Abstract
Sphingomyelin synthase (SMS) produces sphingomyelin while consuming ceramide (a negative regulator of cell proliferation) and forming diacylglycerol (DAG) (a mitogenic factor). Therefore, enhanced SMS activity could favor cell proliferation. To examine if dysregulated SMS contributes to leukemogenesis, we measured SMS activity in several leukemic cell lines and found that it is highly elevated in K562 chronic myelogenous leukemia (CML) cells. The increased SMS in K562 cells was caused by the presence of Bcr-abl, a hallmark of CML; stable expression of Bcr-abl elevated SMS activity in HL-60 cells while inhibition of the tyrosine kinase activity of Bcr-abl with Imatinib mesylate decreased SMS activity in K562 cells. The increased SMS activity was the result of up-regulation of the Sms1 isoform. Inhibition of SMS activity with D609 (a pharmacological SMS inhibitor) or down-regulation of SMS1 expression by siRNA selectively inhibited the proliferation of Bcr-abl-positive cells. The inhibition was associated with an increased production of ceramide and a decreased production of DAG, conditions that antagonize cell proliferation. A similar change in lipid profile was also observed upon pharmacological inhibition of Bcr-abl (K526 cells) and siRNA-mediated down-regulation of BCR-ABL (HL-60/Bcr-abl cells). These findings indicate that Sms1 is a downstream target of Bcr-abl, involved in sustaining cell proliferation of Bcr-abl-positive cells.
Keywords: ceramide, diacylglycerol, imatinib mesylate
Sphingomyelin synthase (SMS) represents an important class of enzymes responsible for the biosynthesis of SM, a critical structural component of the plasma membrane. While producing SM from ceramide and phosphatidylcholine, SMSs also form diacylglycerol (DAG) (1–7). Therefore, the biological importance of SMSs may reside not only in the biosynthesis of SM but also in the regulation, in opposing directions, of the levels of ceramide, a bioactive molecule that mainly exerts a negative effect on cell proliferation, and DAG, a well-established mitogenic lipid (8).
Indeed, increased SMS activity was observed in conditions of enhanced cell proliferation (e.g., regenerating rat liver or stimulation of quiescent astrocytes with basic FGF) (9, 10) and during cell transformation (such as hepatocellular carcinoma vs. normal liver and SV-40-transformed fibroblasts vs. their normal counterpart) (8, 11). Vice versa, inhibition of SMS activity, in a caspase-dependent manner, has been shown to precede the onset of apoptosis induced by either tumor necrosis factor in Kym-1 cells or FAS ligand in Jurkat T cells (12, 13).
Two genes have been identified as responsible for SMS activity in mammalian cells, SMS1 and SMS2 (14, 15). The aminoacidic sequence of the proteins encoded by these two genes mainly differs in their N-termini, with Sms1 carrying a sterile α motif absent in Sms2. In agreement with previous biochemical studies assessing the intracellular distribution of SMS activity, Sms1 localizes at the Golgi, whereas Sms2 localizes at the Golgi and plasma membrane (14, 16–18). Modulation of Sms1 or Sms2 activity in most tested cell types has resulted in an alteration of ceramide levels, whereas changes in DAG mass have been elusive (16, 19–23). On the other hand, in Hela cells it has been shown that, in spite of the absence of significant changes of total DAG levels upon modulation of SMSs, intracellular DAG pools (in particular at the Golgi) appeared to be altered (16).
Whereas inhibition of Sms1 by direct caspase cleavage has been shown after treatment with Fas ligand in Jurkat cells (24), no molecular factors that are able to stimulate Sms1 or Sms2 expression or activity have been identified. One of the conditions in which total SMS activity was found to be increased was a pool of samples isolated from patients with acute lymphoblastic leukemia, acute myeloid leukemia, and chronic myelogenous leukemia (CML) (25).
CML is a myeloproliferative disorder of hemopoietic stem cells and occurs in 1 to 1.5 per 100,000, accounting for 15–20% of adult leukemia. CML is directly linked to a chromosomal abnormality caused by the reciprocal translocation of chromosomes 9 and 22, resulting in a shortened chromosome 22 termed the Philadelphia chromosome. This chromosomal translocation generates a fusion product between the breakpoint cluster region (Bcr) and the Abelson kinase (Abl) genes (26–29). The chimera now encodes for a constitutively active tyrosine kinase, Bcr-abl, which initiates the leukemic transformation (30, 31). In the clinical setting, CML is identified by three distinct phases: a chronic phase, an accelerated phase, and a blast phase (32). The chronic phase is characterized by a slight increase of granulocytic progenitors and a substantial increase of granulocytes in the peripheral blood. At this stage, pharmacological inhibition of the Bcr-abl tyrosine kinase activity by imatinib mesylate has proven key for the containment of the disease (33–35). Nevertheless, the insurgence of mechanisms of resistance to current therapy, most commonly mutations in the ATP-binding pocket of Bcr-abl after long-term treatment with tyrosine kinase inhibitors, are causing disease progression from the chronic phase to the terminal blast phase (36). The blast phase is characterized by a block of differentiation resulting in 30% or more myeloid or lymphoid blast cells in the peripheral blood or bone marrow, from where the blast cells eventually infiltrate other organs such as spleen, liver, or lymph nodes (29). These blast cells are refractory to current treatments, and the survival time for patients affected by blast crisis CML is 3 to 6 months (37–39).
In this study, we provide evidence of a link between the expression of the BCR-ABL oncogene, its activity, and elevated SMS activity. We show that elevated expression of SMS1 and not SMS2 is responsible for the elevated SMS activity and that elevated Sms1 is important for sustaining proliferation of Bcr-abl-positive cells. Inhibition of Bcr-abl, SMS activity, or SMS1 expression results in an alteration of the ceramide to DAG ratio consistent with a negative effect on cell proliferation. Together, these results establish the first molecular link between a well-established oncogene and Sms1 expression.
MATERIALS AND METHODS
Materials
RPMI media, FBS, penicillin/streptomycin, and Trypan Blue dye solution were from Gibco/Invitrogen Corp. (Carlsbad, CA). N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino-C6-ceramide and N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino-C6-sphingomyelin were purchased from Molecular Probes. All other lipids were purchased from Avanti Polar Lipids (Alabaster, AL). D609 was purchased from Biomol International, and Imatinib Mesylate was obtained from Novartis.
Cell culture
K562, HL-60, U937, Jurkat, and MCF-7 cell lines were originally purchased from the American Type Culture Collection (Rockville, MD). Human acute myeloid leukemia HL-60 cells stably expressing p185 bcr-abl (HL-60/bcr-abl) or the empty vector (HL-60 neo) were a generous gift from Dr. K. Bhalla (Medical College of Georgia, Augusta, GA) (40). Human leukemic SupT13 T cells were obtained from Dr. J.S. Thompson at the University of Kentucky. All cell lines were grown in RPMI-1640 supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin and were maintained at 37°C at 5% CO2. Cells were routinely counted after staining with the Trypan blue dye. Pharmacological inhibition of SMS and Bcr-abl was achieved by incubation of cells with D609 and imatinib mesylate as indicated in the individual experiments in complete growth medium (in the absence of antibiotics) using 0.1 × 106 cells/ml cultures.
Down-regulation of SMS1 and SMS2 with siRNA
The siRNA for human SMS1 targeted the sequence CACACTATGGCCAATCAGCAA (16) and the one for SMS2 targeted the sequence ACCGTCATGATCACAGTTGTA (23). In preliminary experiments, the effects on SMSs RNA expression, SMS activity, and cell viability of the AllStars Negative Control siRNA (Qiagen) were compared with those of the transfection reagent by itself, and no difference was found; therefore, the Allstars Negative Control was used in subsequent experimentation as baseline. K562 were transfected with siRNAs by electroporation using the Amaxa Nucleofector device (Amaxa) according to the manufacturer's instructions. The protocol was optimized regarding the concentration of SMS1 siRNA, transfection, and post-transfection conditions in preliminary dose-response experiments (1–10 μg SMS1 siRNA). Consequently, the following optimal conditions were used. Cells (1 × 106 K562 cells or 2 × 106 HL-60 cells) were transfected with 2–5 μg of siRNA. After 30 min of incubation at 37°C, 5% CO2 in 500 μl of complete growth medium, cells were initially seeded at 0.5 × 106 cells/ml and subsequently diluted 1:4 once control cells reached a concentration 1.3 × 106 cells/ml. At the indicated time points after transfection, cells were washed twice with ice-cold PBS and processed according to the required analysis.
RNA isolation and reverse transcription for real-time PCR
To analyze the expression of SMS1 or SMS2 in U937, HL-60/neo, HL-60/Bcr-abl, and K562 cells, total RNA was first isolated from 1 × 106 cells using the RNeasy Mini Kit according to the manufacturer's protocol, including optional DNase digestion (Qiagen, Gmbh, Hilden, Germany). Further DNase digestion was performed using the Turbo DNA free kit according to the manufacturer's protocol (Applied Biosystems). The first strand cDNA synthesis was prepared from 1 μg of RNA using Superscript III (Invitrogen) according to the manufacturer's recommendations. Real-time PCR amplification was carried out as described by Villani et al. (16).
