Abstract
The formation of the atherosclerotic lesion is a complex process influenced by an array of inflammatory and lipid metabolism pathways. We previously demonstrated that NR4A nuclear receptors are highly induced in macrophages in response to inflammatory stimuli and modulate the expression of genes linked to inflammation in vitro. Here we used mouse genetic models to assess the impact of NR4A expression on atherosclerosis development and macrophage polarization. Transplantation of wild-type, Nur77−/−, or Nor1−/− null hematopoetic precursors into LDL receptor (LDLR)−/− recipient mice led to comparable development of atherosclerotic lesions after high-cholesterol diet. We also observed comparable induction of genes linked to M1 and M2 responses in wild-type and Nur77-null macrophages in response to lipopolysaccharides and interleukin (IL)-4, respectively. In contrast, activation of the nuclear receptor liver X receptor (LXR) strongly suppressed M1 responses, and ablation of signal transductor and activator of transcription 6 (STAT6) strongly suppressed M2 responses. Recent studies have suggested that alterations in levels of Ly6Clo monocytes may be a contributor to inflammation and atherosclerosis. In our study, loss of Nur77, but not Nor1, was associated with decreased abundance of Ly6Clo monocytes, but this change was not correlated with atherosclerotic lesion development. Collectively, our results suggest that alterations in the Ly6Clo monocyte population and bone marrow NR4A expression do not play dominant roles in macrophage polarization or the development of atherosclerosis in mice.
Keywords: Nur77, Ly6C, nuclear receptor
Atherosclerosis remains the leading cause of cardiovascular morbidity and mortality in developed countries. The formation of atherosclerotic lesion is a complex process that begins with macrophage scavenging modified LDL species to form foam cells. Further remodeling of the plaque involves the interaction of the endothelium, smooth muscle cells, and a multitude of immune cells. The increased abundance of oxidized sterol in the cellular milieu upregulates a cascade of counter-regulatory responses coordinated by the liver X receptor (LXR), with the principle goal of reducing cholesterol absorption, and increasing cholesterol efflux and excretion (1). Other nuclear receptors implicated in the regulation of atherogenesis include the PPAR family of nuclear receptors as well the NR4A receptors (2–5).
The NR4A receptors consists of three highly homologous members, NR4A1, 2, and 3, otherwise known as Nur77, Nurr1, and NOR1, respectively. Structurally, these receptors have no ligand-binding pockets and are constitutively active. Regulation of these receptors occurs at the level of transcription in response to various environmental stimuli, including increases in cAMP concentration (6). NR4A receptors have been shown to regulate a broad array of biologic processes that are tissue-specific, including dopaminergic neuron development, thymocyte apoptosis, tumor suppression in myeloid cell precursors, and glucose metabolism (7–14). Some degree of functional redundancy exists among these receptors, however, in part related to the ability of all three receptors to transactivate the same Nur-responsive binding element (9).
We previously showed that all three NR4A receptors are rapidly induced in response to various inflammatory signaling, including exposure to lipopolysaccharides (LPS) and oxidized lipids (15). In turn, Nur77 affects the expression of genes linked to inflammatory pathways in macrophage cell lines (16). These findings implicate NR4A receptors as potential regulators of inflammation and suggest that they may play a role in atherosclerosis. In vivo studies examining the role of NR4A receptors in modifying atherosclerotic lesion formation have yielded conflicting results, however. Bruemmer and colleagues showed that NOR1 deletion reduces neointima formation after vascular injury (17). In the atherogenic ApoE−/− setting, NOR1 deletion reduced atherosclerotic lesion, likely due to diminished monocyte adhesion to the endothelium (5). On the other hand, de Vries and colleagues reported that overexpression of NR4A receptors in macrophages reduced lipid loading and proinflammatory response in human macrophages (18) and that transplant of Nur77-deficient bone marrow into LDL receptor (LDLR)−/− mice increased aortic root lesion size (3). Similarly, Hanna et al. observed increased aortic lesions in LDLR−/− mice transplanted with Nur77-null bone marrow, which the authors attributed to polarization of the macrophage inflammatory response toward the M1 phenotype (4). The discrepancy between these various findings highlights the need for additional studies to address the impact of NR4As in macrophage biology and atherogenesis.
In this report, we examined the function of Nur77 and NOR1 receptors in modulating the formation of atherosclerotic lesions. We reconstituted bone marrow of LDLR−/− mice with Nur77- or NOR1-deficient hematopoietic precursors in two independent experiments, and we observed no difference in the formation of atherosclerotic lesions. Cellular analysis of Nur77- and NOR1-null macrophages revealed comparable levels of response to LPS and IL-4, suggesting that Nur77 and NOR1 deficiency does not confer polarization of macrophages toward the M1 response in our system. Finally, we observed reduced numbers of Ly6Clo monocytes in Nur77-null mice in the absence of changes in atherosclerosis. These results suggest that bone marrow expression of Nur77 is not a dominant factor in the formation of atherosclerotic lesions.
