Abstract
This work is a molecular epidemiologic study to detect the incidence of Coxiella burnetii in rodents on Heixiazi Island at the Sino-Russian border of Heilongjiang Province. Liver tissues were extracted and processed to test the incidence of C. burnetii infection using polymerase chain reaction analysis. In total, 18% (66 of 370) of rodents tested positive for infection. The results of logistic regression analysis indicated that infection with C. burnetii was associated significantly with weight and month of capture, and infection was found in all rodent species that were observed; there was no significant difference of sex on the infection of C. burnetii. Though phylogenetic analysis disclosed heterogeneity in the nucleotide sequences isolated from the island rodents, the majority of observed strains were among the most common strains found worldwide. This is the first report on the incidence of C. burnetii in rodents on Heixiazi Island at the Sino-Russian border.
Introduction
Recently, several reported outbreaks of Q fever in European countries have raised public concerns regarding this disease, which for decades has been known to occur throughout the world.1,2 Humans can be infected through inhalation of contaminated aerosols, ingestion of contaminated food, or skin trauma. The manifest symptoms of Q fever in humans include both acute and chronic illnesses, where chronic illness occurs in ∼1–3% of patients. Acute cases occur as an influenza-like illness, hepatitis, and/or pneumonia, which occasionally lead to a lethal respiratory distress syndrome. Endocarditis and hepatitis are the most frequent and serious manifestations of illness in Q fever.3
Q fever is caused by Coxiella burnetii, an intracellular organism with reservoirs in birds, arthropods, wild and domestic mammals.4 The organism has a spore-like morphology that is extremely resistant to heat, pressure, desiccation, and other antiseptic compounds. It can survive in the ambient environment for long periods. Because of its characteristics, C. burnetii aerosols can be used in biological warfare, and it is considered a potential terrorist threat.5
In addition, with economic development, increased habitat loss and fragmentation have made human contact with wild species more frequent. Parks often serve as refuges for wildlife, but they may also be important transmission zones of diseases from wildlife to humans.6 Investigations that attempt to discover wild reservoir species of zoonotic diseases are critically important for understanding the risk of pathogen exchange between wild and human populations.
Heixiazi Island, located at the junction of Heilong River (called Amur in Russia) and the Wusuli River, was once occupied by the former Soviet Union during a 1929 border skirmish. After long years of negotiations, 174 km2, or about half the island, was returned to China after 2008. Because of its special location, Heixiazi Island is of great strategic importance. Its natural environment and cultural history has allowed for the establishment of a distinctive Heixiazi Island tourist attraction, which displays a diverse landscape and abundant wildlife.
Our main objectives for this study were to determine whether the island was the endemic area for Q fever and whether the wild rodents inhabiting the island were naturally infected with C. burnetii; we could then assess the risk of Q fever for visitors to the island.
Materials and Methods
Ethics statement.
The handling of rodents was conducted in compliance with the Animal Welfare Provision of the Chinese Academy of Inspection and Quarantine (CAIQ). All animal experiments were performed following the guidelines of CAIQ.
Trapping of rodents.
Rodents were collected from April through October, 2011, on a monthly basis, from different landscapes, including woodland, grassland, and edge of bush in Heixiazi Island.
Using fresh peanuts as bait, rodents were trapped by snap traps. Traps were set in the evening and checked early in the morning.7 Generally, each rodent was placed in an individual cloth bag in the field and then transported to the local laboratory. A zoologist identified each rodent according to morphologic features specific to its species and developmental stage. After identification of their species and sex and developmental stage, the rodents were dissected. Liver tissues from the rodents were removed and stored immediately in liquid nitrogen and then transported to CAIQ laboratory for further processing.
Detection of C. burnetii infection.
Total genomic DNA was extracted from the liver tissue samples by using a Tissue DNA Extract kit (Tiangen Biotech Inc., Beijing, China), following the instructions of the manufacturer. Nested polymerase chain reaction (PCR) was performed to amplify the com1 gene as previously described.8 The primers used in the first- and second-round PCR reactions are listed in Table 1.
Table 1.