Lentiviral-mediated down-regulation of SMS1
For lenti-viral experiments, K562 cells were transduced with nontargeting control particles (shRNA-NTCP) or Mission lentiviral transduction particles (both from Sigma-Aldrich) that contained the pLKO.1-puro vector including the SMS1 targeting sequence 5′-CCGGCCAACTGCGAAGAATAATGAACTCGAGTTCATTATTCTTCGCAGTTGGTTTTTTG-3′ (sh-RNA-SMS1). During transduction, 3 × 104 K562 cells/ml resuspended in RPMI-1640 containing 10% FBS and 10 μg/ml of Hexabromide (Sigma) were infected at a multiplicity of infection of 10 with the appropriate lentiviral particles. Cells were incubated at 37°C, 5% CO2, and 3 days after transduction 10 μg/ml of puromycin (Sigma) was added to the media to select for pLK0.1-puro positive clones and re-added every 3–4 days to maintain selection. The concentration of puromycin needed to select for vector expressing clones was based on a kill-curve analysis showing 70–80% cell death after 3 days in nontransduced K562 cells (data not shown). SMS1 down-regulation (approximately 50%) was confirmed by quantitative real-time PCR.
[3H]Thymidine incorporation assay
K562 cells stably transduced with shRNA-SMS1 or shRNA-NTCP lentivirus particles were used for transient siRNA transfections. K562/shRNA-SMS1 and K562/shRNA-NTCP were transiently transfected with SMS1 siRNA or Allstars Negative control, respectively, and their cellular proliferation rates were compared by thymidine incorporation. The transient transfections were accomplished by the same electroporation protocol as described above and were diluted 48 h after transfection by 1.25 dilution factor in complete medium. The thymidine incorporation assay was optimized for K562 cells to determine the cell concentration and total cell number as well as the amount of [3H]thymidine (American Radiolabeled Chemicals; 1 μCi/μl specific activity) needed for maximum [3H]thymidine incorporation within the linear range of the assay (data not shown). At the indicated time points after transient transfection, 1 μCi of [3H]thymidine was added to 500 μl of serum-free RPMI-1640 containing 2 × 105 K562 cells in each of the experimental groups (as above) and incubated for 4 h at 37°C and 5% CO2. After incubation, cells were centrifuged at 1,000 g for 5 min at 4°C, and the supernatant was removed. The cell pellets were washed twice with ice-cold PBS to remove all excess unincorporated [3H]thymidine followed by a 30 min incubation in 5% ice-cold trichloroacetic acid (700 μl). The suspensions were centrifuged as above, trichloroacetic acid was removed, and pellets were subsequently solubilized in 0.5 N NaOH containing 0.5% SDS (500 μl). For analysis, 150 μl of the solubilized pellets were counted in 4 ml of scintillation fluid (MP Biomedicals), and incorporated [3H]thymidine was measured as counts per minute in a Beckman-Coulter scintillation counter. CPM values were normalized to the total 500 μl of solubilized pellets.
Down-regulation of BCR-ABL
Bcr-abl was transiently down-regulated with a validated siRNA, targeting the sequence 5′-AAGCAGAGTTCAAAAGCCCTT-3′ (41). HL-60/Bcr-abl cells were electroporated with 2 μg of BCR-ABL targeting siRNA or the Allstar Negative Control siRNA according to the conditions described for SMS1 and SMS2 down-regulation with the following modifications. Twelve hours after transfection, cells were diluted to 0.2 × 106 cells/ml in complete medium, and 36 h after transfection, cells were harvested, washed twice with ice-cold PBS, and processed for lipid analysis. Effective down-regulation of Bcr-abl was determined by Western blotting, and an approximately 70% reduction of Bcr-abl in BCR-ABL siRNA treated cells was observed (data not shown).
SMS activity assay
SMS activity was performed as previously described with a few modifications (16). Briefly, three to four million cells were harvested on ice, washed in PBS, and resuspended in 150 μl of homogenization buffer containing 250 mM Tris, 50 mM EDTA (pH 7.4), Pierce Halt phosphatase inhibitor, Pierce Halt protease inhibitor (Pierce, Rockville, IL), and 5 mM PMSF. Cells were then lysed by sonication at 11% power for 20 s (preliminary optimization experiments showed 95% cell lysis with these conditions). Unbroken cells were pelleted by centrifugation at 400 g for 5 min at 4°C, and the supernatant was collected and assayed for protein concentration using the Bio-Rad protein determination assay reagent (Bio-Rad, Hercules, CA). Fifty micrograms of proteins were used for SMS activity, which was carried out and analyzed as previously described (16).
Total membrane preparation and Sms1 Western blotting
Twenty million cells were harvested on ice, washed in PBS, and resuspended in 800 μl of homogenization buffer containing 250 mM Tris, 50 mM EDTA (pH 7.4), Pierce Halt phosphatase inhibitor, Pierce Halt protease inhibitor (Pierce), and 5 mM PMSF. Cells were then lysed as indicated for SMS activity and centrifuged at 1,000 g for 10 min at 4°C. Supernatants were collected and centrifuged at 120,000 g for 1 h at 4°C. Membrane pellets were resuspended in 200 μl of homogenization buffer and passed through a 21 gauge needle until uniform. Membrane protein concentration was determined using the Bio-rad protein determination assay reagent (Bio-rad). Membranes were diluted with 4× sample buffer and incubated at 37°C for 30 min. Cell membrane proteins (80 μg) were separated by 10% SDS-PAGE under reducing conditions and transferred to nitrocellulose membranes. Membranes were blocked with 5% milk PBS-Tween 0.1% for 1 h at room temperature. Membranes were then incubated with Sms1 antibodies raised against the full-length protein (1:1,000) (Ex-α Biologics, Maynard, MA) for 4 h at room temperature in 5% milk PBS-Tween 0.1%. After extensive washings, membranes were incubated with peroxidase-conjugated goat-anti-rabbit (1:3,500; Santa Cruz Biotechnology) in 5% milk PBS-Tween 0.1% for 1 h at room temperature. Signals were visualized using Super Signal (Pierce Chemical Co.) and exposure to Kodak BioMax MR Film (Eastman Kodak Co., Rochester, NY).
Detection of poly(ADP-ribose) polymerase cleavage as an indicator of apoptosis
Analysis of poly(ADP-ribose) polymerase (PARP) cleavage was conducted on total lysates (20 μg) on a 7.5% SDS-PAGE under reducing conditions. After transfer to nitrocellulose membranes and blocking as indicated above, PARP and its cleaved fragment were visualized using primary rabbit polyclonal antibodies (1:1,500; Santa Cruz Biotechnology) and secondary peroxidase-conjugated goat-anti-rabbit (1:10,000; Santa Cruz) both in 5% milk PBS-Tween 0.1%. Signals were visualized using ECL (GE Healthcare) and exposure to Kodak BioMax MR Film (Eastman Kodak Co.).
Lipid analysis
Ceramide, DAG, and SM species were measured by the Lipidomics Core Facility at MUSC (http://www.musc.edu/BCMB/lipidomics/index.shtml). For the lipid measurements, 1 × 106 K562 cells or 2 × 106 HL-60/neo or HL-60/Bcr-abl cells were used for each experimental condition. Aliquots of lipid extracts were used to measure inorganic phosphate, which was used as normalization factor (42).
Statistical analysis
Statistical analysis of the data was performed using Student's t-test, and P < 0.05 was considered statistically significant.
RESULTS
Expression of Bcr-abl increased SMS activity
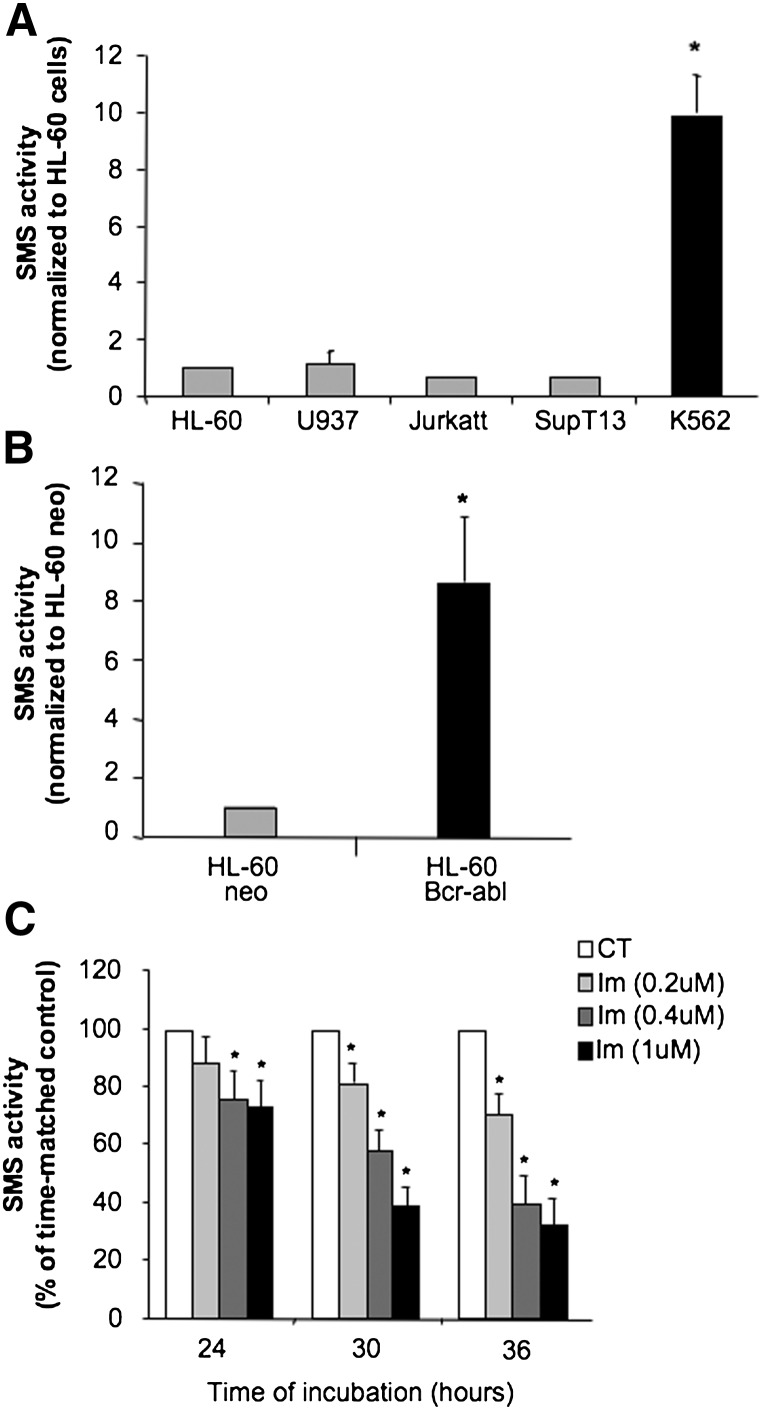
It has been shown that a pool of mononuclear cells isolated from peripheral blood or bone marrow of chemoresistant patients with different leukemias presented elevated SMS activity along with decreased ceramide levels (25). Therefore, to assess if increased SMS activity was associated with specific hematological malignancies, a screening was carried out in different leukemic cell lines in vitro. K562 cells exhibited a significantly higher SMS activity (by approximately 10-fold) compared with the other cell lines (Fig. 1A). The enzymatic activity of SMS was linear with the amount of cell lysates and time of reaction (data not shown), indicating that the differences were not an artifact of the assay conditions. K562 cells originate from a pleural effusion of a patient with chronic myelogenous leukemia (CML) in blast crisis and are characterized by the expression of the BCR-ABL oncogene.
