Abstract
In human cells, cytosolic citrate is a major precursor for the synthesis of fatty acids, triacylglycerols, cholesterol and low-density lipoprotein. Cytosolic citrate further regulates the cell’s energy balance by activating the fatty acid synthesis pathway while down-regulating both the glycolysis and fatty acid β-oxidation pathways (Supplementary Fig. 1) 1–4. The rate of fatty acid synthesis in liver and adipose cells, the two major tissue types for such synthesis, correlates directly with the concentration of citrate in the cytosol 2–5. The cytosolic citrate concentration partially depends on direct import across the plasma membrane via the Na+-dependent citrate transporter (NaCT) 6,7. Mutations of the homologous fly gene (INDY, I’m Not Dead Yet) result in reduced fat storage through calorie restriction 8. More recently, NaCT-knockout mice have been found to have increased hepatic mitochondrial biogenesis, higher lipid oxidation and energy expenditure, and reduced lipogenesis, which taken together protect the mice from obesity and insulin resistance 9. To understand the transport mechanism of NaCT/INDY proteins, here we report the 3.2 Å crystal structure of a bacterial INDY homolog. One citrate molecule and one sodium ion are bound per protein, and their binding sites are defined by conserved amino acid motifs, forming the structural basis for understanding the transporters’ specificity. Comparison of the structures of the two symmetrical halves of the transporter suggests conformational changes that propel substrate translocation.
NaCT is a member of the mammalian solute carrier family 13 (SLC13), which also includes two dicarboxylate transporters (NaDC1 and NaDC3, Supplementary Figs. 2&3) 10,11. Whereas these three plasma membrane proteins transport both tricarboxylates (citrate) and dicarboxylates, such as succinate, malate and fumarate, they also display distinct substrate specificity 12–17. While NaCT transports primarily citrate, NaDC1 and NaDC3 have a higher affinity for succinate, with NaDC3 being the high affinity transporter. Intriguingly, the SLC13 family also contains two additional highly homologous proteins, NaS1 and NaS2 (Supplementary Figs. 2&3). Instead of carboxylates, they transport sulfate 10,11. Transport by SLC13 proteins is Na+-driven, with one substrate molecule being co-transported together with 3 or 4 Na+ ions per cycle 11. Along with the homologous fly INDY, the mammalian SLC13 proteins belong to the divalent anion/Na+ symporter (DASS) family, which also contains numerous bacterial members (Supplementary Figs. 2&3) 11,18. Several of these bacterial DASS proteins have been shown to catalyze Na+-coupled dicarboxylate uptake, with two Na+ ions being co-transported with one substrate molecule 19–22. In turn, the DASS family belongs to the Ion Transporter (IT) superfamily 18, which comprises 16 transporter families, with over 32,000 members identified thus far.
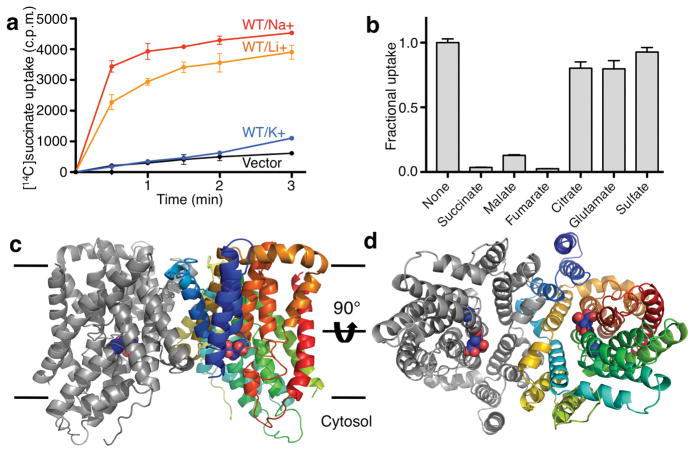
In order to understand the transport mechanism of INDY proteins, we functionally characterized the INDY homolog from Vibrio cholerae (vcINDY). vcINDY consists of 462 amino acids and shares 26 – 33% sequence identity with the three human SLC13 transporters (Supplementary Fig. 3). We first tried to verify whether vcINDY was a Na+-driven carboxylate transporter and to identify its substrate. In E. coli whole cells transformed with the Vibrio gene 13,22, vcINDY catalyzed uptake of succinate (Fig. 1a). The transport was driven by a Na+ gradient, but K+ had no effect. Interestingly, a Li+ gradient also drove transport, although at a slightly slower rate. The uptake reached saturation within 3 minutes, similar to what had been observed for its homologs 10,11. The transport of succinate by vcINDY could be inhibited by malate and fumarate, slightly inhibited by glutamate, but not inhibited by sulfate (Fig. 1b). This suggests that malate and fumarate, two other dicarboxylates, are also substrates of vcINDY, as observed in other mammalian and bacterial INDY homologs 10,11. Citrate also slightly inhibited succinate transport by vcINDY, presumably in a competitive manner. vcINDY is found to be a dimer in detergent, judged by size exclusion chromatography (Supplementary Fig. 4). Although its dimeric state was unaffected by the presence of Na+, dicarboxylate or citrate, its peak height at elevated temperatures depended on the presence of carboxylate (Supplementary Fig. 5). While succinate or malate stabilized the protein modestly, the presence of citrate markedly improved the protein’s thermostability, indicating specific interaction between vcINDY and citrate.