MATERIALS AND METHODS
Generation of aP2-Nur77 transgenic mice and animal husbandry
We modified the pCK4800 expression plasmid by first cloning Nur77 cDNA into the EcoRI site, and subsequently replacing the 4,800 bp MCK enhancer with the −5.4 kb aP2 promoter element excised from pGL3-aP2-Luc (19). The linearized transgene was injected into pronuclei of C57BL/6 embryo by UCSD DERC Transgenic and Knock-out Core. Transgene genotype was confirmed by PCR amplification of a 691 bp product, using the following primers: forward 5′ CCACAATGAGGCAAATCCAT 3′ and reverse 5′ TCCTCAAACTTGAAGGACGCCGAA 3′. With the exception of Nur77 and NOR1 knockout fetal liver cell transplant (cells harvested from mice on 129SvEv/C57BL6 background), all other mice were on the C57BL/6J background. The NOR1 knockout mice from the mixed background were further backcrossed 10 generations onto C57BL/6J background prior to using them for the inflammation and flow cytometry experiments. Nur77 knockout mice were originally provided by Dr. Pinchas Cohen (UCLA). Signal transducer and activator of transcription 6 (STAT6) knockout and LDLR knockout mice on C57BL/6J mice were originally purchased from the Jackson Laboratory. Mice were age- and gender-matched for all experiments. Mice were fed ad libitum and maintained on a 12 h light-dark cycle. Animal studies were conducted in conformity with the Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals and in accordance with UCLA Animal Research Committee guidelines, the Salk Institute Animal Research Committee, Baylor College of Medicine Institutional Animal Care and Use Committee (IACUC), and UCSF IACUC.
Fetal liver cell transplant
Seven-week-old male LDLR−/− mice were irradiated with two doses of 600 rad 4 h apart at 100 cm. 24 h later, 2 × 106 fetal liver cells from Nur77−/− or NOR1−/− mice and wild-type littermates in 129SvEv/C57BL6 background (20, 21) were injected retro-orbitally. After four weeks of marrow reconstitution, mice were fed the atherogenic diet (Harlan Teklad, TD94059) for 16 weeks. Fasting plasma cholesterol and triglycerides were determined by enzymatic assays (Wako Chemicals and Thermo). The following primers sequences were used for genotyping reconstituted bone marrow: Nur77−/−: 5′ GTACTCCCAGGAAGTGACTG, 3′ CGGAATAGCTCTCCCCCTCC, Neo CTCGTGCTTTACGGTATCGC (expected band sizes: WT = 1,100 bp, KO = 700 bp); NOR1−/−: 5′GGCCGCAGCTGCACTCAGTC, 3′GTTCTGCCACCACAGAGCATC TTG, Neo GTGGCGGACCGCTATCAGGAC (expected band sizes: WT = 960 bp, KO = 1,200 bp).
Bone marrow transplant
Bone marrow transplant was performed as previously described (22). Briefly, eight-week-old male LDLR−/− mice were lethally irradiated with 900 rad one day prior to tail-vein injection with bone marrow cells collected from male wild-type, Nur77 knockout, or aP2-transgenic mice. After four weeks of marrow reconstitution, mice were fed the Western diet (Research Diets, D12079B) for 13 weeks (aP2-Nur77 transgenic) or 15 weeks (Nur77−/−) prior to terminal collection of aorta for lesion analysis. Difference in duration of Western diet between transgenic and Nur77-knockout groups was related to logistical availability. Each group was internally controlled with wild-type mice fed the same duration. Mice were fed ad lib prior to sacrifice. We measured plasma glucose using the Accu-Chek glucometer (Roche) and triglycerides using the Sigma Triglyceride Reagent (T2449/F6428). WAKO enzymatic kits were used to measure total cholesterol (Chol-E) and nonesterified free fatty acids (NEFA-HR) (2). Bone marrow reconstitution was determined by Q-PCR measurement of mRNA expression of total Nur77: 5′ ATGCCTCCCCTACCAATCTTC, 3′ CACCAGTTCCTGGAACTTGGA.
Analysis of aortic atherosclerotic lesion
Mice were euthanized and perfused with 7.5% sucrose in paraformaldehyde. Aortas were dissected, pinned, and stained with Sudan IV. Images were captured with a CCD camera. Computer-assisted image analysis of the aortic root and descending aorta (en face) has been previously described. Lesion development is expressed as the percentage of total aortic surface covered by lesions (23).
Cell culture
Primary peritoneal macrophages were collected four days after thioglycollate injection and prepared as described (24). Bone marrow cells were differentiated into macrophages by incubating cells in L929 cell-conditioned media for seven days. For inflammation studies, macrophages were incubated in 0.5% FBS in DMEM, with 5 μM simvastatin and 100 μM mevalonic acid. Five to seven hours later, cells were pretreated with DMSO or 1 μM LXR ligand GW3965 overnight (provided by Tim Willson and Jon Collins at GlaxoSmithKline), prior to stimulation with either 100 ng/ml lipopolysaccharides (Axxora, ALX-581-008-L002) for 4 h or 10 ng/ml IL-4 for 30 h (Sigma, I1020).