Fig. 1.
Expression of Bcr-abl increased SMS activity. A: In vitro SMS activity was measured in a panel of exponentially growing leukemic cell lines as described in the Materials and Methods section. The SMS activity was normalized to that of HL-60 cells. B: In vitro SMS activity was measured in exponentially growing HL-60 cells stably expressing the BCR-ABL oncogene (p185) (HL-60 Bcr-abl) and control cells carrying the empty vector (HL-60 neo) as described in the Materials and Methods section. C: K562 cells (0.1 × 106 cell/ml) were incubated in the presence of the indicated concentrations of imatinib (Im) or vehicle control (CT) for different periods of time. At the reported time points, 3 × 106 cells were collected for SMS activity. Data represent the average and standard deviation of at least three separate experiments. Where standard deviations are not visible, the standard deviation is too small compared with the scale of the y axis. Asterisk indicates significance to HL-60/neo or to control cells (CT) (P < 0.05).
To determine if the increase of SMS in K562 was a direct result of BCR-ABL expression, SMS activity was measured in HL-60 cells stably transfected with the BCR-ABL gene (HL-60/bcr-abl) and compared with cells transfected with the empty vector (HL-60/neo) (40) (Fig. 1B). It was found that the expression of Bcr-abl in HL-60 cells induced a 9-fold increase of SMS activity but not due to the assay experimental conditions (data not shown). As a complementary approach, the effect of inhibition of Bcr-abl tyrosine kinase activity on SMS activity was determined in K562 cells. Inhibition of Bcr-abl activity by imatinib mesylate induced a dose- and time-dependent inhibition of SMS activity, which became significant by 24 h of incubation and continued through 36 h of treatment (Fig. 1C). The inhibition of SMS activity by imatinib at 24 h and 30 h was observed when no significant effect of the drug on cell proliferation could be appreciated. Inhibition of cell proliferation by imatinib started at 36 h of treatment with all concentrations tested (supplementary Fig. I), and it was accompanied by the presence of 3.7% of dead cells at 0.2 μM, 8.3% at 0.4 μM, and 10% at 1 μM (vs. 2% of dead cells in the vehicle control sample). Together, these results suggest that, in K562 cells, SMS activity is regulated by the tyrosine kinase activity of Bcr-abl.
Bcr-abl-induced increase of SMS activity is attributable to the up-regulation of Sms1
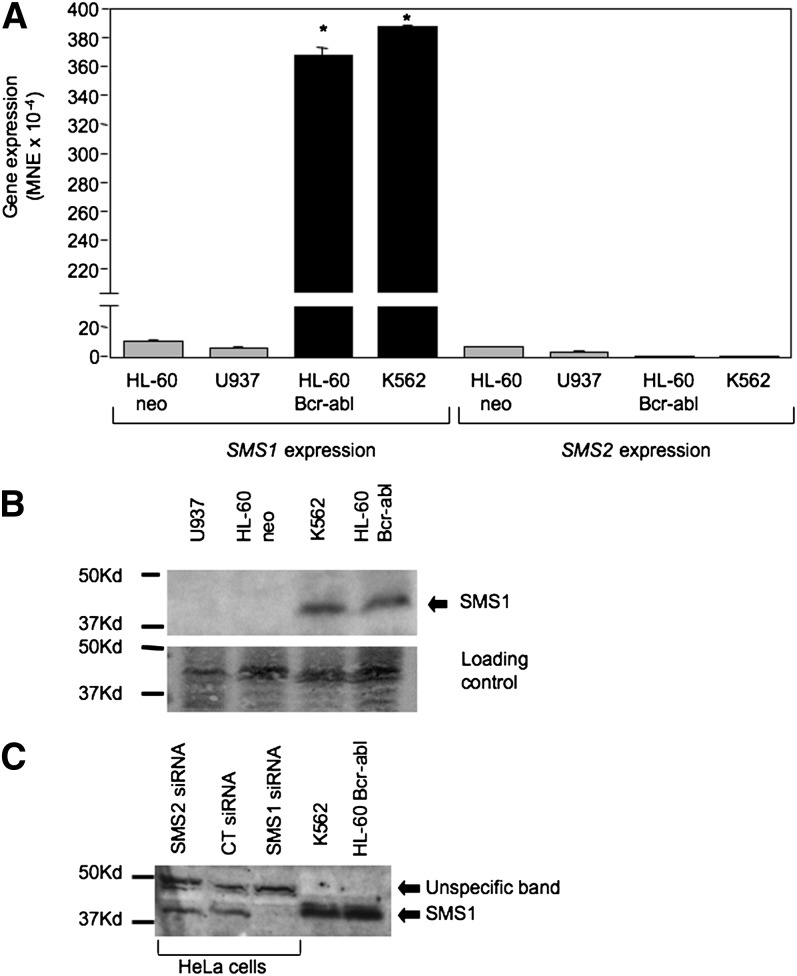
Two SMSs have been identified in mammalian cells. To determine which of these SMSs is responsible for the observed increase in SMS activity, the effect of Bcr-abl on the expression of SMS1 and SMS2 was determined by real-time PCR. The expression of SMS1 mRNA in K562 and HL-60/bcr-abl was significantly higher than that in Bcr-abl-negative cells (HL-60/neo or U937) (Fig. 2A). The results obtained by real-time PCR were also confirmed by Northern blot analysis of total RNA collected from HL-60/neo and HL-60/bcr-abl cells, using the full-length SMS1 cDNA as probe (data not shown). On the other hand, the expression of Bcr-abl caused a reproducible and significant drop of SMS2 mRNA level (Fig. 2A).The absolute expression level of SMS2 in HL-60/neo is significantly lower than that of SMS1 (1.32 × 10−3 vs. 6.24 × 10−4 MNE), and, by dropping in Bcr-abl-positive cells, it became minimal.
Fig. 2.
Bcr-abl induced up-regulation of Sms1. A: Expression of BCR-ABL (HL-60/Bcr-abl and K562) caused an approximately 35-fold increase of SMS1 expression compared with Bcr-abl-negative cells (HL-60 neo and U937), whereas it caused a drop of SMS2 expression compared with Bcr-abl-negative cells. Expression of SMS1 and SMS2 were determined by real-time PCR as indicated in the Materials and Methods section and normalized against β-actin. B: Total membrane fractions were collected from exponentially growing cells, and Western blotting analysis using 80 μg of proteins was performed as described in the Materials and Methods section using nonpurified polyclonal antibodies raised against recombinant full-length Sms1 protein (ExAlpha Biologicals). Equal loading was confirmed by Ponceau staining (loading control panel). C: The identity of the Sms1 band was determined based on the specific disappearance of the same molecular weight band in Hela cells after siRNA-mediated SMS1 down-regulation as compared with cells transfected with a scrambled siRNA (CT siRNA) or a siRNA against SMS2 (SMS2 siRNA). These results are representative of two separate experiments. Where error bars are not visible, the error bar is too small compared with the scale of the y axis. Asterisk indicates significance to HL-60/neo (P < 0.05). MNE, mean of normalized expression.
To confirm that increased expression of SMS1 in Bcr-abl-positive cells led to elevated Sms1 protein, Western blotting was performed on the total membrane fraction isolated from Bcr-abl-negative and -positive cell lines. As shown in Fig. 2B and in line with the expression pattern (Fig. 2A), the levels of Sms1 in Bcr-abl-negative cells were barely detectable, whereas they were significantly elevated in bcr-abl-positive cells (K562 and HL-60/bcr-abl). The identification of the Sms1 band in leukemia cells was based on the comparison with Hela cells treated with SMS1 siRNA (Fig. 2C). On the other hand, no commercially available antibodies for Sms2 (ExAlpha or Santa Cruz) were able to detect any band in any of these cell lines, whereas they were able to detect Sms2 in Hela cells; data not shown. Together, these data suggest that Sms1 may indeed be responsible for the increase in SMS activity observed in Bcr-abl-positive cells.