Fig. 1.
Functional characterization and structure determination of the Na+-dependent dicarboxylate transporter vcINDY from Vibrio cholerae. a, Na+-driven succinate transport by vcINDY measured in whole-cells 13,22. The succinate uptake was measured in vcINDY-transformed E. coli in buffers that contained 5 μM [14C]succinate and either Na+, Li+ or K+. Thecontrol experiment was carried out in Na+ buffer using cells that were transformed with empty vector. b, Uptake of [14C]succinate in the presence of various di- and tri-carboxylates and sulfate (at 1 mM concentration). For a and b, N = 3. c, Crystal structure of the vcINDY dimer at 3.2 Å resolution viewed from within the membrane. A citrate and a Na+ ion are adjacently bound to each vcINDY protomer at the cytosolic basin of the protein dimer. d, Crystal structure of the vcINDY dimer viewed from the cytosol. The bound citrate is exposed to the cytosolic space whereas the Na+ ion is buried. In c and d, the polypeptide in one protomer is colored using the standard rainbow scheme.
We then crystallized vcINDY in the presence of citrate, Na+ and Li+, and the crystals diffracted X-ray to 3.2 Å resolution. The crystal structure was determined using single-wavelength anomalous diffraction from data merged from four separate selenomethionyl datasets (Supplementary Fig. 6; Supplementary Tables 1&2) 23. In the crystal structure, the vcINDY protein formed a dimer, which has the shape of the letter “M” when viewed from within the membrane plane, with a concave aqueous basin (Figs. 1c&d, Supplementary Figs. 7&8). Each protein protomer comprises eleven transmembrane α-helices, TMs 1–11 (Fig. 2a). As the N- and C-terminal of vcINDY proteins from other species have been shown to be in the cytosol and the extracellular space, respectively 24, the extramembraneous extrusions of the protein and the concave aqueous basin are inferred to be the cytosolic side. The interface between the two protein protomers is formed by TM3, TM4a and TM9b, interacting with TM4b, TM8 and TM9a of the neighboring protomer (Fig. 1d, Supplementary Fig. 7). The interface between the two protomers has an area of ~2,500 Å2, a large interface area in agreement with the observed stable protein dimer in detergent solution (Supplementary Fig. 4).
Fig. 2.
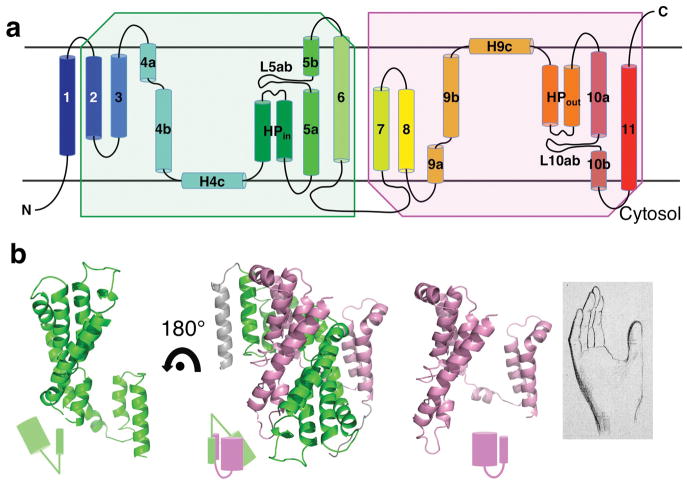
Structure of the vcINDY protomer. a, Transmembrane topology of vcINDY. The two halves of the protein, TMs2-6 and TMs7-11, are related by a repeat in amino acid sequence, resulting in a transmembrane topology that displays an inverted twofold symmetry. b, The N-and C-terminal halves of the protomer each forms a hand-shaped structure, and the two hands are related by an inverted twofold symmetry. TMs2&3 form the thumb, and the helical bundle of TM4b – TM6 takes the shape of the palm in the N-terminal half; in the C-terminal half, the thumb is formed by TMs7&8 and the palm by TM9a – TM11. Note the linker helix between the palm and the thumb in the N-terminal half is at a larger angle from the membrane plane than that of the linker in the C-terminal hand, giving the former a V-shape and the latter a U-shape. The structures of two helical bundles, the palms, are similar and their superposition yields an r.m.s.d. of 2.9 Å for backbone Cα atoms. The N- and C-terminal halves of the protein are colored green and purple, respectively.
Among the eleven transmembrane α-helices, TM4, TM5, TM9 and TM10 are each broken into two segments within the membrane, and each pair is named “a” and “b”, respectively (Figs. 2a, Supplementary Fig. 8). The loops between TM5a and TM5b (L5ab), and between TM10a and TM10 (L10ab) are each 8 amino acids long. In addition, vcINDY also contains several other secondary structure elements. A helical hairpin (HPin) inserts into the membrane from the cytosolic side, which is connected to TM4 via helix H4c on the membrane surface and by a loop to TM5. Similarly, on the opposite side of the membrane, a helical hairpin (HPout) inserts into the protein from the periplasm and connects to TM9 via helix H9c. Such helical hairpins and intramembrane loops within a broken helix are often found to play a major mechanistic role in membrane transport proteins 25–28.