RNA isolation and quantitative real-time PCR
Total RNA was prepared by Trizol (Invitrogen) per manufacturer protocol. RNA (0.5 μg) was reverse transcribed using iScript cDNA synthesis kit (Bio-Rad). Q-PCR was performed with Sybrgreen 2× master mix (Diagenode) on the Applied Biosystems 7900HT sequence detector. Gene expression was normalized to 36B4 and represents averages of duplicate samples. See supplementary Table I for primer sequences.
Flow cytometry
Blood samples were collected by retro-orbital sampling under isoflurane anesthesia from age-matched female mice. Red blood cells were depleted by hypotonic lysis. Cells were resuspended in PBS with 0.2% BSA and 0.1% sodium azide. Single-cell suspensions were incubated for 15 min with anti-CD16/32 (Fc block) and stained for 30 min at 4°C with antibodies described below. Anti-mouse CD3ϵ (145-2C11), Ly6G (1A8), CD19 (1D3), CD49b (Dx5), and CD115 (AFS98) antibodies were purchased from eBioscience. Anti-mouse Ly6C (AL21) and CD11b (M1/70) antibodies were purchased from BD Biosciences. Cells were analyzed on FACSCalibur (Becton Dickinson) with FlowJo software v.9.5.2 (Tree Star).
Statistical analysis
Nonpaired Student t-test was used to determine statistical significance, defined at P-value < 0.05. Unless otherwise noted, error bars represent standard deviations. For Q-PCR analysis of macrophages, each condition represents averages of two independent samples.
RESULTS
Loss of Nur77 or NOR1 does not alter atherosclerotic plaque area
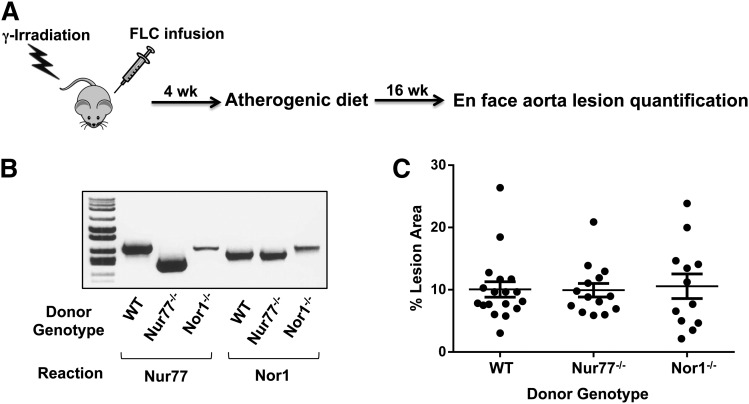
We previously showed that all three NR4A transcripts were induced acutely by lipopolysaccharides (LPS) and nuclear factor-kappa B (NF-κB), and that Nur77 modulates the expression of genes linked to inflammatory signaling in RAW267.4 and J774 macrophage cell lines (15, 16). These findings led us to hypothesize that NR4A deletion in monocyte/macrophages may alter arterial wall inflammation and affect the development of atherosclerosis. To examine the impact of nuclear receptor NR4A subfamily members Nur77 and NOR1 on the development of atherosclerosis, we reconstituted the bone marrow of irradiated LDLR−/− male mice with wild-type, Nur77−/−, or NOR1−/− fetal liver cells and analyzed plaque area after 16 weeks of exposure to atherogenic diet (Fig. 1A). At time of sacrifice, we harvested the LDLR−/− bone marrow and performed PCR genotyping using primers specific for Nur77−/− or NOR1−/− constructs. Engraftment was confirmed based on size of PCR product matching that of the donor genotype (Fig. 1B). The extent of atherosclerosis was expressed as the percentage of surface area of the entire aorta covered by lesions. Similar to LDLR−/− mice reconstituted with wild-type fetal liver cells, mice with NR4A-deficient fetal liver cells displayed plaques covering an average of approximately 10% of the total aorta area analyzed (Fig. 1C: wild-type mean 10.1%, Nur77−/− mean 9.9%, NOR1−/− mean 10.6%). As expected, the animal-to-animal variability in lesion development was very high, but a comparable range was observed in both wild-type and NR4A-transplanted mice, and there was no statistically significant difference in plaque area between cohorts. Cholesterol and triglycerides were elevated as expected after 8 and 16 weeks of exposure to atherogenic diet, but they did not vary between wild-type and NR4A-deficient cohorts overall (Table 1). There was also no significant difference in white blood cell count at the time mice were euthanized (Table 1).
Fig. 1.
Transplantation of NR4A-deficient hematopoietic precursors into LDLR−/− mice. A. Schematic of fetal liver cell (FLC) transplantation. B. PCR-genotyping of LDLR−/− bone marrow collected at time of sacrifice. Donor genotype was confirmed using primers flanking sequences deleted in the Nur77−/− or NOR1−/− constructs. C. Percentage of aorta surface area with atherosclerotic plaque in transplanted LDLR−/− mice. N = 12–18.
TABLE 1.