To prove that Bcr-abl regulates SMS activity by promoting SMS1 expression and activity, SMS1 was down-regulated by transient treatment with siRNA in K562 cells. Down-regulation was achieved by electroporation of SMS1-specific siRNA followed by both expression and activity analysis at various time points. Down-regulation of SMS1 caused a significant reduction of the expression of SMS1 after 24 h (Fig. 3A) , and the SMS1 down-regulation caused a 50–60% of total SMS activity (Fig. 3B). The SMS1 siRNA showed target specificity because no change of SMS2 expression was observed (Fig. 3C). Similarly, treatment of K562 cells with a validated SMS2 siRNA (23) caused the already minimal SMS2 expression to become barely detectable (at 48 h after siRNA, SMS2 expression went from 5.1 × 10−6 ± 7.9 × 10−7 to 1.8 × 10−6 ± 1.9 × 10−7 MNE) while not affecting SMS1 expression (data not shown). In contrast to SMS1, down-regulation of SMS2 did not affect SMS activity (Fig. 3D). Together, these observations demonstrate that Sms1 is the main contributor to the Bcr-abl-induced increase of SMS activity.
Fig. 3.
Sms1 is the main contributor to SMS activity in K562 cells. SMS1 (SMS1-) and SMS2 (SMS2-) were down-regulated in K562 cells by treatment with siRNA (40 nM up to 72 h). Treatment with SMS1 siRNA significantly decreased SMS1 expression as measured by real-time PCR (A) without significantly affecting SMS2 expression (C). The siRNA-induced SMS1 decrease caused a decrease of total in vitro SMS activity (B). SMS2 siRNA did not affect in vitro SMS activity (D). These results are representative of at least three separate experiments. AU, arbitrary fluorescence units; MNE, mean of normalized expression. Asterisk indicates significance to SCR control (P < 0.05).
Inhibition of SMS activity caused accumulation of ceramide and a decrease of DAG
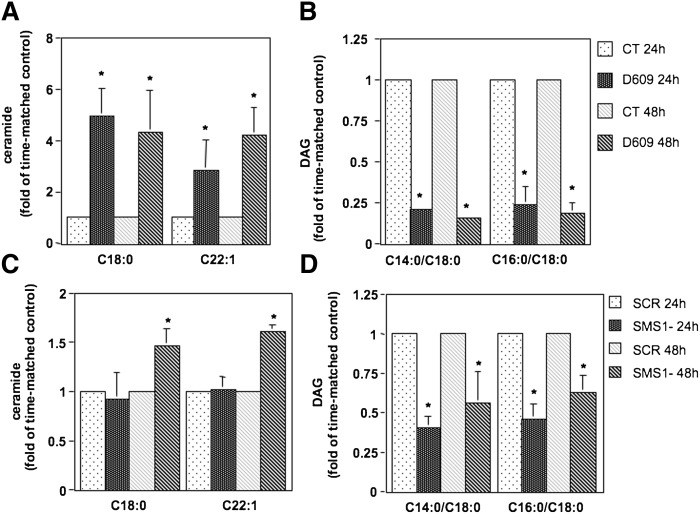
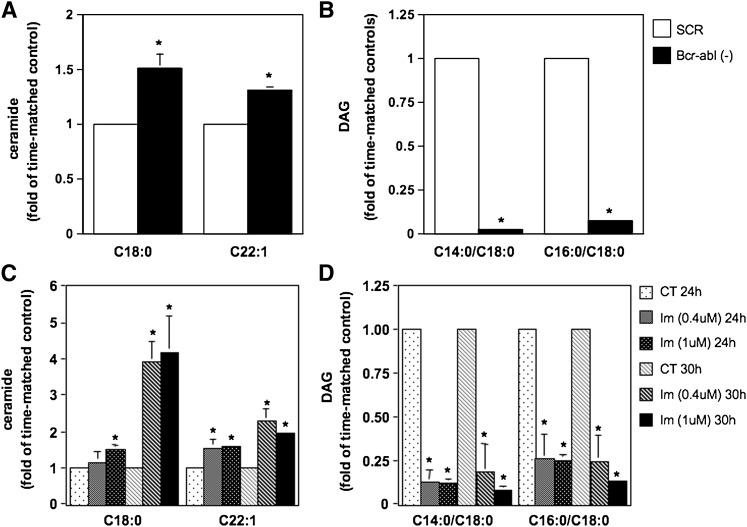
Because SMS promotes the formation of DAG by consuming ceramide and DAG sustains cell proliferation (whereas ceramide has generally a negative effect on cell growth) (43–53), lipid analysis upon modulation of SMS activity was conducted to determine its impact on the levels of these bioactive lipids. To this aim, lipid levels were determined upon inhibition of SMS activity by treatment with the SMS pharmacological inhibitor D609 (50 μg/ml) (Fig. 4A, B) or siRNA-mediated down-regulation of SMS1 (Fig. 4C, D) in K562 cells. Cells for each experimental condition were collected after 24 h and 48 h of treatment. Because the basal levels of the lipids (for these and all subsequent lipid measurements) were variable among separate experiments (whereas the pattern of changes for each experiment was similar as compared with other repeat experiments), results are reported as fold of time-matched controls. Absolute values of all detected ceramide, SM, and DAG molecular species for a representative experiment are provided in supplementary Tables I and II. Pharmacological inhibition of SMS induced a significant accumulation of representative ceramide species C18:0 and C22:1 at 24 h and 48 h of treatment (Fig. 4A). On the other hand, the levels of representative DAG species C14:0/C18:0 and C16:0/C18:0 significantly decreased upon inhibition of SMS after 24 h and 48 h of treatment (Fig. 4B). The overall lipid changes cause a shift of the ratio between total ceramide levels over total DAG levels from 0.11 of control cells to 0.45 of D609-treated cells at 24 h and from 0.09 of control cells to 0.50 of D609-treated cells at 48 h (supplementary Table I). Similarly, although in a less pronounced manner, C18:0 and C22:1 ceramide levels accumulated after 48 h of down-regulation of SMS1 (Fig. 4C), whereas C14:0/C18:0 and C16:0/C18:0 DAG levels decreased significantly after 24 h (Fig. 4D). In this case, the ratio between total ceramide levels over total DAG levels goes from 0.17 of control cells to 0.26 of SMS1 cells after 24 h and from 0.46 to 0.64 after 48 h (supplementary Table II).
Fig. 4.
Effect of inhibition of SMS on ceramide and DAG levels. K562 cells were treated with D609 (50 μg/ml) (A and B) or with control siRNA (SCR) or SMS1 siRNA (SMS1-) (C and D) for 24 h and 48 h. One million cells were collected for each time point and experimental condition and analyzed by mass spectrometry for ceramide (A and C) and DAG (B and D) levels and species by the Lipidomics Core Facility at MUSC. Reported in the graphs are representative ceramide and DAG species. Levels are reported as fold changes versus time-matched control because of the large variation in baseline values. A: C18:0 ceramide [pmoles/nmole Pi], CT 24 h: 0.023 ± 0.013, CT 48 h: 0.015 ± 0.004; C22:1 ceramide (pmoles/nmole Pi): CT 24 h: 0.026 ± 0.016, CT 48 h: 0.02 ± 0.005. B: C14:0/C18:0 DAG (pmoles/nmole Pi): CT 24 h: 0.14 ± 0.1, CT 48 h: 0.36 ± 0.06; C16:0/C18:0 DAG (pmoles/nmole Pi): CT 24 h: 2.57 ± 1.62, CT 48 h: 4.16 ± 0.47. C: C18:0 ceramide (pmoles/nmole Pi), CT 24 h: 0.11 ± 0.05, CT 48 h: 0.13 ± 0.01; C22:1 ceramide (pmoles/nmole Pi): CT 24 h: 0.037 ± 0.009, CT 48 h: 0.07 ± 0.003. D: C14:0/C18:0 DAG (pmoles/nmole Pi): CT 24 h: 0.54 ± 0.22, CT 48 h: 0.084 ± 0.033; C16:0/C18:0 DAG (pmoles/nmole Pi): CT 24 h: 8.91 ± 3.43, CT 48 h: 2.27 ± 0.71. Where standard deviations are not visible, the standard deviation is too small compared with the scale of the y axis. CT, vehicle control. Asterisk indicates significance to time-matched control (P < 0.05).
Together, these results indicate that inhibition of SMS/Sms1 induced changes in some ceramide and DAG molecular species that shift the ceramide/DAG ratio in favor of ceramide.