Oligonucleotides used for the detection of Coxiella burnetii by nested polymerase chain reaction (PCR)
| Target gene | Primer | Nucleotide sequence | Size of PCR product | |
|---|---|---|---|---|
| Com1 | 1st round | omp1 | AGTAGAAGCATCCCAAGCATTG | 501 bp |
| omp2 | TGCCTGCTAGCTGTAACGATTG | |||
| 2nd round | omp3 | GAAGCGCAACAAGAAGAACAC | 438 bp | |
| omp4 | TGGAAGTTATCACGCAGTTG | |||
To avoid possible contamination, DNA extraction, the reagent setup, amplification, and agarose gel electrophoresis were performed in separate rooms, and negative control samples (distilled water) was included in all amplifications.
Amplifications were performed in a total volume of 50 μL containing 5 μL of DNA template, 0.5 μM MgCl2, 0.2 μM dNTPs, and 1 μM of each primer pair, and 1U of rTaq DNA polymerase (TaKaRa Bio Inc., Dalian, China). The amplification program for the primer OMP1/OMP2 was 36 cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 1 min. The second amplification with the primer OMP3/OMP4 consisted of 36 cycles at 94°C for 1 min, 56°C for 1 min, and 72°C for 1.5 min. Amplification was conducted using a GeneAmp PCR System 9700 (Applied Biosystems, Carlsbad, CA). The PCR-amplified products were detected by electrophoresis in a 1.5% agarose gel, stained with Goodview. The PCR products were purified using the Omega Gel extraction kit (BioTek Instruments, Inc., Winooski, VT). The purified products were sent to Sangon Biotech (Shanghai) Co., Ltd., for sequencing. The resulting sequences were analyzed using Clustal X (version 1.83) followed by phylogenetic analysis using MEGA (version3.1). The statistical significance of the inferred phylogenies was estimated using bootstrap analysis with 1,000 pseudo-replicate data sets. Some of the nucleotide sequences generated in the study were deposited in GenBank under accession nos. JX522479–JX522489.
Statistical analysis.
The differences in proportions were analyzed using either the χ2 test or Fisher's exact test; P < 0.05 was considered significant. Multivariable logistic regression analysis was used to detect the relationship between C. burnetii infection and risk factors. A two-tailed test with P < 0.05 was considered statistically significant. The SPSS version 18.0 software package was used for all analyses (SPSS, Inc., Chicago, IL).
Results
In total, 370 rodents belonging to six species of three families were captured on Heixiazi Island over a continuous seven-month period from April to October 2011 (Table 2).
Table 2.
Species of rodents captured on Heixiazi Island.
| Family | Species | No. | No. positive for Coxiella burnetii |
|---|---|---|---|
| Sciuridae | Eutamias sibiricus | 5 | 2 (40%) |
| Cricetidae | Clethrionomys rutilus | 141 | 25 (18%) |
| Microtus fortis | 68 | 9 (13%) | |
| Muridae | Apodemus agrarius | 138 | 26 (19%) |
| Rattus norvegicus | 3 | 1 (33%) | |
| Apodemus speciosus | 15 | 3 (20%) | |
| Total | 370 | 66 (18%) |
The majority of rodents captured on Heixiazi Island included members of the species Clethrionomys rutilus (38.1%), Apodemus agrarius (37.3%), and Microtus fortis (18.4%), whereas the minority of rodents included members from Apodemus peninsulae (4.1%), Eutamias sibiricus (1.4%), and Rattus norvegicus (0.8%), respectively. In total, 66 rodents were positive for C. burnetii infection by nested PCR on a partial Com1 gene, with an overall positive rate of 18%, all observed species included rodents infected with the organism, and the infection rates of C. burnetii among various species were not significantly different (χ2 = 3.294, P > 0.05). The average weight of C. burnetii infection in rodents was 31.71 ± 17.86 g, and was not significantly higher than 30.57 ± 15.24 g in non-infection rodents (F = 0.280, P > 0.05). Rodents were captured in seven continuous months from April to October. There was a significant difference in the prevalence of C. burnetii between the months of capture (χ2 = 73.768, P < 0.05). The highest prevalence of C. burnetii infection in rodents was observed in September, followed by July and May; no positive samples were detected in August (see in Table 3).