Metabolic profile of LDLR−/− mice transplanted with Nur77−/− and NOR1−/− fetal liver cells
| Bone Marrow Genotype | WT | Nur77−/− | NOR1−/− |
| N | 19 | 14 | 12 |
| Final weight (g) | 26.8 ± 2.7 | 27.5 ± 2.4 | 23.4 ± 1.7* |
| Glucose (mg/dl), 16 weeks fasted | 171 ± 31 | 146 ± 39 | 159 ± 45 |
| Cholesterol (mg/dl), baseline | 347 ± 43 | 340 ± 83 | 288 ± 70* |
| Cholesterol (mg/dl), 8-week diet | 2075 ± 605 | 2054 ± 505 | 1436 ± 385* |
| Cholesterol (mg/dl), 16-week diet | 2509 ± 816 | 2087 ± 710* | 2099 ± 727 |
| TG (mg/dl), baseline | 256 ± 102 | 253 ± 72 | 240 ± 54 |
| TG (mg/dl), 8-week diet | 739 ± 315 | 564 ± 341 | 385 ± 144* |
| TG (mg/dl), 16-week diet | 765 ± 350 | 644 ± 271 | 638 ± 228 |
| WBC (K/μl), 16-week diet | 3.65 ± 2.29 | 3.52 ± 1.92 | 4.43 ± 2.88 |
* P < 0.05 for comparison with wild-type.
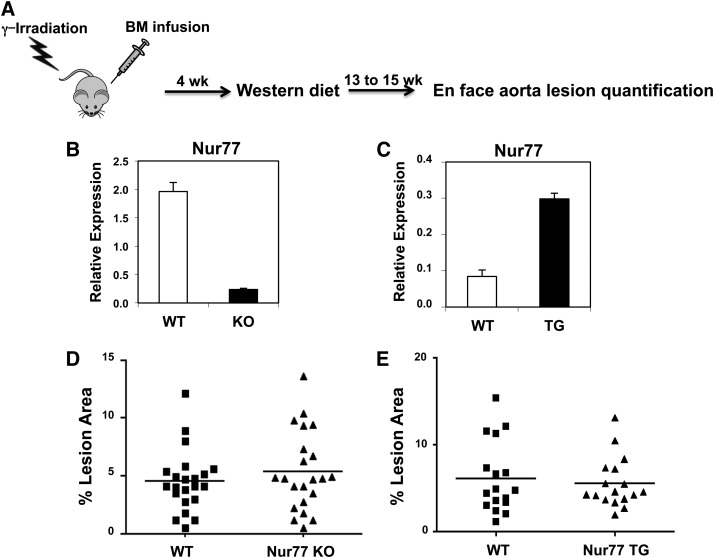
We considered the possibility that the very high cholesterol content of the atherogenic diet (1.25%) might have masked small phenotypic differences between control and NR4A-deficient mice. We therefore repeated the study with a moderate level of cholesterol (Western diet containing 0.21% cholesterol, compared with a standard mouse chow diet with 0.0014% cholesterol) (Fig. 2A). In this study, the source of hematopoietic precursor cells was wild-type or Nur77-null bone marrow. Engraftment was confirmed by Q-PCR analysis of bone marrow from recipient mice collected at time of sacrifice. As expected, recipients of the Nur77-knockout bone marrow showed a markedly reduced level of Nur77 expression (Fig. 2B). The extent of atherosclerotic plaque formation was lower than that observed with the 1.25% cholesterol diet, but again, there was no difference between control and Nur77-null cohorts (Fig. 2D: wild-type 4.52%, Nur77−/− 5.38%). We have previously observed marked differences in lesion formation between LDLR−/− mice transplanted with wild-type and LXR-deficient bone marrow using this protocol (25). Similar to findings from the fetal liver cell transplants, the levels of plasma glucose and lipid profiles in these LDLR−/− recipients were comparable irrespective of donor genotype (Table 2).
Fig. 2.
Transplantation of Nur77-null or Nur77-overexpressing bone marrow into LDLR−/− mice. A. Schematic of bone marrow transplantation. B, C. Expression of Nur77 in LDLR-/- bone marrow posttransplantation. D, E. Percentage of aorta surface area with atherosclerotic plaque in LDLR−/− mice transplanted with Nur77-knockout (KO) (D) or Nur77-overexpressing transgenic (TG) (E) marrow. N = 21–22 for D, N = 17 for E.
TABLE 2.