The N- and C-terminal halves of vcINDY share a 26.2% identity in amino acid sequence. Closer inspection of its crystal structure reveals that the protein consists of a twofold repeat (Fig. 2). The N-terminal half of the protein, TM2 – TM6, is related to the C-terminal half, TM7- TM11, by an inverted, twofold symmetry, with the symmetry axis parallel to the membrane plane and the two halves of the protein inserted into the membrane from opposite directions. Each half of the protein has the shape of a hand (Fig. 2b). For the N-terminal half, TM2 and TM3 form a thumb, and the palm is formed by a five-helix bundle that consists of TM4b, TM5 and TM6 as well as the helical hairpin, HPin. The thumb and the palm are connected by TM4a, which is at a 45° angle from the membrane plane, yielding a “V” shaped hand. The entire N-terminal hand inserts into the membrane from the periplasmic side. Related by the inverted twofold symmetry, the C-terminal hand its—thumb formed by TM7 and TM8 and its palm formed by TM9b, helical hairpin HPout, TM10 and TM11—inserts from the cytosolic side. The helix that connects the thumb and the palm, TM9a, occurs at an angle to the membrane plane of about 25°. Thus, the angle between the thumb and palm in the C-terminal hand has the shape of a letter “U.” When superimposed by their thumbs, the two helical bundles are at different heights in the membrane (Supplementary Figs. 9&10). As expected from the crystallization conditions containing Na+ and citrate, we identified a citrate molecule and one Na+ ion bound to each of the transporter protomers in the structure (Figs. 1c&d, Supplementary Fig. 8). The citrate and Na+ ion are located near each other in a cleft at the inner end of the dimer basin, directly exposed to the cytosolic space (Fig. 3a). It follows that the crystal structure of vcINDY represents an inward-facing conformation.
Fig. 3.
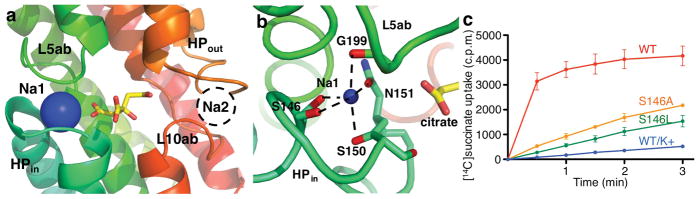
Na+ ion-binding sites in vcINDY. a, Structure of the Na+-binding site (Na1) formed by the tip of HPin and the L5ab loop. The binding site has the shape of a clamshell, which we named the “hairpin tip – capping loop motif” for sodium binding. A second, putative Na+-binding site (Na2), is suggested to be located between the tip of HPout and the L10ab loop formed by the C-terminal hairpin tip – capping loop motif. However, no electron density for Na+ was found at this site in the crystal structure. In the current inward-facing transporter structure, the Na2 site is directly exposed to the cytosolic space. b, Coordination of the Na+ ion at the Na1 site. Both side chains of amino acid residues and backbone carboxyl oxygen atoms are involved in the Na+ coordination. c, Succinate transport activity of Na1-site mutants. N = 3.
The transport of substrates into the cell by vcINDY is driven by the inward sodium gradient (Fig. 1a). In the vcINDY protomer, a Na+ ion sits in a clamshell formed by the helical hairpin HPin tip and the loop L5ab, and is separated from the cytosolic space by the bound citrate molecule (Fig. 3a, Supplementary Fig. 11). We named this structural element the “hairpin tip – capping loop motif” for sodium binding. The Na+ ion interacts directly with both the amino acid side chains and backbone carbonyl oxygen atoms of these residues (Fig. 3b). Specifically, the Na+ ion is coordinated by the Ser146 side chain, its backbone oxygen, the Ser150 backbone oxygen and the Asn151 side chain, all from the helical hairpin HPin, and by the backbone oxygen of Gly199 from the loop L5ab (Fig. 3b, Supplementary Table 3). When Ser146 was mutated into an alanine or a leucine, the transport rate of vcINDY decreased markedly (Fig. 3c, Supplementary Fig. 12), supporting the critical role of this residue in the coordination of the Na+ ion. We named this Na+ ion Na1.