Table 3.
The relationship of month of capture and prevalence of Coxiella burnetii in rodents
| Month of capture | Prevalence of C. burnetii in rodents |
|---|---|
| April | 3% (2/72) |
| May | 26% (19/72) |
| June | 16% (10/63) |
| July | 28% (11/39) |
| August | 0% (0/51) |
| September | 58% (19/33) |
| October | 13% (5/40) |
Multivariable logistic regression analysis was used to detect the risk factors of C. burnetii, including month of capture, weight, gender, and species. To detect whether there was a difference of prevalence among the species, Eutamias sibiricus was used for the indicator index and compared with the other species (shown in Table 4), the results showed there was not a significant difference among the rodent species. In this model, month of capture and weight were found to be associated with the infection of C. burnetii in the rodents (P < 0.05).
Table 4.
Logistic regression analysis of the risk factors for Coxiella burnetii infection
| Variables | P | OR and 95% CI |
|---|---|---|
| Month of capture | 0.002* | 1.298 (1.097, 1.536) |
| Species | 0.449 | |
| Species (1) | 0.747 | 1.494 (0.130, 17.166) |
| Species (2) | 0.814 | 1.344 (0.114, 15.812) |
| Species (3) | 0.536 | 0.360 (0.014, 9.098) |
| Species (4) | 0.403 | 0.413 (0.052, 3.286) |
| Species (5) | 0.828 | 1.331 (0.101, 17.505) |
| Sex | 0.378 | 0.741 (0.381, 1.443) |
| Weight | 0.048* | 1.034 (1.000, 1.068) |
Means P < 0.05.
OR = odds ratio; CI = confidence interval.
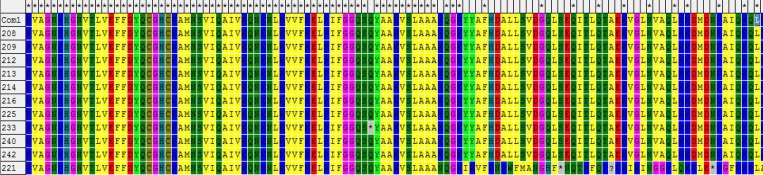
The 438-nt fragment corresponding to the Com1 gene of the positive species was sequenced. Considering sequences of some positive samples from the same study site were nearly identical, 11 representative sequences of the positive samples were used for alignment and phylogenetic analysis. The identities of the nucleotide sequences isolated from rats on the island ranged from 85.9% to 100% (see Figure 1 ). Figure 2 shows the variations of amino acid sequences of C. burnetii isolated in the Island. The results of phylogenetic analysis showed that 10 of the strains clustered in a clade with others isolated from the United States, Japan, China, and Russia, and No.221 were placed in a separate clade, which is distinct from the other sequences (see Figure 3).
Figure 1.
Identities of the 11 sequences of Coxiella burnetii isolated in Heixiazi Island in the study.
Figure 2.
The variation of amino acid sequences of the isolated strains of Coxiella burnetii in Heixiazi Island compared with the Com1 gene.
Figure 3.
Phylogenetic analysis of the com1 gene of Coxiella burnetii detected in the study from April to October 2011. The accession nos. included in the tree were 208(JX522479), 209(JX522480), 212(JX522481), 213(JX522482), 214(JX522483), 216(JX522484), 221(JX522485), 225(JX522486), 233(JX522487), 240(JX522488), 242(JX522489), JX131364, HM804027, AF317646, AF318148, AB004709, HM237793, and GU797241, respectively.