Metabolic profile of LDLR−/− mice transplanted with Nur77−/− and aP2-Nur77 transgenic bone marrow
| Bone Marrow Genotype | WT | Nur77−/− | WT | aP2-Nur77 |
| N | 10 | 10 | 10 | 10 |
| Glucose (mg/dl) | 295 ± 29 | 262 ± 25 | 198 ± 19 | 186 ± 19 |
| Cholesterol (mg/dl) | 596 ± 28 | 599 ± 40 | 629 ± 32 | 631 ± 35 |
| TG (mg/dl) | 44 ± 6 | 64 ± 13 | 138 ± 20 | 152 ± 17 |
| Non-esterified free fatty acids (mmol/l) | 0.87 ± 0.07 | 0.86 ± 0.07 | 1.00 ± 0.05 | 0.99 ± 0.06 |
The NR4A receptors are highly homologous and functionally redundant (6), raising the possibility that compensation by other NR4A members may explain the lack of phenotypic difference in donor Nur77−/− or NOR1−/− bone marrow in atherosclerotic plaque formation. As a complement to our loss-of-function studies, we used a gain-of-function approach. We generated the aP2-Nur77 transgenic mice that overexpressed Nur77 in adipose tissue and macrophages. Nur77-overexpressing bone marrow was transplanted into LDLR−/− recipients as described above. Engraftment was confirmed by increased expression of Nur77 in the bone marrow from recipient mice (Fig. 2C). As shown in Fig. 2E and Table 2, overexpression of Nur77 in macrophages did not alter atherosclerotic lesion formation (wild-type mean 6.14%, aP2-Nur77 transgenic mean 5.56%) or metabolic parameters, such as plasma glucose and lipid profiles. We conclude from our gain- and loss-of-function mouse models that bone marrow expression of Nur77 and NOR1 is not a dominant factor in atherosclerotic plaque formation in mice, at least under the conditions employed here.
Preserved response to inflammatory stimuli in Nur77-deficient macrophages
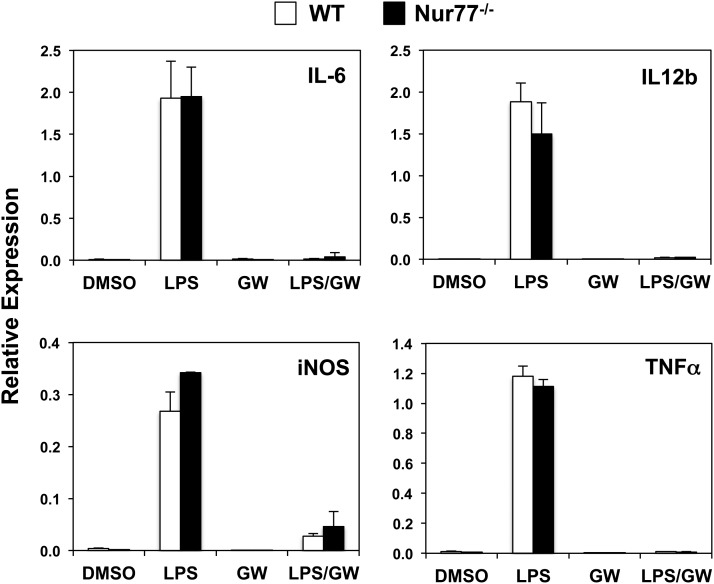
The formation of atherosclerotic lesion is subject to complex regulation involving multiple cell types, including endothelial cells, smooth muscle cells, and a heterogeneous population of monocyte/macrophages. Although we did not observe any differences in plaque formation, we proceeded with cellular analysis to determine whether there were intrinsic differences in the inflammatory response between wild-type and Nur77-null macrophages. We tested this hypothesis by stimulating wild-type and Nur77-null thioglycollate-elicited peritoneal macrophages with LPS. Four h after LPS stimulation, the expression of several genes known to mediate the inflammatory response, including IL-6, IL-12b, inducible NO synthase (iNOS), and tumor necrosis factor alpha (TNFα), was robustly induced in both control and Nur77-null macrophages. Consistent with our previous work, LPS-induced inflammatory gene expression was strongly suppressed by activation of the LXR with the synthetic agonist GW3965, indicating that nuclear receptor transrepression pathways are functional under the conditions employed in these studies (Fig. 3) (26).
Fig. 3.
Expression of inflammatory response genes in LPS-treated Nur77-null peritoneal macrophages. Cells were pretreated with DMSO or 1 μM GW3965 (LXR agonist) overnight prior to the addition of 100 ng/ml LPS. Gene expression was analyzed by real-time Q-PCR and normalized to 36B4 control.
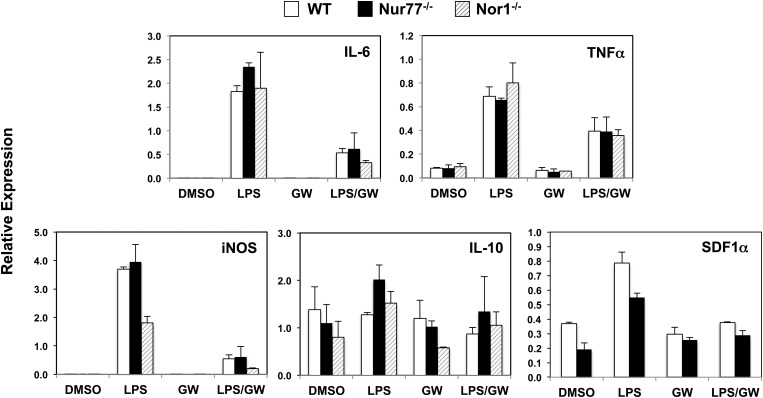
We considered the possibility that the activated state of thioglycollate-elicited peritoneal macrophages could mask subtle changes in inflammatory responses. We therefore repeated the LPS stimulation with bone marrow-derived macrophages. Similar to our findings with peritoneal macrophages, LPS elicited comparable levels of IL-6, iNOS, TNFα, and IL-10 expression between control and Nur77-null bone marrow-derived macrophages (Fig. 4). This result differs from previous reports that Nur77-null macrophages exhibit increased expression of proinflammatory cytokines and reduced expression of the protective cytokine IL-10 in response to LPS (3, 4). In addition, Nur77-null bone marrow-derived macrophages did not express higher level of stromal-derived factor 1 alpha (SDF1α), a chemokine postulated to be suppressed by Nur77 (3), either in the basal or LPS-stimulated state in our studies.