Modulation of Bcr-abl expression and activity caused lipid changes consistent with downstream regulation of Sms1
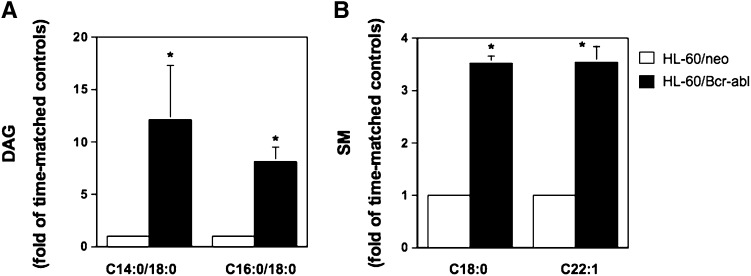
If Bcr-abl promotes SMS1 expression and activity, then modulation of Bcr-abl activity should cause lipid changes consistent with changes in SMS activity. To investigate this relationship, we first determined the effect of Bcr-abl expression on basal levels of DAG, ceramide, and SM by mass spectrometry. HL-60/neo and HL-60/Bcr-abl cells (Fig. 5) were grown under standard conditions and collected once the cells reached the logarithmic phase of growth. Cells with similar passage numbers were used to control for lipid changes resulting from cell culture aging, potentially disguising the lipid changes specifically mediated by Bcr-abl expression. The levels of several DAG and SM species were increased in HL-60/Bcr-abl cells as compared with HL-60/neo (Fig. 5 and supplementary Table III). In particular, the representative DAG species that decreased upon down-regulation of SMS1, C14:0/C16:0, and C16:0/C18:0 were also elevated in BCR-ABL-expressing cells (Fig. 5A). Moreover, the majority of SM species were significantly elevated in HL-60/Bcr-abl cells as compared with HL-60/neo (Fig. 5B and supplementary Table IIIC). This is in agreement with the enhanced SMS activity observed in HL-60/Bcr-abl cells as compared with HL-60/neo (Fig. 1B). BCR-ABL expression did not decrease ceramide levels (supplementary Table IIIA).
Fig. 5.
Effect of overexpression of Bcr-abl in HL-60 cells on the lipids regulated by Sms1. Two million HL-60/Bcr-abl or HL-60/neo cells of similar passage were allowed to reach the logarithmic phase of growth and were harvested at the same time for lipid analysis. The levels and species of DAG (A) and SM (B) were analyzed from two independent experiments by mass spectrometry at the Lipidomics Core Facility at MUSC. Reported in the graphs are representative DAG and SM species. Levels are reported as fold change versus time-matched control (HL-60/neo) because of the large variation in baseline values. A: C14:0/C18:0 DAG (pmoles/nmole Pi): HL-60/neo: 2.22 ± 1.08; HL-60/Bcr-abl: 23.32 ± 7.01; C16:0/C18:0 DAG (pmoles/nmole Pi): HL-60/neo: 0.96 ± 0.31, HL-60/Bcr-abl: 7.61 ± 2.01. B: C18:0 SM (pmoles/nmole Pi): HL-60/neo: 1.27 ± 0.21; HL-60/Bcr-abl: 4.99 ± 0.18; C22:1 SM (pmoles/nmole Pi): HL-60/neo: 0.98 ± 0.14, HL-60/Bcr-abl: 3.55 ± 0.14. Asterisk indicates significant difference (P < 0.05) of HL-60/Bcr-abl versus control cells (HL-60/neo) once the fold of time-matched control was calculated.
In a complementary approach, the effect of siRNA-mediated down-regulation of BCR-ABL was determined on SMS-regulated lipids. The amount of siRNA, time of incubation, and cell culture conditions were optimized in preliminary experiments, and conditions were selected on the basis of maximum BCR-ABL down-regulation, as measured by Western blotting, and decreased SMS activity (data not shown). HL-60/bcr-abl cells were treated with siRNA targeting BCR-ABL (Bcr-abl−) or with the scrambled siRNA Allstar control (SCR), and cells from each experimental condition were harvested 36 h after siRNA treatment for lipid analysis. Absolute lipid levels from one representative experiment are reported in supplementary Table IV, and representative lipid species are reported in Fig. 6. These results showed that reduction of BCR-ABL expression induced a significant increase of the levels of representative ceramide species C18:0 and C22:1 and a decrease of representative DAG species C14:0/C18:0 and C16:0/C18:0 (Fig. 6A, B and supplementary Table IV). These changes resulted in an overall shift of the total ceramide over total DAG ratio from 0.34 of control cells to 1.34 of BCR-ABL down-regulated cells. Furthermore, Bcr-abl inhibition resulted in a significant decrease of total SM levels because the levels of several molecular species were affected (supplementary Table 4C).
Fig. 6.
Inhibition of Bcr-abl induced accumulation of ceramide and reduction of DAG levels. A and B: HL-60/Bcr-abl cells were treated with scrambled siRNA control (SCR) or BCR-ABL siRNA (Bcr-abl-) for 36 h. Two million cells were collected from each experimental condition, and the levels and species of ceramide (A) and DAG (B) were analyzed from three independent experiments. C and D: K562 cells were treated with imatinib (Im, 0.4 or 1 μM) for 24 h and 30 h, and the levels of different species of ceramide (C) and DAG (D) were measured by mass spectrometry. Levels are reported as fold changes versus time-matched control (SCR or CT) because of the large variation in baseline values. A: C18:0 ceramide (pmoles/nmole Pi), SCR: 0.069 ± 0.009; C22:1 ceramide (pmoles/nmole Pi): SCR: 0.11 ± 0.004. B: C14:0/C18:0 DAG (pmoles/nmole Pi): SCR: 0.526 ± 0.027; C16:0/C18:0 DAG (pmoles/nmole Pi): SCR: 7.04 ± 0.24. C: C18:0 ceramide (pmoles/nmole Pi), CT 24 h: 0.043 ± 0.006, CT 30 h: 0.031 ± 0.001; C22:1 ceramide (pmoles/nmole Pi): CT 24 h: 0.083 ± 0.030, CT 30 h: 0.092 ± 0.017. D: C14:0/C18:0 DAG (pmoles/nmole Pi): CT 24 h: 0.28 ± 0.067, CT 30 h: 0.60 ± 0.073; C16:0/C18:0 DAG (pmoles/nmole Pi): CT 24 h: 4.87 ± 1.023, CT 30 h: 9.66 ± 1.35. Where standard deviations are not visible, the standard deviation is too small compared with the scale of the y axis. Asterisk indicates significant difference between HL-60/Bcr-abl(-) and SCR control or imatinib-treated and control cells (P < 0.05). CT, control; Im, imatinib-treated cells. Reported in the graphs are representative ceramide and DAG species.
The effect of loss of active Bcr-abl on DAG, ceramide, and SM levels was also determined in K562 cells after pharmacological inhibition with imatinib. K562 cells were treated with 0.4 μM and 1 μM imatinib for 24 h and 30 h (concentrations of imatinib and time of incubation that effectively inhibited SMS activity before affecting cell proliferation; supplementary Fig. 1). Cells for each experimental condition were collected, and lipids were extracted for mass spectrometry analysis. Accumulation of the representative C18:0 and C22:1 ceramide species was observed after 24 h and in a more pronounced manner after 30 h of treatment with both concentrations (Fig. 6C). On the other hand, C14:0/C18:0 and C16:0/C18:0 DAG levels were greatly suppressed after 24 h of treatment with both imatinib concentrations, and they remained low after 30 h of incubation. In the presence of 1 μM imatinib, the reported lipid changes cause a shift of the ratio between total ceramide over total DAG from 0.64 of control cells to 1.21 of imatinib-treated cells after 24 h and from 0.34 to 1.03 after 48 h. Absolute lipid values from one representative experiment are reported in supplementary Table 5. Together, these results are consistent with modulation of SMS activity by Bcr-abl.
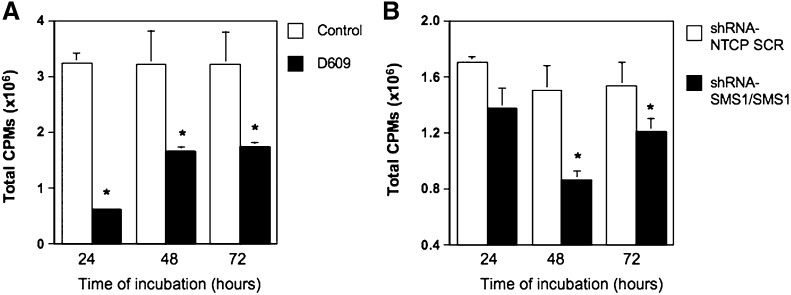
Inhibition of Sms1 caused a significant defect in cell growth of Bcr-abl-positive cells
Because down-regulation of SMS1 caused a decrease of DAG and an increase of ceramide and because DAG and ceramide are involved in the regulation of cellular proliferation and/or cell death (43–53), the effect of Sms1 inhibition was determined on these cellular processes. K562 cells were first treated with D609, and its effect on cell proliferation was tested by incorporation of tritiated thymidine (Fig. 7A). A significant inhibitory effect of D609 was observed after 24 h, and it continued after 48 h and 72 h of incubation. Next, the effect of specific down-regulation of SMS1 was determined. Initially, transient down-regulation of SMS1 with siRNA delivered variable results depending on the extent of SMS1 reduction (inhibition of cellular proliferation was only observed in the case of suppression of SMS1 by more than 75%). We reasoned that this might be due to the high basal SMS1 expression in these cells and to a possible threshold effect. Indeed, in K562 cells, the levels of SMS1 expression after SMS1 siRNA-mediated knock down (equal approximately to one third of basal SMS1 expression [Fig. 3A] and corresponding to approximately one third of SMS activity [Fig. 3B]) were still significantly higher than those of bcr-abl-negative cells (SMS1 expression in HL-60 is ∼1/35th of K562 cells [Fig. 2A], and SMS activity in HL-60 cells is ∼1/10 of SMS activity in K562 [Fig. 1A]). Thus, to investigate the biological effect of SMS1 in K562 cells and to obtain a more reproducible effect on cell proliferation, we aimed at achieving down-regulation of SMS1 that would approximate as much as possible the levels of bcr-abl-negative cells (HL-60 neo or U937 cells). To this aim, K562 cells were first transduced with viral particles containing short hairpin RNA targeting SMS1 (shRNA-SMS1) followed by selection of stably expressing cells (this ensured a basal 50% reduction of SMS1 expression [supplementary Fig. II). To further down-regulate Sms1, K562 cells stably transduced with shRNA-SMS1 were transfected with SMS1 siRNA (shRNA-SMS1/SMS1), and those transduced with control shRNA (shRNA-NTCP) were transiently transfected with Allstar Scrambled siRNA (shRNA-NTCP/SCR) (supplementary Fig. II). After double SMS1 down-regulation (shRNA-SMS1/SMS1), SMS1 was approximately one tenth of the expression in control shRNA-NTCP/SCR cells (90% down-regulation). Sustained inhibition of SMS1 in K562 cells (shRNA-SMS1/SMS1) resulted in reduced [3H]thymidine incorporation as compared with control cells (shRNA-NTCP/SCR) at 24 h and significantly after 48 h and 72 h (Fig. 7B). On the other hand, inhibition of SMS1 expression by more than 75% in HL-60/neo cells did not affect their cell growth (supplementary Fig. III). These results suggest that sustained inhibition of SMS1 expression/activity in K562 cells significantly impairs cell proliferation.