For bacterial INDY proteins, biochemical experiments have shown that, typically, two Na+ ions are co-transported with one substrate molecule 19,20,22. In the vcINDY structure, there is a second hairpin tip – capping loop motif, located in the C-terminal half of the protein that is related to the N-terminal site by inverted twofold symmetry (Fig. 3a). This motif comprises the helix hairpin HPout tip and loop L10ab (Fig. 2a). Both the amino acid sequences for the two motifs and the HPin and HPout segments are highly conserved among various INDY proteins (Supplementary Fig. 3). We therefore hypothesize that the hairpin tip – capping loop motif in the C-terminal half of vcINDY forms the second Na+-binding site (Fig. 3a). However, no electron density for a Na+ ion is observed at this region. There are two possible explanations. One is that this Na2 site is occupied by a Li+ ion, which is too light to visualize at the current resolution. Another possibility, which we favor, is that the Na2 ion has already been released. As this site is directly exposed to the cytosolic space, it is logical to be the first Na+ ion to escape before release of the substrate molecule itself. This is further supported by the observation that the distance between the helix hairpin HPout tip and loop L10ab in the empty Na2 clamshell is larger than that of the occupied Na1 clamshell (Supplementary Fig. 13), indicating an open structure following Na+ release. Interestingly, when the equivalent glutamate residue of vcINDY-Glu374 at the HPout tip in the Na2 clamshell was mutated in the mammalian NaDC1 (Glu475), the Michaelis constant Km for substrate transport increased markedly 14, in agreement with a Na+ site being at this location. Finally, in the L10ab loop, the equivalent residue of vcINDY-Cys413 in human NaCT, Phe500, has been shown to be essential for Li+ binding and its stimulation of citrate transport 29. As the Na1 ion is buried and inaccessible to the cytoplasmic space until the bound substrate is released, the Na1 and the tentative Na2 ions are mechanistically non-equivalent.
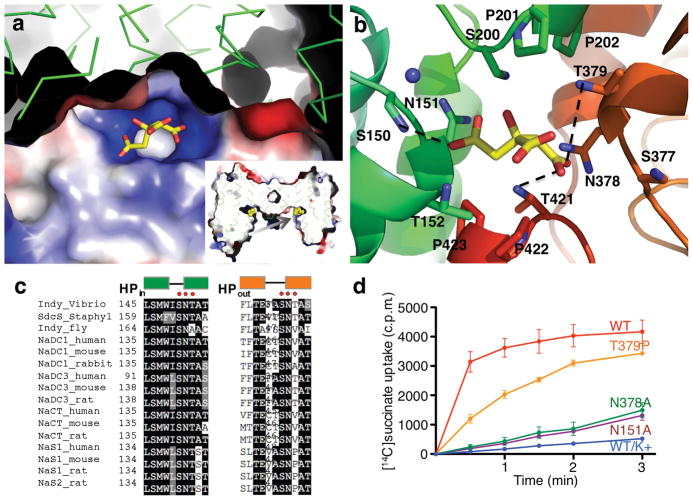
Between the Na1 and Na2 sites, a citrate molecule is found to bind in the middle of the vcINDY protomer (Fig. 4, Supplementary Fig. 14). This binding pocket displays a strong positive electrostatic surface potential (Fig. 4a). It is formed by residues from HPin, TM5, HPout and TM10. Just like the twofold symmetry of the citrate molecule, the binding pocket is also symmetrical. Citrate’s 5-carboxyl group points to Ser150, Asn151 and Thr152 from HPin, while the 1-carboxyl group directly interacts with an inverted triangle formed by Ser377, Asn378 and T379 from HPout (Fig. 4b). The cytosolic and periplasmic sides of the binding pocket are formed by Thr421 and Pro422 from L10ab, and by Pro201 and Ser202 for L5ab, respectively. Additional interaction to the citrate is mediated via hydrogen bonds with the side chains of Ser150, Thr379 and Thr421.
Fig. 4.
Substrate-binding site in vcINDY. a, Electrostatic surface potential of the substrate-binding site. Insert: cross section of the electrostatic surface potential of vcINDY dimer. The plane of this central cross section is perpendicular to the membrane and is at a small angle from the long axis of the dimer in order to show both citrate molecules bound to the transporter dimer. The arrow points in the direction for the view in a. b, Structure of the substrate-binding site with a citrate bound, showing the coordination of the substrate analog. Three hydrogen bonds are indicated by dashed lines. The citrate lies at a small angle to the membrane plane, and its long axis is parallel to the protomer-protomer interface. The central 6-hydroxyl-, carboxyl-groups are exposed to the cytosolic space. While the side chain of Ser150 forms a hydrogen bond with the 5-carboxyl group of the citrate, its backbone carbonyl oxygen atom participates in the coordination of the Na1 ion. Similarly, the side chain of Asn151 interacts with both Na1 and the bound citrate. c, Amino acid sequence alignment of vcINDY and its homologs, showing the two SNT carboxylate-binding motifs. d, Succinate transport activity of substrate-binding site mutants. N = 3.