Fig. 4.
Expression of inflammatory response genes in LPS-treated Nur77-null and NOR1-null bone marrow-derived macrophages. Macrophages were treated as described in Fig. 3. Gene expression was analyzed by real-time Q-PCR and normalized to 36B4 control.
We further tested whether deletion of NOR1, another member of the NR4A family, alters the LPS-induced inflammatory response. As shown in Fig. 4, we observed subtly reduced expression of iNOS in NOR1-null bone marrow-derived macrophages 4 h after LPS stimulation, relative to wild-type and Nur77-null macrophages. However, the expression of other inflammatory genes measured (IL-6 and TNFα) was unchanged compared with wild-type macrophages, suggesting that NOR1 deletion did not cause a global shift in the inflammatory response. We conclude that, in contrast to LXR activation, loss of Nur77 does not exert a major effect on the induction of macrophage M1 responses under the conditions used here.
Preserved alternative macrophage activation in the absence of Nur77
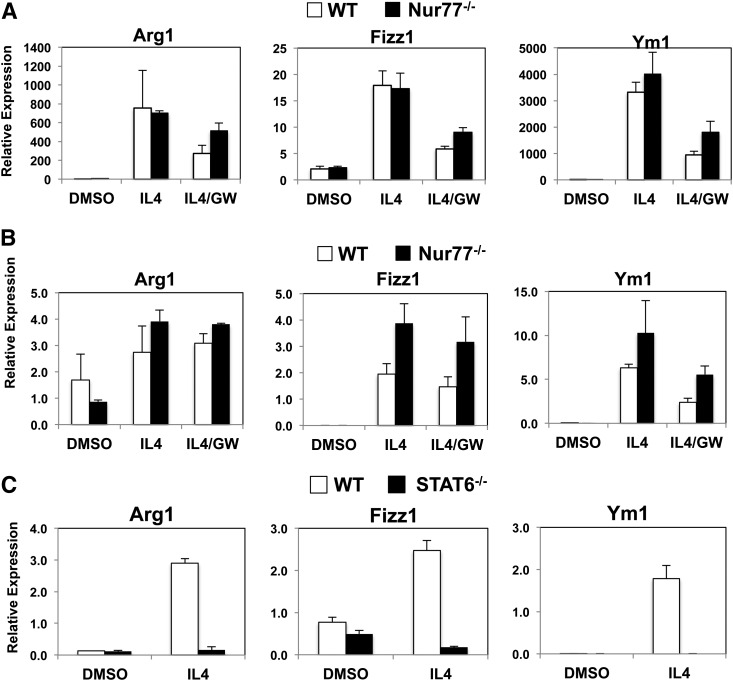
During immune responses, macrophages may become polarized toward either the classical “proinflammatory” M1- or the alternative TH2-helper cell-mediated M2 phenotype (27). Derangement of the delicate balance of M1 and M2 responses has been proposed to contribute to a wide range of diseases, including insulin resistance, asthma, and cancer (28). Recent studies suggested that the genetic absence of Nur77 shifts the balance of macrophage activation toward the M1 phenotype with a concomitant reduction of M2 response (3, 4). We tested this hypothesis by activating the M2 response in both peritoneal macrophages and bone marrow-derived macrophages with IL-4. As shown in Fig. 5A, IL-4 promoted comparable level of expression of classic alternative activation markers arginase 1 (Arg1; found in inflammatory zone), Fizz1 (also known as Retnla), and chitinase 3-like protein 3 (Ym1), in wild-type and Nur77-null peritoneal macrophages. IL-4 induced the expression of Fizz1 more robustly in Nur77-null bone marrow-derived macrophages, although there was no difference in the expression of Arg1 and Ym1 (Fig. 5B). Overall, we did not observe a reduction in the M2 response from Nur77-null macrophages. Unlike the potent suppression of LPS-induced inflammatory markers, LXR ligand GW3965 had little effect on antagonizing IL-4 induced responses. By comparison, the IL-4-induced response (expression of Arg1, Fizz1, and Ym1) was virtually abolished in peritoneal macrophages lacking expression of STAT6 (Fig. 5C). Upon IL-4 stimulation, STAT6 dimerizes and activates downstream effector genes of alternative activation and is known to act as a key mediator of the M2 phenotype in macrophages (29–31). We conclude that, unlike STAT6, Nur77 is not a dominant determinant of IL-4-induced activation of the macrophage M2 response.
Fig. 5.