Fig. 7.
Inhibition of SMS activity in K562 cells significantly reduced cell proliferation. Changes in proliferation after pharmacological (A) or siRNA-mediated inhibition of Sms1 (B) were measured by 3H-thymidine incorporation after 24, 48, and 72 h. A: 3H-Thymidine incorporation was determined after inhibition of total SMS with D609 (50 μg/ml). B: Cells were transduced with lentivirus containing control particles (shRNA-NTCP) or viral particles containing sequences to induce partial stable down-regulation of SMS1 (shRNA-SMS1). 3H-thymidine incorporation was then measured in shRNA-NTCP transiently transfected with Allstar scrambled siRNA control (shRNA-NTCP/SCR) and shRNA-SMS1 cells transiently transfected with SMS1 siRNA. Results represent the average and standard deviation of a minimum of four separate experiments. Where standard deviations are not visible, the standard deviation is too small compared with the scale of the y axis. Asterisk indicates significant difference versus control or shRNA-NTCP/SCR (P < 0.05). CPMs, counts per minute; NTCP, nontransducing control particles; ShRNA, short-hairpin RNA.
On the other hand, down-regulation of SMS1 did not cause cytotoxicity or apoptosis. In fact, no significant increase in trypan blue-positive cells and no positivity to propidium iodide were detected by fluorescence-activated cell sorter analysis (data not shown). Moreover, no evidence of caspase-induced apoptosis was observed in K562 cells upon treatment with D609 or after down-regulation of SMS1 (supplementary and data not shown). Together, these results implicate Sms1 in specific regulation of cell proliferation, and not cell death, of Bcr-Abl-positive cells.
DISCUSSION
In this study, we provide evidence of a link between the expression of the Bcr-abl oncogene, its activity, and elevated SMS activity. We further show that enhanced expression of SMS1 and not SMS2 is responsible for the elevated SMS activity and that elevated Sms1 is important for sustaining proliferation of Bcr-abl-positive cells. Moreover, modulation of Bcr-abl, SMS activity, or SMS1 expression results in a similar alteration of the ratio of the bioactive lipids ceramide and DAG. Finally, inhibition of SMS1 significantly affects cell proliferation of Bcr-abl-positive cells, consistent with the shift of the ceramide to DAG ratio in favor of ceramide.
These results are significant because no molecular factors that are able to stimulate Sms1 or Sms2 expression/activity have been identified. In particular, increased SMS activity has been associated with some transformed phenotypes (8, 11, 25); thus, the regulation of Sms1 by Bcr-abl reported herein represents the first molecular link between a well-known oncogene and an SMS to support that association.
The involvement of Sms1 as a potential downstream target of Bcr-abl is unknown, and in general the role of ceramide in Bcr-abl-mediated signaling is unclear. In particular, whereas Bcr-abl expression was shown to impart total or partial resistance to treatment with exogenous short-chain ceramide analogs (C2 or C6 ceramide), Bcr-abl was also found to accelerate ceramide-mediated apoptosis by these same analogs (40, 54, 55). Recently, accumulation of endogenous ceramide and a decrease of the antiapoptotic sphingolipid sphingosine 1 phosphate (S1P) were shown to be in part responsible for the cytotoxic effect exerted by imatinib treatment in CML-derived cell lines (56). It was also shown that a shift of the balance between endogenous ceramide and S1P in favor of S1P might be of critical relevance for imparting imatinib resistance to CML cells (56, 57). In our experimental model, we observed similar changes in ceramide levels after treatment with imatinib as those reported in the literature (56), and these lipid changes were also observed in the case of pharmacological or siRNA-mediated inhibition of SMS1. Together with the fact that inhibition of Bcr-abl causes a significant block of SMS activity (Fig. 1C), this suggests that, at least in part, ceramide accumulation upon inhibition of Bcr-abl might be due to inhibition of Sms1 activity. Down-regulation of SMS1 induced accumulation of specific ceramide molecular species such as C16:0, C18:0, C20:0, and C22:1, which were also accumulated upon pharmacological inhibition with D609 (supplementary Tables IA and IIA). D609 treatment also caused the accumulation of other ceramide species, and this may be due to the more rapid and sustained inhibition of SMS activity compared with siRNA and/or the activation of other ceramide-generating pathways, such as the de novo pathway (58). C18:0, C20:0, and C22:1 ceramides were also increased upon pharmacological inhibition of Bcr-abl, and this is consistent with Sms1 being a downstream target of Bcr-abl. Also in the case of Bcr-abl inhibition, other ceramide species were increased, and this may be the result of activation of certain ceramide synthases, such as CerS1 (56).
The fact that ceramide accumulation upon inhibition of Bcr-abl might be in part due to Sms1 inhibition is also supported by the analysis of SM and DAG levels. Indeed, we found that, upon inhibition of Bcr-abl or SMS/Sms1, there was a tendency for several species of SM to decrease, although some changes were not statistically significant (supplementary Tables IC, IIC, and VC). Additionally, overexpression of Bcr-abl in HL-60 cells induced a significant accumulation of various species of SM (supplementary Table IIIC), and siRNA-mediated down-regulation of the oncogene significantly lowered SM levels (supplementary Table IVC).
Inhibition of Bcr-abl or SMS/Sms1 induced a significant drop of several DAG species, and these changes were even more pronounced than the changes in ceramide levels (supplementary Tables IB, IIB, and IIIB). Pharmacological and siRNA-mediated inhibition of SMS1 induced a significant decrease of specific DAG molecular species, such as low abundant C14:0/C16:0 and C14:0/18:0 DAGs and highly represented C16:0/18:0, C16:0/18:1, C18:0/18:1, and Di-C16:0 DAGs. These same species were decreased upon inhibition of Bcr-abl (supplementary Table VB), suggesting that there might be a specific pattern of DAG changes induced by modulation of SMS1. These results were also observed upon siRNA mediated down-regulation of BCR-ABL in HL-60/bcr-abl cells (supplementary Table IVB).
The relationship between Bcr-abl and DAG has not been documented in the literature. Indirect evidence of the involvement of DAG as a downstream signaling molecule of Bcr-abl comes from reports showing that activation of phospholipase C (PLC) γ1 and of the PI3K/Akt pathway is required for Bcr-abl signaling through mTOR/p70S6-kinase (59, 60). The regulation of mTOR/p70S6-kinase by PLCγ1 seems to occur via modulation of calcium signaling (CaMKII) (presumably through formation of IP3) and classical and novel protein kinase C (likely through production of DAG). In our system, we observed a very significant drop of DAG levels when Bcr-abl or SMS/Sms1 was inhibited. Being Sms1 regulated by Bcr-abl, it is possible that part of the observed drop of DAG levels after inhibition of Bcr-abl is due to the decrease of SMS/Sms1 activity, in addition to inhibition of PLCγ1.
The changes of ceramide (accumulation) and DAG (decrease) levels observed upon inhibition of Bcr-abl or SMS/Sms1 are consistent with the inhibition of cell proliferation observed in response to these treatments, and these concomitant lipid changes may work in concert to cause this phenotype. Down-regulation of SMS1 did not cause a change in proliferation of the Bcr-abl-negative cell line HL-60 (supplementary Fig. III), indicating a specific role for Sms1 in supporting proliferation of Bcr-abl-positive K562 cells. The difference in the requirement of SMS1 expression for proliferation between Bcr-abl-positive and -negative cells may be linked to the expression of SMS2 and the ability of Sms2 to compensate for the loss of Sms1. In fact, SMS2 is barely detectable in Bcr-abl-positive K562 but is expressed in HL-60 to a similar level as SMS1 (Fig. 2A). This scenario implies that whenever Sms1 is the only SMS expressed in a certain cell type, then its activity is crucial for the survival of those cells.
An interesting observation is the fact that SMS1 mRNA is increased in Bcr-abl-positive cells, opening up the question of the mechanism by which SMS1 expression is stimulated by Bcr-abl (transcriptional regulation or mRNA stability). Preliminary results support the first mechanism; thus, in the near future it is our intention to characterize the transcriptional regulation of SMS1 by Bcr-abl to elucidate the signaling pathway, down-stream of Bcr-abl, responsible for up-regulation of SMS1.
Even if K562 cells are blast phase CML, and therefore characterized by a block of differentiation potentially due to the accumulation of mutations in addition to the formation of the Bcr-abl chimaera, Sms1 activity is still under the control of the Bcr-abl tyrosine kinase activity. Therefore, it is unlikely that Sms1 represents a “druggable” target to contain newly diagnosed blast-phase CML. On the other hand, in spite of the success obtained in controlling the chronic phase of CML, the increase in resistance to Bcr-abl inhibitors is starting to represent a significant medical challenge. Thus, establishing SMS1 as an important down-stream regulator of Bcr-abl may provide the basis for future work set to test Sms1 as a potential novel pharmacological target for CML, in particular for cases that arise from resistance to bcr-abl inhibitors.