As citrate inhibits the transport of succinate (Fig. 1b), it is reasonable to assume that the observed citrate-binding pocket in vcINDY (Fig. 4) is also the binding site for dicarboxylate substrates and that a substrate molecule binds to the transporter in a similar manner. In fact, when either Asn151 or Asn378 was mutated into an alanine, the affinity of vcINDY to succinate was found to be markedly reduced and the transport rate decreased (Figs. 4c&d, Supplementary Fig. 12). Just like the amino acid sequence conservation for the Na+-binding motifs, the Ser-Asn-Thr (SNT) motif in the N-terminal half is highly conserved among INDY proteins of various species, from bacteria to human, while the C-terminal motif allows for variation between a threonine and a valine only in the third position (Fig. 4c). Therefore, these two SNT motifs can be regarded as the signature sequences of Na+-dependent tri-,dicarboxylate transporters in the DASS family. In agreement with this notion, it was previously observed that when the serine and asparagine residues in the C-terminal SNT motif were mutated into a cysteine in the rabbit NaDC1 protein, the transport rate for succinate dropped to 25% and 0% of the wild-type, respectively 16. Alternatively, when the equivalent of vcINDY-Pro422 or Pro423 in rabbit NaCT1 was mutated into a glycine or alanine 17, the protein still transported succinate, further supporting that the determinant for substrate specificity is primarily mediated via 1,5-carboxyl groups of the substrate to the two SNT motifs of the protein. The highly positively charged nature of the substrate-binding site explains why dicarboxylates such as glutamate, which has an HN+ group, are not a substrate for mammalian SLC13 proteins 12. Finally, the substrate-binding pocket in the structure also explains the substrate preference between carboxylate and sulfate transporters among SLC13 proteins (Fig. 4c, Supplementary Fig. 15, Supplementary Discussion) 10,11.
Although the bound Na1 ion does not directly coordinate the substrate molecule as in some other transporters 26, the substrate-binding site shares residues with both the Na1 and the putative Na2 binding sites (Fig. 3a). While the side chain of Ser150 forms a hydrogen bond with the 5-carboxyl group of the citrate, its backbone carbonyl oxygen atom participates in the coordination of the Na1 ion. Similarly, the side chain of Asn151 binds to citrate on one side and coordinates Na1 on its opposite side. Such close proximity and, especially, the residue-sharing nature of the substrate- and the Na+-binding sites immediately suggests an ion-coupling mechanism of substrate transport. As previous biochemical experiments have shown that Na+ ions bind to INDY proteins before a substrate can bind 13,15,19,20,22, it follows that the binding of Na+ ions creates an optimal binding site for the substrate via an induced-fit mechanism. Our mechanism is supported by previous studies on the single-nucleotide polymorphism of human NaDC1 30. The change of Val477, at the third position of the C-terminal SNT motif, to a methionine markedly lowers affinity to Na+ and simultaneously abolishes succinate transport.
The core of the vcINDY protomer structure formed by the two helical bundles (the palms) resembles that of the recently determined concentrative nucleoside transporter, CNT 28, which has no detectable homology with vcINDY and is not a member of the IT superfamily 18. The palms of the vcINDY protein, TM4-TM6 and TM9-TM11, are equivalent to H3-H5 and H6-H8 in CNT, respectively (Supplementary Fig. 16). The substrate binding sites in the two transporters are also located at approximately the same position. With only one clamshell Na+-binding motif, CNT has one Na+ ion bound, which is located at the equivalent site of the vcINDY Na1. As vcINDY is a dimer and CNT a trimer, the scaffoldings and the manners of expected conformational changes in the two transporters are different (Supplementary Discussion).
Our crystal structure also suggests a model for conformational changes needed in vcINDY to propel substrate across the membrane (Supplementary Fig. 17, Supplementary Discussion). In the Co state, the two halves of a protomer adopt a N(U)-C(V) conformation. Upon Na+ and substrate binding, it converts to a N(V)-C(U) conformation, followed by Na+ and substrate release to the cytosol. Such a transport mechanism, along with the structural basis of substrate and ion specificity and ion-coupling to substrate transport, provides a direct frame for understanding its mammalian counterpart 10,11. As the human NaCT protein may be a particularly attractive drug target for obesity, diabetes and cardiovascular diseases, the identification of the substrate-binding motifs may aid in the development of such agents.
Methods
Expression and purification
A transporter protein (Q57486) from Haemophilus influenzae, a homolog of human NaDC-1 and Drosophila INDY, was expressed, purified and characterized using standard protocols 31–33. The Haemophilus protein was then nominated to the cloning core of the New York Consortium of Membrane Protein Structure for the cloning of its homologs 34. Among the 31 clones tested, the homologous protein from Vibrio Cholera (AAF95939, vcINDY) was found to give the highest expression levels. For overexpression, E. coli BL21AI (Invitrogen) cells were transformed with a modified pET vector 34 encoding N-terminal 10x His-tagged vcINDY. After harvesting of the cells, membranes were solubilized in 1.2 % n-decyl-β-maltoside (DM) and the protein was purified on a cobalt affinity column (TALON, Clontech), followed by preparative size exclusion chromatography in buffer containing 50 mM Tris pH 7.5, 100 mM NaCl, 50 mM lithium citrate, 5% glycerol and 0.15% DM. SeMet protein was produced and purified in the same way only in E.coli B834DE3 (Novagen) cells grown in minimal media containing seleno-L-methionine. For the determination of the oligomeric state of purified vcINDY protein in detergent solution, protein samples were injected onto an analytical size exclusion chromatography column (Shodex KW804, Thomson) on HPLC (Shimadzu) in buffer containing 200 mM Na2SO4, 50 mM Tris pH 8.0, 3 mM NaN3 and 0.05 % n-dodecyl-β-maltoside (DDM) 31. Transporter proteins with similar molecular weights and well-characterized oligomeric states, the monomeric glycerol-3-phosphate transporter from E. coli (GlpT) 31 and the dimeric tetracycline transporter from Bacillus subtilis (TetL) 35, were used as standards. To measure the effects of various compounds in thermostabilizing vcINDY, purified protein samples were incubated in the presence of each of these compounds (50 mM concentration) at 44 °C for 10 minutes, followed by SEC analysis on HPLC 31,36.