Comparable expression of IL-4-responsive genes in wild-type and Nur77-null macrophages. (A) Peritoneal macrophages and (B) bone marrow-derived macrophages were treated overnight with DMSO or 1 μM GW3965. Cells were then treated with IL-4 10 ng/ml for 30 h. C. Expression of IL-4-responsive genes in STAT6-null peritoneal macrophages. Cells were incubated with IL-4 10 ng/ml for 30 h. Gene expression was analyzed by real-time Q-PCR and normalized to 36B4 control.
Reduced Ly6Clo population in Nur77-null macrophages
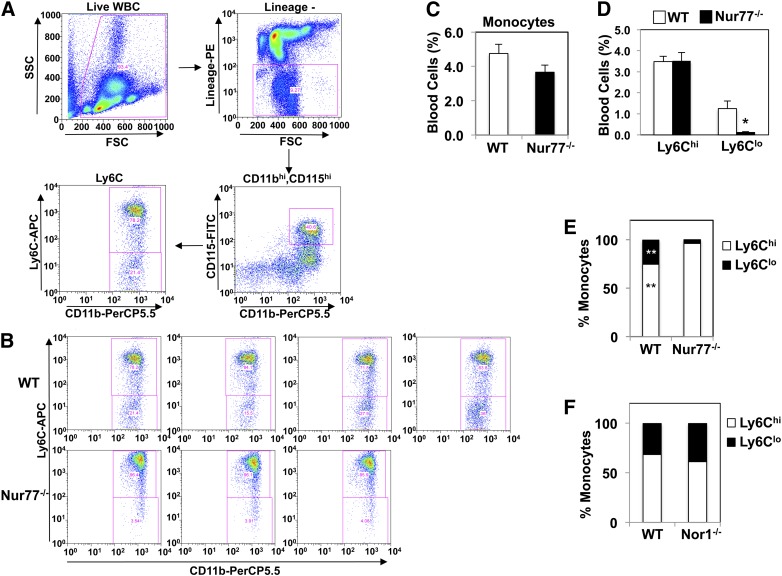
Previous studies showed that Nur77 deletion impaired the differentiation of Ly6Clo monocytes from macrophage dendritic precursors in bone marrow (32). Hanna et al. postulated that loss of Ly6Clo monocytes polarizes macrophage response toward the M1-inflammatory pathway, thereby promoting atherosclerotic lesion formation (4). To investigate whether the absence of a proinflammatory phenotype in our system was due to the “preservation” of the Ly6Clo population in our particular cohort of Nur77-deficient mice, we analyzed the abundance of Ly6Clo population in Nur77-null and NOR1-null mice (Fig. 6A). The prevalence of CD11bhiCD115hi monocytes in blood was comparable between wild-type and Nur77-null mice (Fig. 6C: wild-type 4.75 ± 0.53%, Nur77−/− 3.66 ± 0.41%, P = 0.19). However, there was a clear reduction in the Ly6Clo population in Nur77-null mice (Fig. 6B, D: wild-type 1.25 ± 0.36%, Nur77−/− 0.14 ± 0.01%, P = 0.046), resulting in a relative shift in the proportion of Ly6Chi versus Ly6Clo monocytes (Fig. 6E). By comparison, there was no difference in the ratio of Ly6Chi to Ly6Clo monocytes in NOR1-null mice (Fig. 6F). These findings confirm the previous report of reduced Ly6Clo monocytes in Nur77-null mice. However, the lack of correlation between atherosclerotic lesion development, inflammatory phenotype, and reduction of the Ly6Clo population in our study suggests that marked reduction in the numbers of Ly6Clo monocytes alone is not sufficient to alter macrophage polarization and atherosclerosis in mice.
Fig. 6.
Quantitation of Ly6Clo population in NR4A-deficient mice. A. Gating strategy for blood monocyte subsets. Cells were plotted for forward scatter (FSC) by side scatter (SSC). Live, lineage− (CD3e−, CD19−, CD49b−, Ly6G−) cells were plotted for CD115 and CD11b expression. CD11bhi, CD115hi monocytes were then sorted by Ly6C expression. B. FACS plot of Ly6C expression among CD11bhi, CD115hi monocytes. C. Abundance of CD11bhi, CD115hi monocytes among live wild-type and Nur77-null white blood cells. N = 3–4. D. Percentage of Ly6Chi versus Ly6Clo monocytes in wild-type and Nur77-null monocytes. *P < 0.05. E. Relative proportion of Ly6Chi versus Ly6Clo in wild-type and Nur77-null monocytes. **P < 0.01. F. Relative proportion of Ly6Chi versus Ly6Clo in wild-type and NOR1-null monocytes. N = 4. Error bars represent standard errors.
DISCUSSION
Previous studies demonstrating NR4A receptors regulating inflammatory gene expression in macrophages suggested that these receptors might play a role in development of atherosclerotic lesion formation (15, 16). Using a well-established protocol, we tested this hypothesis by reconstituting bone marrow of LDLR−/− mice with either Nur77-null fetal liver cells or bone marrow and subjecting the mice to a high-cholesterol dietary challenge. We observed no statistically significant difference in the aortic lesion size in either experiment. Similarly, bone marrow reconstitution with NOR1-null fetal liver cell did not alter the progression of atherosclerosis in LDLR−/− mice. Transplant with Nur77-overexpressing bone marrow cells likewise had no effect on lesion size. Analysis of wild-type and Nur77-null peritoneal and bone marrow-derived macrophages revealed comparable levels of expression of markers of classical (M1) and alternative (M2) activation in response to LPS and IL-4 stimulation, respectively. We confirmed that Nur77-null mice have a diminished population of Ly6Clo monocytes as previously reported (32), without affecting macrophage polarization and lesion size. Our failure to observe a difference in atherosclerosis using both gain- and loss-of-function mouse models suggests that Nur77 deficiency alone is not sufficient to affect the formation of atherosclerotic lesions in mice, at least under the conditions used here.