In summary, the discovery of the Bcr-abl/Sms1 connection provides the first molecularly characterized model for a better understanding of potential modes of regulation and downstream functions of the elusive Sms1.
Supplementary Material
Acknowledgments
The authors thank Drs. Yusuf Hannun, Lina Obeid, and Maurizio Del Poeta for enthusiastic support and critical discussion.
Footnotes
Abbreviations:
- CML
- chronic myelogenous leukemia
- DAG
- diacylglycerol
- PARP
- poly (ADP-ribose) polymerase
- PLC
- phospholipase C
- S1P
- sphingosine-1-phosphate
- SMS
- sphingomyelin synthase
This work was supported in part by a Hollings Cancer Center/Medical University of South Carolina Department of Defense grant “Translational Research on Cancer Control and Related Therapy” (Subcontract GC-3319-05-4498CM), by the National Institutes of Health Grant P20 RR-017677 from the National Center for Research Resources, and by Grant IRG 97-219-08 from the American Cancer Society (C.L.), and by a scholarship from the Abney Foundation (T.A.B.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies. Tara A. Burns is supported by the Graduate Assistance in Areas of National Need (GAANN) training grant in “Lipidology and New Technologies” (to Dr. Maurizio Del Poeta) from the US Department of Education.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of seven figures.
REFERENCES
- 1.Bernert J. T., Ullman M. D. 1981. Biosynthesis of sphingomyelin from erythro-ceramides and phosphatidylcholine by a microsomal cholinephosphoesterase. Biochim. Biophys. Acta. 666: 99–109 [DOI] [PubMed] [Google Scholar]
- 2.Hatch G. M., Vance D. E. 1992. Stimulation of sphingomyelin biosynthesis by brefeldin A and sphingomyelin breakdown by okadaic acid treatment of rat hepatocytes. J. Biol. Chem. 267: 12443–12451 [PubMed] [Google Scholar]
- 3.Marggraf W. D., Anderer F. A., Kanfer J. 1981. The formation of sphingomyelin from phosphatidylcholine in plasma membrane preparations from mouse fibroblasts. Biochim. Biophys. Acta. 664: 61–73 [DOI] [PubMed] [Google Scholar]
- 4.Marggraf W. D., Zertani R., Anderer F. A., Kanfer J. N. 1982. The role of endogenous phosphatidylcholine and ceramide in the biosynthesis of sphingomyelin in mouse fibroblasts. Biochim. Biophys. Acta. 710: 314–323 [DOI] [PubMed] [Google Scholar]
- 5.Merrill A. H., Jr, Jones D. D. 1990. An update of the enzymology and regulation of sphingomyelin metabolism. Biochim.Biophys.Acta Lipids Lipid Metab. 1044: 1–12 [DOI] [PubMed] [Google Scholar]
- 6.Ullman M. D., Radin N. S. 1974. The enzymatic formation of sphingomyelin from ceramide and lecithin in mouse liver. J. Biol. Chem. 249: 1506–1512 [PubMed] [Google Scholar]
- 7.Voelker D. R. K., Kennedy E. P. 1982. Cellular and enzymic synthesis of sphingomyelin. Biochemistry. 21: 2753–2759 [DOI] [PubMed] [Google Scholar]
- 8.Luberto C., Hannun Y. A. 1998. Sphingomyelin synthase, a potential regulator of intracellular levels of ceramide and diacylglycerol during SV40 transformation. Does sphingomyelin synthase account for the putative phosphatidylcholine-phospholipase C? J. Biol. Chem. 273: 14550–14559 [DOI] [PubMed] [Google Scholar]
- 9.Miro-Obradors M.-J., Osada J., Aylagas H., Sanchez-Vegazo I., Palacios-Alaiz E. 1993. Microsomal sphingomyelin accumulation in thioacetamide-injured regenerating rat liver: involvement of sphingomyelin synthase activity. Carcinogenesis. 14: 941–946 [DOI] [PubMed] [Google Scholar]
- 10.Riboni L., Viani P., Bassi R., Giussani P., Tettamanti G. 2001. Basic fibroblast growth factor-induced proliferation of primary astrocytes. evidence for the involvement of sphingomyelin biosynthesis. J. Biol. Chem. 276: 12797–12804 [DOI] [PubMed] [Google Scholar]
- 11.van den Hill A., van Heusden G. P., Wirtz K. W. 1985. The synthesis of sphingomyelin in the Morris hepatomas 7777 and 5123D is restricted to the plasma membrane. Biochim. Biophys. Acta. 833: 354–357 [DOI] [PubMed] [Google Scholar]
- 12.Bourteele S., Hausser A., Doppler H., Horn-Muller J., Ropke C., Schwarzmann G., Pfizenmaier K., Muller G. 1998. Tumor necrosis factor induces ceramide oscillations and negatively controls sphingolipid synthases by caspases in apoptotic Kym-1 cells. J. Biol. Chem. 273: 31245–31251 [DOI] [PubMed] [Google Scholar]
- 13.Watanabe M., Kitano T., Kondo T., Yabu T., Taguchi Y., Tashima M., Umehara H., Domae N., Uchiyama T., Okazaki T. 2004. Increase of nuclear ceramide through caspase-3-dependent regulation of the “sphingomyelin cycle” in Fas-induced apoptosis. Cancer Res. 64: 1000–1007 [DOI] [PubMed] [Google Scholar]
- 14.Huitema K., Van Den Dikkenberg J., Brouwers J. F., Holthuis J. C. 2004. Identification of a family of animal sphingomyelin synthases. EMBO J. 23: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaoka S., Miyaji M., Kitano T., Umehara H., Okazaki T. 2004. Expression cloning of a human cDNA restoring sphingomyelin synthesis and cell growth in sphingomyelin synthase-defective lymphoid cells. J. Biol. Chem. 279: 18688–18693 [DOI] [PubMed] [Google Scholar]
- 16.Villani M., Subathra M., Im Y. B., Choi Y., Signorelli P., Del Poeta M., Luberto C. 2008. Sphingomyelin synthases regulate production of diacylglycerol at the Golgi. Biochem. J. 414: 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeang C., Varshney S., Wang R., Zhang Y., Ye D., Jiang X. C. 2008. The domain responsible for sphingomyelin synthase (SMS) activity. Biochim. Biophys. Acta. 1781: 610–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tani M., Kuge O. 2009. Sphingomyelin synthase 2 is palmitoylated at the COOH-terminal tail, which is involved in its localization in plasma membranes. Biochem. Biophys. Res. Commun. 381: 328–332 [DOI] [PubMed] [Google Scholar]
- 19.Separovic D., Hanada K., Maitah M. Y., Nagy B., Hang I., Tainsky M. A., Kraniak J. M., Bielawski J. 2007. Sphingomyelin synthase 1 suppresses ceramide production and apoptosis post-photodamage. Biochem. Biophys. Res. Commun. 358: 196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Separovic D., Semaan L., Tarca A. L., Awad Maitah M. Y., Hanada K., Bielawski J., Villani M., Luberto C. 2008. Suppression of sphingomyelin synthase 1 by small interference RNA is associated with enhanced ceramide production and apoptosis after photodamage. Exp. Cell Res. 314: 1860–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding T., Li Z., Hailemariam T., Mukherjee S., Maxfield F. R., Wu M. P., Jiang X. C. 2008. SMS overexpression and knockdown: impact on cellular sphingomyelin and diacylglycerol metabolism, and cell apoptosis. J. Lipid Res. 49: 376–385 [DOI] [PubMed] [Google Scholar]
- 22.Tafesse F. G., Huitema K., Hermansson M., van der Poel S., van den Dikkenberg J., Uphoff A., Somerharju P., Holthuis J. C. 2007. Both sphingomyelin synthases SMS1 and SMS2 are required for sphingomyelin homeostasis and growth in human HeLa cells. J. Biol. Chem. 282: 17537–17547 [DOI] [PubMed] [Google Scholar]
- 23.Li Z., Hailemariam T. K., Zhou H., Li Y., Duckworth D. C., Peake D. A., Zhang Y., Kuo M. S., Cao G., Jiang X. C. 2007. Inhibition of sphingomyelin synthase (SMS) affects intracellular sphingomyelin accumulation and plasma membrane lipid organization. Biochim. Biophys. Acta. 1771: 1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lafont E., Milhas D., Carpentier S., Garcia V., Jin Z. X., Umehara H., Okazaki T., Schulze-Osthoff K., Levade T., Benoist H., et al. 2009. Caspase-mediated inhibition of sphingomyelin synthesis is involved in FasL-triggered cell death. Cell Death Differ. 17: 642–654 [DOI] [PubMed] [Google Scholar]
- 25.Itoh M., Kitano T., Watanabe M., Kondo T., Yabu T., Taguchi Y., Iwai K., Tashima M., Uchiyama T., Okazaki T. 2003. Possible role of ceramide as an indicator of chemoresistance: decrease of the ceramide content via activation of glucosylceramide synthase and sphingomyelin synthase in chemoresistant leukemia. Clin. Cancer Res. 9: 415–423 [PubMed] [Google Scholar]
- 26.Wong S., Witte O. N. 2004. The BCR-ABL story: bench to bedside and back. Annu. Rev. Immunol. 22: 247–306 [DOI] [PubMed] [Google Scholar]