Transport assays
Transport activity of vcINDY at 25 °C was characterized using a whole-cell assay of [14C]succinate uptake 13,22,37. E. coli BL21 p-Lys cells were transformed with either the wild type gene, mutants or empty vector. 60 ml cultures were grown to OD660 0.7, induced with isopropyl-b-thiogalactoside for 2 – 2.5 hours and harvested. Cells were washed twice with wash buffer (50 mM K-Phosphate, pH 7.5) and resuspended in the same buffer to OD660~10. Transport was initiated by mixing the cell suspension with 10 fold concentrated assay buffer at a ratio of 9:1 to yield a final concentration of 5 mM NaCl (LiCl or KCl at the same concentration), 95 mM Tris pH 7.5 and 5 μM [14C]succinate (stock, 54 mCi/mmol, Moravec Biochemicals). Aliquots of 100 μl were taken at various time points covering the range from 0 to 5 mins and the transport reaction was terminated by harvesting the cells on 0.45 μm nitrocellulose filters under vacuum, followed by washing with 4 ml of ice-cold wash buffer. The filters were incubated for 10 mins in scintillation fluid prior to measuring radioactivity with a liquid scintillation counter 38,39. For the competition assay, the final buffer also included 1 mM of the test compound. For the activity measurements of mutants, their expression levels in E. coli cells were verified by Western blot using India HisProbe-HRP (Pierce).
Crystallization
Crystals were grown at 18 °C in hanging-drop vapor diffusion by mixing equal amounts of sizing column-purified protein at 4 – 6 mg/ml (supplemented with 0.12% of n-nonyl-β-D-glucoside) and reservoir solution (29% (v/v) polyethylene glycol 1000, 50 mM lithium citrate and 50 mM MOPS, pH 6.5).
Crystallography
Crystal screening was carried out at the beamlines X25 and X29 of the National Synchrotron Light Source (NSLS) in the Brookhaven National Laboratory and at 23ID at the Advanced Photon Source at the Argonne National Laboratory. X-ray diffraction data were collected at NSLS beamline X4A with a Quantum Q4R CCD detector. There are four protein molecules in the asymmetric unit, and each contains 23 methionine residues. To collect anomalous diffraction data from SeMet crystals, the wavelength was tuned to 0.9792 Å as verified by fluorescence scan on crystals. Inverse-beam mode data collection was used and four complete diffraction data sets were collected from 3 crystals, with two data sets from each end of a long crystal. The phases were obtained using a multi-crystal single-wavelength anomalous dispersion (SAD) phasing method 23,40. Briefly, anomalous diffraction data sets were indexed and integrated using XDS 41. To enhance anomalous signals, integrated intensities were scaled, analyzed and merged by SCALA 42 to 3.2 Å resolution. Se substructure was determined by SHELXD 43 from the merged data. Attempts were made with various numbers of expected Se sites, various high resolution cutoffs and various Emin cutoffs. The 92-site Se substructure, corresponding to 4 molecules in the crystallographic asymmetric unit cell, was identified from the merged data that was truncated at 4.1 Å for high resolution cutoff and 1.5 for Emin cutoff. The substructure was refined and completed for SAD phasing by PHASER 44 with the merged data. The SAD phases were then density modified by DM 45 to break the phase ambiguity, resulting in electron density maps at 3.5 Å resolution of sufficient quality for model building. Model building was done in Coot 46. The first 18 residues at the N-terminus and a fragment in a central loop (a.a. 240 – 251) were disordered in the crystals. For residues 252 – 259, only the backbone was visible, so a poly-alanine model was constructed in that part. Model refinement to 3.2 Å resolution was accomplished using PHENIX 47 and CCP4 48 packages. For the bound citrate molecule in each of the four protein protomers in the crystallographic asymmetric unit, two have a B-factor of 90–100 Å 2, similar to that of the protein, whereas the other two have a B-factor of 120–137 Å 2. Structural figures were prepared using Pymol 49 and Coot.