Discussion
Emerging zoonoses are linked to increasing globalization. These diseases impact not only animal populations but also humans who are in close contact with animal species. Q fever is a zoonotic disease caused by C. burnetii that has been widely distributed in nature. Coxiella burnetii in domestic animals, such as cattle, sheep, and goats, has been widely reported.9–12 However, there is little detailed epidemiological data regarding the distribution and determinants of C. burnetii infection in rodents. Recent findings in Germany indicated regular sightings of wild rodents as risk factors for C. burnetii infection in humans, suggesting wild rodents as a direct source for human infection.13 In this study, C. burnetii was identified by PCR in 18% of the rodents captured on Heixiazi Island at the Sino-Russian border, from April to October in 2011. However, it is difficult to compare these findings with those reported in other works, which present information on the presence of antibodies to C. burnetii in rodents.13–16 The statistical results showed that weight was associated with C. burnetii infection, possibly because larger rodents become matured and move widely and have more chances to be infected with C. burnetii. The reason for the association between month of capture and C. burnetii infection may be the temperature differences between months. Previous studies showed that Hantaviruses coevolved with specific rodents17 which means certain pathogens dominate in certain species. However, in this study, no significant association of specific rodent species and C. burnetii infection was found, and all observed rodent species incidences of infection with C. burnetii. This result was partly consistent with the previous study that identified Rattus norvegicus as a host for C. burnetii infection. In a human-based study,3 it was found that sex was related with C. burnetii infection, where men were 2.5 times more likely to be infected with the pathogen than women; the same result was also reported in mice in a laboratory-based study.16 However, the results of our study were different; no significant association was found between C. burnetii infection and sex in wild rodents.
The results of the molecular epidemiologic analysis showed that although discrepancy existed in the nucleotide sequences of C. burnetii in the tested rodents, the majority of sequences showed high shared identity in the region of the com1 gene, and the sequences aligned with other strains isolated elsewhere in the world. This sequence heterogeneity led us to continue surveillance of the pathogen to describe the variants of the pathogen that occur in the island rodent population.
In conclusion, this study confirms that rodents are reservoirs of C. burnetii, and it is the first evidence of C. burnetii infection in rodents on Heixiazi Island at the Sino-Russian border of Heilongjiang Province, China. As reservoirs, however, rodents have not yet been shown as a vector for transmission of Q fever to humans, and further studies need to address pathogen maintenance in and transmission to humans.
Disclaimer: There is no conflict of interest for the authors with the present study.
Footnotes
Financial support: This study was supported by Basic Research Expenditure of CAIQ 2012JK038, and the National Special Project of International Cooperation in Science and Technology 2012DFA30540 and 2010DFA34250.
Disclosure: Dr. Liu is an epidemiologist at the Institute of Health Quarantine of CAIQ. Her primary research interest is public health in the field of Quarantine.
Authors' addresses: Lijuan Liu, Xu Baoliang, Guo Tianyu, Yang Yu, Sun Xiaohong, and Wang Jing, Institute of Health Quarantine, Chinese Academy of Inspection and Quarantine, Beijing, PRC, E-mails: lljyhxx@126.com, xubaol@yahoo.com.cn, gty200411@sina.com, redyy99@gmail.com, dotsunny@hotmail.com, and wangjing0115@126.com. Fu Yingqun, Li Ming, and Hou Yong, Department of Health and Quarantine, Heilongjiang Entry-Exit Inspection and Quarantine Bureau, Harbin, PRC, E-mails: fuyingqun109@yahoo.com.cn, chfi2002@163.com, lemon_hy2003@126.com. Jiang Chao and Wang Shasha, Department of Zoology, Shenyang Agriculture University, Shenyang, PRC, E-mails: jc_0043@163.com and wangsha_327@163.com. Hu Manxia, Department of Health and Quarantine, Suifenhe Entry-Exit Inspection and Quarantine Bureau, PRC, E-mail: hmx1@163.com.
