LH, Villumsen S, Markussen MDK, Jensen JS, Leirs H, Heiberg AC. High prevalence of Leptospira spp. in sewer rats (Rattus norvegicus) Epidemiol Infect. 2009;137:1586–1592. doi: 10.1017/S0950268809002647. [DOI] [PubMed] [Google Scholar]
- 36.Perez J, Brescia F, Becam J, Mauron C, Goarant C. Rodent abundance dynamics and leptospirosis carriage in an area of hyper-endemicity in New Caledonia. PLoS Negl Trop Dis. 2011;5:e1361. doi: 10.1371/journal.pntd.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carter ME, Cordes DO. Leptospirosis and other infections of Rattus rattus and Rattus norvegicus. N Z Vet J. 1980;28:45–50. doi: 10.1080/00480169.1980.34688. [DOI] [PubMed] [Google Scholar]
- 38.Easterbrook JD, Kaplan JB, Vanasco NB, Reeves WK, Purcell RH, Kosoy MY, Glass GE, Watson J, Klein SL. A survey of zoonotic pathogens carried by Norway rats in Baltimore, Maryland, USA. Epidemiol Infect. 2007;135:1192–1199. doi: 10.1017/S0950268806007746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agudelo-Florez P, Londono AF, Quiroz VH, Angel JC, Moreno N, Loaiza ET, Munoz LF, Rodas JD. Prevalence of Leptospira spp. in urban rodents from a groceries trade center of Medellin, Colombia. Am J Trop Med Hyg. 2009;81:906–910. doi: 10.4269/ajtmh.2009.09-0195. [DOI] [PubMed] [Google Scholar]
- 40.Wagenaar J, Zuerner RL, Alt D, Bolin CA. Comparison of polymerase chain reaction assays with bacteriologic culture, immunofluorescence, and nucleic acid hybridization for detection of Leptospira borgpetersenii serovar hardjo in urine of cattle. Am J Vet Res. 2000;61:316–320. doi: 10.2460/ajvr.2000.61.316. [DOI] [PubMed] [Google Scholar]
- 41.Fornazari F, Costa da Silva R, Richini-Pereira VB, Beserra HEO, Luvizotto MCR, Langoni H. Comparison of conventional PCR, quantitative PCR, bacteriological culture and the Warthin Starry technique to detect Leptospira spp. in kidney and liver samples from naturally infected sheep from Brazil. J Microbiol Methods. 2012;90:321–326. doi: 10.1016/j.mimet.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Shafei MN, Sulong MR, Yaacob NA, Hassan H, Wan Mohd Zahiruddin WM, Daud A, Ismail Z, Abdullah MR. Seroprevalence of leptospirosis among town service workers on northeastern State of Malaysia. International Journal of Collaborative Research on Internal Medicine & Public Health (IJCRIMPH) 2012;4:395–403. [Google Scholar]
- 43.Vedhagiri K, Natarajaseenivasan K, Prabhakaran SG, Selvin J, Narayanan R, Shouche YS, Vijayachari P, Ratnam S. Characterization of Leptospira borgpetersenii isolates from field rats (Rattus norvegicus) by 16S rRNA and lipL32 gene sequencing. Braz J Microbiol. 2010;41:150–157. doi: 10.1590/S1517-838220100001000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romero EC, Blanco RM, Galloway RL. Application of pulsed-field gel electrophoresis for the discrimination of leptospiral isolates in Brazil. Lett Appl Microbiol. 2009;48:623–627. doi: 10.1111/j.1472-765X.2009.02580.x. [DOI] [PubMed] [Google Scholar]