Supplementary Material
Acknowledgments
We are grateful to M. Punta and B. Rost for bioinformatics analysis of membrane transporters, to J. Love and B. Kloss for assistance in cloning, and to the staff at beamlines X4, X25 and X29 of the National Synchrotron Light Source in the Brookhaven National Laboratory and at the 23ID at the Advanced Photon Source at the Argonne National Laboratory for assistance in X-ray diffraction experiments, and to J. Llodra for help with artwork. We thank B.K. Czyzewski, W.A. Hendrickson, N.K. Karpowich, F. Mancia and J.J. Marden for helpful discussions and for participating in synchrotron trips. This work was financially supported by the NIH (U54-GM075026, R01-DK073973, R01-GM093825 and R01-MH083840).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions R.M. and D.N.W. designed the project. R.M. did all the experiments, with assistance from G.G. in diffraction data processing, phasing and structure refinement and from Q.L. in phasing. R.M. and D.N.W. wrote the manuscript.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under access codes 4F35.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Spencer AF, Lowenstein JM. Supply of precursors for synthesis of fatty acids. J Biol Chem. 1962;237:3640–3648. [PubMed] [Google Scholar]
- 2.Bloch K, Vance D. Control mechanisms in synthesis of saturated fatty-acids. Annu Rev Biochem. 1977;46:263–298. doi: 10.1146/annurev.bi.46.070177.001403. [DOI] [PubMed] [Google Scholar]
- 3.Ruderman NB, Saha AK, Vavvas D, Witters LA. Malonyl-CoA, fuel sensing, and insulin resistance. Am J Physiol-Endoc M. 1999;276:E1–E18. doi: 10.1152/ajpendo.1999.276.1.E1. [DOI] [PubMed] [Google Scholar]
- 4.Sul HS, Smith S. Fatty acid synthesis in eukaryotes. In: Vance DE, Vance JE, editors. Biochemistry of Lipids, Lipoproteins and Membranes. Elsevier; New York: 2008. pp. 155–190. [Google Scholar]
- 5.Nishikor K, Iritani N, Numa S. Levels of acetyl coenzyme a carboxylase Its effectors in rat-liver after short-term fat-free refeeding. Febs Lett. 1973;32:19–21. doi: 10.1016/0014-5793(73)80725-2. [DOI] [PubMed] [Google Scholar]
- 6.Inoue K, Zhuang L, Maddox DM, Smith SB, Ganapathy V. Structure, function, and expression pattern of a novel sodium-coupled citrate transporter (NaCT) cloned from mammalian brain. J Biol Chem. 2002;277:39469–39476. doi: 10.1074/jbc.M207072200. [DOI] [PubMed] [Google Scholar]
- 7.Gopal E, et al. Expression and functional features of NaCT, a sodium-coupled citrate transporter, in human and rat livers and cell lines. Am J Physiol-Gastr L. 2007;292:G402–G408. doi: 10.1152/ajpgi.00371.2006. [DOI] [PubMed] [Google Scholar]
- 8.Rogina B, Reenan RA, Nilsen SP, Helfand SL. Extended life-span conferred by cotransporter gene mutations in Drosophila. Science. 2000;290:2137–2140. doi: 10.1126/science.290.5499.2137. [DOI] [PubMed] [Google Scholar]
- 9.Birkenfeld AL, et al. Deletion of the mammalian INDY homolog mimics aspects of dietary restriction and protects against adiposity and insulin resistance in mice. Cell Metab. 2011;14:567–567. doi: 10.1016/j.cmet.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markovich D, Murer H. The SLC13 gene family of sodium sulphate/carboxylate cotransporters. Pflug Arch Eur J Phy. 2004;447:594–602. doi: 10.1007/s00424-003-1128-6. [DOI] [PubMed] [Google Scholar]
- 11.Pajor AM. Molecular properties of the SLC13 family of dicarboxylate and sulfate transporters. Pflug Arch Eur J Phy. 2006;451:597–605. doi: 10.1007/s00424-005-1487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright SH, Kippen I, Klinenberg JR, Wright EM. Specificity of the transport system for tricarboxylic acid cycle intermediates in renal brush borders. J Membr Biol. 1980;57:73–82. doi: 10.1007/BF01868987. [DOI] [PubMed] [Google Scholar]
- 13.Wright SH, Hirayama B, Kaunitz JD, Kippen I, Wright EM. Kinetics of sodium succinate cotransport across renal brush-border membranes. J Biol Chem. 1983;258:5456–5462. [PubMed] [Google Scholar]
- 14.Griffith DA, Pajor AM. Acidic residues involved in cation and substrate interactions in the Na+/dicarboxylate cotransporter, NaDC-1. Biochemistry. 1999;38:7524–7531. doi: 10.1021/bi990076b. [DOI] [PubMed] [Google Scholar]
- 15.Yao X, Pajor AM. The transport properties of the human renal Na+-dicarboxylate cotransporter under voltage-clamp conditions. Am J Physiol Renal Physiol. 2000;279:F54–64. doi: 10.1152/ajprenal.2000.279.1.F54. [DOI] [PubMed] [Google Scholar]
- 16.Pajor AM. Conformationally sensitive residues in transmembrane domain 9 of the Na+/dicarboxylate co-transporter. J Biol Chem. 2001;276:29961–29968. doi: 10.1074/jbc.M011387200. [DOI] [PubMed] [Google Scholar]