References
- 1.Parker NR, Barralet JH, Bell AM. Q fever. Lancet. 2006;367:679–688. doi: 10.1016/S0140-6736(06)68266-4. [DOI] [PubMed] [Google Scholar]
- 2.Raoult D, Marrie T, Mege J. Natural history and pathophysiology of Q fever. Lancet Infect Dis. 2005;5:219–226. doi: 10.1016/S1473-3099(05)70052-9. [DOI] [PubMed] [Google Scholar]
- 3.Andoh M, Naganawa T, Hotta A, Yamaguchi T, Fukushi H, Masegi T, Hirai K. SCID mouse model for lethal Q fever. Infect Immun. 2003;71:4717–4723. doi: 10.1128/IAI.71.8.4717-4723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masala G, Porcu R, Sanna G, Chessa G, Cillara G, Chisu V, Tola S. Occurrence, distribution, and role in abortion of Coxiella burnetii in sheep and goats in Sardinia, Italy. Vet Microbiol. 2004;99:301–305. doi: 10.1016/j.vetmic.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Madariaga MG, Rezai K, Trenholme GM, Weinstein RA. Q fever: a biological weapon in your backyard. Lancet Infect Dis. 2003;3:709–721. doi: 10.1016/s1473-3099(03)00804-1. [DOI] [PubMed] [Google Scholar]
- 6.Potasman I, Rzotkiewicz S, Pick N, Keysary A. Outbreak of Q fever following a safari trip. Clin Infect Dis. 2000;30:215–217. doi: 10.1086/313613. [DOI] [PubMed] [Google Scholar]
- 7.Jiang JF, Zuo SQ, Zhang WY, Wu XM, Tang F, De Vlas SJ, Zhao WJ, Zhang PH, Dun Z, Wang RM, Cao WC. Prevalence and genetic diversities of hantaviruses in rodents in Beijing, China. Am J Trop Med Hyg. 2008;78:98–105. [PubMed] [Google Scholar]
- 8.Akihiko HI, Akiko NA, Takashi CH, Akemi KA. Development of a method for detecting Coxiella burnetii in cheese samples. J Vet Med Sci. 2012;74:175–180. doi: 10.1292/jvms.11-0023. [DOI] [PubMed] [Google Scholar]
- 9.McCaughey C, Murray LJ, McKenna LP, Menzies FD, McCullough SJ, O'Neill HJ, Wyatt DE, Cardwell CR, Coyle PV. Coxiella burnetii (Q fever) seroprevalence in cattle. Epidemiol Infect. 2009;138:21–27. doi: 10.1017/S0950268809002854. [DOI] [PubMed] [Google Scholar]
- 10.Tilburg JJ, Roest H-J, Buffet S, Nabuurs-Franssen MH, Horrevorts AM, Raoult D, Klaassen CH. Epidemic genotype of Coxiella burnetii among goats, sheep, and humans in the Netherlands. Emerg Infect Dis. 2012;18:887–889. doi: 10.3201/eid1805.111907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodolakis A. Q fever, state of the art: epidemiology, diagnosis and prophylaxis. Small Rumin Res. 2006;62:121–124. [Google Scholar]
- 12.Mediannikov O, Fenollar F, Socolovschi C, Diatta G, Bassene H, Molez JF, Sokhna C, Trape JF, Raoult D. Coxiella burnetii in humans and ticks in rural Senegal. PLoS Negl Trop Dis. 2010;4:e654. doi: 10.1371/journal.pntd.0000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reusken C, van der Plaats R, Opsteegh M, de Bruin A, Swart A. Coxiella burnetii (Q fever) in Rattus norvegicus and Rattus rattus at livestock farms and urban locations in the Netherlands; could Rattus spp. represent reservoirs for (re)introduction? Pre Vet Med. 2011;101:124–130. doi: 10.1016/j.prevetmed.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Riemann HP, Behymer DE, Franti CE, Crabb C, Schwab RG. Survey of Q-fever agglutinins in birds and small rodents in northern California, 1975–76. J Wildl Dis. 1979;15:515–523. doi: 10.7589/0090-3558-15.4.515. [DOI] [PubMed] [Google Scholar]
- 15.Webster JP, Lloyd G, Macdonald DW. Q fever (Coxiella burnetii) reservoir in wild brown rat (Rattus norvegicus) populations in the UK. Parasitology. 1995;110:31–35. doi: 10.1017/s0031182000081014. [DOI] [PubMed] [Google Scholar]
- 16.Textoris J, Ban LH, Capo C, Raoult D, Leone M, Mege JL. Sex-related differences in gene expression following Coxiella burnetii infection in mice: potential role of circadian rhythm. PLoS ONE. 2010;5:e12190. doi: 10.1371/journal.pone.0012190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plyusnin A, Morzunov SP. Virus evolution and genetic diversity of Hantaviruses and their rodent hosts. Curr Top Microbiol Immunol. 2001;256:47–75. doi: 10.1007/978-3-642-56753-7_4. [DOI] [PubMed] [Google Scholar]