- 27.Quintas-Cardama A., Cortes J. 2009. Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood. 113: 1619–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marley S. B., Gordon M. Y. 2005. Chronic myeloid leukaemia: stem cell derived but progenitor cell driven. Clin. Sci. (Lond.). 109: 13–25 [DOI] [PubMed] [Google Scholar]
- 29.Karbasian Esfahani M., Morris E. L., Dutcher J. P., Wiernik P. H. 2006. Blastic phase of chronic myelogenous leukemia. Curr. Treat. Options Oncol. 7: 189–199 [DOI] [PubMed] [Google Scholar]
- 30.Ren R. 2005. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat. Rev. Cancer. 5: 172–183 [DOI] [PubMed] [Google Scholar]
- 31.Deininger M. W., Goldman J. M., Melo J. V. 2000. The molecular biology of chronic myeloid leukemia. Blood. 96: 3343–3356 [PubMed] [Google Scholar]
- 32.Sessions J. 2007. Chronic myeloid leukemia in 2007. Am. J. Health Syst. Pharm. 64: S4–S9 [DOI] [PubMed] [Google Scholar]
- 33.Kantarjian H. M., Cortes J., O'Brien S., Giles F. J., Albitar M., Rios M. B., Shan J., Faderl S., Garcia-Manero G., Thomas D. A., et al. 2002. Imatinib mesylate (STI571) therapy for Philadelphia chromosome-positive chronic myelogenous leukemia in blast phase. Blood. 99: 3547–3553 [DOI] [PubMed] [Google Scholar]
- 34.Lydon N. B., Druker B. J. 2004. Lessons learned from the development of imatinib. Leuk. Res. 28(Suppl 1): S29–S38 [DOI] [PubMed] [Google Scholar]
- 35.Marin D., Marktel S., Bua M., Armstrong L., Goldman J. M., Apperley J. F., Olavarria E. 2002. The use of imatinib (STI571) in chronic myelod leukemia: some practical considerations. Haematologica. 87: 979–988 [PubMed] [Google Scholar]
- 36.Apperley J. F. 2007. Part I: mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 8: 1018–1029 [DOI] [PubMed] [Google Scholar]
- 37.Kantarjian H. M., Talpaz M., Kontoyiannis D., Gutterman J., Keating M. J., Estey E. H., O'Brien S., Rios M. B., Beran M., Deisseroth A. 1992. Treatment of chronic myelogenous leukemia in accelerated and blastic phases with daunorubicin, high-dose cytarabine, and granulocyte-macrophage colony-stimulating factor. J. Clin. Oncol. 10: 398–405 [DOI] [PubMed] [Google Scholar]
- 38.Kantarjian H. M., Walters R. S., Keating M. J., Talpaz M., Andersson B., Beran M., McCredie K. B., Freireich E. J. 1988. Treatment of the blastic phase of chronic myelogenous leukemia with mitoxantrone and high-dose cytosine arabinoside. Cancer. 62: 672–676 [DOI] [PubMed] [Google Scholar]
- 39.Axdorph U., Stenke L., Grimfors G., Carneskog J., Hansen J., Linder O., Ljungman P., Lofvenberg E., Malm C., Simonsson B., et al. 2002. Intensive chemotherapy in patients with chronic myelogenous leukaemia (CML) in accelerated or blastic phase: a report from the Swedish CML Group. Br. J. Haematol. 118: 1048–1054 [DOI] [PubMed] [Google Scholar]
- 40.Amarante-Mendes G. P., Naekyung Kim C., Liu L., Huang Y., Perkins C. L., Green D. R., Bhalla K. 1998. Bcr-Abl exerts its antiapoptotic effect against diverse apoptotic stimuli through blockage of mitochondrial release of cytochrome C and activation of caspase-3. Blood. 91: 1700–1705 [PubMed] [Google Scholar]
- 41.Scherr M., Battmer K., Winkler T., Heidenreich O., Ganser A., Eder M. 2003. Specific inhibition of bcr-abl gene expression by small interfering RNA. Blood. 101: 1566–1569 [DOI] [PubMed] [Google Scholar]
- 42.Ames B. N., Dubin D. T. 1960. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J. Biol. Chem. 235: 769–775 [PubMed] [Google Scholar]
- 43.Kishimoto A., Takai Y., Mori T., Kikkawa U., Nishizuka Y. 1980. Activation of calcium and phospholipid-dependent protein kinase by diacylglycerol, its possible relation to phosphatidylinositol turnover. J. Biol. Chem. 255: 2273–2276 [PubMed] [Google Scholar]
- 44.Takai Y., Kishimoto A., Kikkawa U., Mori T., Nishizuka Y. 1979. Unsaturated diacylglycerol as a possible messenger for the activation of calcium-activated, phospholipid-dependent protein kinase system. Biochem. Biophys. Res. Commun. 91: 1218–1224 [DOI] [PubMed] [Google Scholar]
- 45.Griner E. M., Kazanietz M. G. 2007. Protein kinase C and other diacylglycerol effectors in cancer. Nat. Rev. Cancer. 7: 281–294 [DOI] [PubMed] [Google Scholar]
- 46.Obeid L. M., Linardic C. M., Karolak L. A., Hannun Y. A. 1993. Programmed cell death induced by ceramide. Science. 259: 1769–1771 [DOI] [PubMed] [Google Scholar]
- 47.Senkal C. E., Ponnusamy S., Rossi M. J., Bialewski J., Sinha D., Jiang J. C., Jazwinski S. M., Hannun Y. A., Ogretmen B. 2007. Role of human longevity assurance gene 1 and C18-ceramide in chemotherapy-induced cell death in human head and neck squamous cell carcinomas. Mol. Cancer Ther. 6: 712–722 [DOI] [PubMed] [Google Scholar]
- 48.Marchesini N., Osta W., Bielawski J., Luberto C., Obeid L. M., Hannun Y. A. 2004. Role for mammalian neutral sphingomyelinase 2 in confluence-induced growth arrest of MCF7 cells. J. Biol. Chem. 279: 25101–25111 [DOI] [PubMed] [Google Scholar]
- 49.Kroesen B. J., Jacobs S., Pettus B. J., Sietsma H., Kok J. W., Hannun Y. A., de Leij L. F. 2003. BcR-induced apoptosis involves differential regulation of C16 and C24-ceramide formation and sphingolipid-dependent activation of the proteasome. J. Biol. Chem. 278: 14723–14731 [DOI] [PubMed] [Google Scholar]
- 50.White-Gilbertson S., Mullen T., Senkal C., Lu P., Ogretmen B., Obeid L., Voelkel-Johnson C. 2009. Ceramide synthase 6 modulates TRAIL sensitivity and nuclear translocation of active caspase-3 in colon cancer cells. Oncogene. 28: 1132–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bikman B. T., Summers S. A. 2011. Ceramides as modulators of cellular and whole-body metabolism. J. Clin. Invest. 121: 4222–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henry B., Moller C., Dimanche-Boitrel M. T., Gulbins E., Becker K. A. 2011. Targeting the ceramide system in cancer. Cancer Lett. [DOI] [PubMed] [Google Scholar]
- 53.Mullen T. D., Obeid L. M. 2011. Ceramide and apoptosis: exploring the enigmatic connections between sphingolipid metabolism and programmed cell death. Anticancer. Agents Med. Chem. [DOI] [PubMed] [Google Scholar]
- 54.Maguer-Satta V. 1998. CML and apoptosis: the ceramide pathway. Hematol. Cell Ther. 40: 233–236 [PubMed] [Google Scholar]
- 55.Nica A. F., Tsao C. C., Watt J. C., Jiffar T., Kurinna S., Jurasz P., Konopleva M., Andreeff M., Radomski M. W., Ruvolo P. P. 2008. Ceramide promotes apoptosis in chronic myelogenous leukemia-derived K562 cells by a mechanism involving caspase-8 and JNK. Cell Cycle. 7: 3362–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baran Y., Salas A., Senkal C. E., Gunduz U., Bielawski J., Obeid L. M., Ogretmen B. 2007. Alterations of ceramide/sphingosine 1-phosphate rheostat involved in the regulation of resistance to imatinib-induced apoptosis in K562 human chronic myeloid leukemia cells. J. Biol. Chem. 282: 10922–10934 [DOI] [PubMed] [Google Scholar]
- 57.Bonhoure E., Lauret A., Barnes D. J., Martin C., Malavaud B., Kohama T., Melo J. V., Cuvillier O. 2008. Sphingosine kinase-1 is a downstream regulator of imatinib-induced apoptosis in chronic myeloid leukemia cells. Leukemia. 22: 971–979 [DOI] [PubMed] [Google Scholar]
- 58.Perry R. J., Ridgway N. D. 2004. The role of de novo ceramide synthesis in the mechanism of action of the tricyclic xanthate D609. J. Lipid Res. 45: 164–173 [DOI] [PubMed] [Google Scholar]
- 59.Gotoh A., Miyazawa K., Ohyashiki K., Toyama K. 1994. Potential molecules implicated in downstream signaling pathways of p185BCR-ABL in Ph+ ALL involve GTPase-activating protein, phospholipase C-gamma 1, and phosphatidylinositol 3′-kinase. Leukemia. 8: 115–120 [PubMed] [Google Scholar]
- 60.Markova B., Albers C., Breitenbuecher F., Melo J. V., Brummendorf T. H., Heidel F., Lipka D., Duyster J., Huber C., Fischer T. 2010. Novel pathway in Bcr-Abl signal transduction involves Akt-independent, PLC-gamma1-driven activation of mTOR/p70S6-kinase pathway. Oncogene. 29: 739–751 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.