- 17.Joshi AD, Pajor AM. Role of conserved prolines in the structure and function of the Na+/dicarboxylate cotransporter 1, NaDC1. Biochemistry. 2006;45:4231–4239. doi: 10.1021/bi052064y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prakash S, Cooper G, Singhi S, Saier MH. The ion transporter superfamily. Bba-Biomembranes. 2003;1618:79–92. doi: 10.1016/j.bbamem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Hall JA, Pajor AM. Functional characterization of a Na+-coupled dicarboxylate carrier protein from Staphylococcus aureus. J Bacteriol. 2005;187:5189–5194. doi: 10.1128/JB.187.15.5189-5194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall JA, Pajor AM. Functional reconstitution of SdcS, a Na+-coupled dicarboxylate carrier protein from Staphylococcus aureus. J Bacteriol. 2007;189:880–885. doi: 10.1128/JB.01452-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Youn JW, Jolkver E, Kramer R, Marin K, Wendisch VF. Identification and characterization of the dicarboxylate uptake system DccT in Corynebacterium glutamicum. J Bacteriol. 2008;190:6458–6466. doi: 10.1128/JB.00780-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strickler MA, Hall JA, Gaiko O, Pajor AM. Functional characterization of a Na+-coupled dicarboxylate transporter from Bacillus licheniformis. Bba-Biomembranes. 2009;1788:2489–2496. doi: 10.1016/j.bbamem.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q, Zhang Z, Hendrickson WA. Multi-crystal anomalous diffraction for low-resolution macromolecular phasing. Acta Crystallogr D Biol Crystallogr. 2011;67:45–59. doi: 10.1107/S0907444910046573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang FF, Pajor AM. Topology of the Na+/dicarboxylate cotransporter: the N-terminus and hydrophilic loop 4 are located intracellularly. Biochim Biophys Acta. 2001;1511:80–89. doi: 10.1016/s0005-2736(00)00385-0. [DOI] [PubMed] [Google Scholar]
- 25.Hunte C, et al. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 26.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 27.Faham S, et al. The crystal structure of a sodium galactose transpoter reveals mechanic insights into Na+/sugar symport. Science. 2008;321:810–814. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson ZL, Cheong CG, Lee SY. Crystal structure of a concentrative nucleoside transporter from Vibrio cholerae at 2.4 Å. Nature. 2012;483:489–493. doi: 10.1038/nature10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inoue K, Zhuang L, Maddox DM, Smith SB, Ganapathy V. Human sodium-coupled citrate transporter, the orthologue of Drosophila Indy, as a novel target for lithium action. Biochem J. 2003;374:21–26. doi: 10.1042/BJ20030827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pajor AM, Sun NN. Single nucleotide polymorphisms in the human Na+-dicarboxylate cotransporter affect transport activity and protein expression. Am J Physiol Renal Physiol. 2010;299:F704–711. doi: 10.1152/ajprenal.00213.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auer M, et al. High-yield expression and functional analysis of Escherichia coli glycerol-3-phosphate transporter. Biochemistry. 2001;40:6628–6635. doi: 10.1021/bi010138+. [DOI] [PubMed] [Google Scholar]
- 32.Li XD, et al. Monomeric state and ligand binding of recombinant GABA transporter from Escherichia coli. FEBS Lett. 2001;494:165–169. doi: 10.1016/s0014-5793(01)02334-1. [DOI] [PubMed] [Google Scholar]
- 33.Wang DN, et al. Practical aspects of overexpressing bacterial secondary membrane transporters for structural studies. Biochim Biophys Acta. 2003;1610:23–36. doi: 10.1016/s0005-2736(02)00709-5. [DOI] [PubMed] [Google Scholar]
- 34.Love J, et al. The New York Consortium on Membrane Protein Structure (NYCOMPS): a high-throughput platform for structural genomics of integral membrane proteins. Journal of structural and functional genomics. 2010;11:191–199. doi: 10.1007/s10969-010-9094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Safferling M, et al. The TetL tetracycline efflux protein from Bacillus subtilis is a dimer in the membrane and in detergent solution. Biochemistry. 2003;42:13969–13976. doi: 10.1021/bi035173q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boulter JM, Wang DN. Purification and characterization of human erythrocyte glucose transporter in decylmaltoside detergent solution. Prot Expr Purif. 2001;22:337–348. doi: 10.1006/prep.2001.1440. [DOI] [PubMed] [Google Scholar]
- 37.Hirato T, Shinagawa M, Ishiguro N, Sato G. Polypeptide involved in the Escherichia coli plasmid-mediated citrate transport system. J Bacteriol. 1984;160:421–426. doi: 10.1128/jb.160.1.421-426.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Law CJ, Yang Q, Soudant C, Maloney PC, Wang DN. Kinetic evidence is consistent with the rocker-switch mechanism of membrane transport by GlpT. Biochemistry. 2007;46:12190–12197. doi: 10.1021/bi701383g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law CJ, et al. Salt-bridge dynamics control substrate-induced conformational change in the membrane transporter GlpT. J Mol Biol. 2008;378:828–839. doi: 10.1016/j.jmb.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Q, et al. Structures from anomalous diffraction of native biological macromolecules. Science. 2012;336:1033–1037. doi: 10.1126/science.1218753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Read RJ, McCoy AJ. Using SAD data in Phaser. Acta Crystallogr D Biol Crystallogr. 2011;67:338–344. doi: 10.1107/S0907444910051371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cowtan KD, Zhang KY. Density modification for macromolecular phase improvement. Prog Biophys Mol Biol. 1999;72:245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeLano WL. The PyMOL User’s Manual. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.