Interestingly, NR4A receptors have been shown to be both pro- and anti-inflammatory in other diseases involving chronic inflammation. In arthritis models, NR4A receptors are robustly expressed in synoviocytes and macrophages isolated from chronically inflamed joints (33). In fact, Nurr1 (NR4A2) has been shown to promote synoviocyte proliferation and induce the expression of matrix metalloproteinase 13 (MMP13), consistent with its function in executing inflammatory signaling and extracellular matrix remodeling (34). At the same time, Nurr1 suppresses the expression of MMP1 in cartilage (35), consistent with a biological system with checks and balances built-in to curtail progression of unrestrained inflammation. Nurr1 is also thought to exert anti-inflammatory effects in microglia cells by docking the p65 subunit of NFκB on promoters of inflammatory genes and recruiting the CoREST corepressor complex (36). On balance, these findings suggest that acute induction of NR4A receptors by inflammatory signals can trigger effector pathways in a tissue- and cell type-specific fashion that may help to contain the infection/insult and mitigate the proinflammatory processes to restore homeostasis.
Our findings differ from two recent studies reporting that Nur77-deletion polarized macrophages toward an inflammatory phenotype and promoted increased atherosclerotic lesion formation (3, 4). We speculate that one contributing factor to this difference may be the control cohorts used in Hanna et al. (4). Given the biologic variation in lesion formation in mice, a wide distribution is expected in quantification of plaque area. Although the Nur77−/− cohorts exhibited the expected broad distribution of lesion area, the control cohorts used in Hanna et al. displayed an unexpectedly tight distribution and very low lesion burden. We also consider variability in intestinal microbiota of the mouse colonies as a potential difference between the experimental systems. Resident intestinal bacterial flora is a rich source of proinflammatory signals, including LPS and peptidoglycans (37). The abundance and type of microbiota have been proposed as contributing factors to systemic inflammation of the host and can potentially modulate the progression of atherosclerosis (38). We suspect that small differences in proinflammatory signaling may be amplified depending on the particular host-environmental interactions. Finally, we cannot exclude the possibility that technical differences in the various approaches to lesion quantification may contribute to the differences between studies.
Previous studies demonstrated that Nur77-null mice have reduced abundance of Ly6Clo monocytes (32), although the in vivo function of these cells is unclear. The Ly6Clo monocytes are analogous to human CD14loCD16hi monocytes, whereas murine Ly6Chi monocytes correspond to human CD14hi CD16lo monocytes (32, 39). Ly6Chi monocytes are robustly induced by LPS and thought to be proinflammatory. Ly6Clo monocytes have been ascribed the role of patrolling endothelium during homeostasis, and may activate either an “alternative activation” (M2-like) or proinflammatory (M1-like) transcriptional program, depending on the site of action (40). Gautier et al. recently reported that Ly6Clo blood and peritoneal macrophages express a higher level of PPARγ and are more responsive to PPARγ-ligand activation, consistent with a putative role of PPARγ in tissue repair (41). Despite these findings, clarifying the in vivo function of Ly6lo monocytes remains a subject of active investigation for the field. Hanna et al. have proposed that loss of Ly6Clo monocytes in Nur77-null bone marrow polarizes macrophages toward the proinflammatory phenotype and contributes to development of atherosclerosis (4). That we did not observe the correlation between Ly6Clo monocytes and plaque size suggests that loss of Ly6Clo monocytes alone does not impair modulate macrophage phenotype and atherosclerotic lesion progression. Additional studies would be required to delineate the developmental lineage of Ly6Clo monocytes and their in vivo relevance to immune function.
If loss of Nur77 and NOR1 from bone marrow has little effect on lesion size, what role, if any, do these receptors play in atherosclerosis? Bruemmer and colleagues have demonstrated that NR4A3 (NOR1) deletion exerts its atheroprotective effects at the level of smooth muscle and endothelial cells by limiting monocyte adhesion and neointima formation (5). It is also worth noting that compound deletion of multiple NR4A family members has uncovered redundant function for these transcription factors in other contexts. For example, compound NR4A null mice develop myelodysplastic/myeloproliferative neoplasms (14). Unfortunately, development of blood dyscrasias in mice devoid of NR4A1 and NR4A3 precludes the evaluation of NR4A double deficiency in atherogenesis (13). Collectively, we conclude that expression of individual NR4A receptors in macrophages does not appear to be a major determinant of macrophage polarization, systemic inflammation, or the development of atherogenic lesions in mice.
Supplementary Material
Acknowledgments
The authors are grateful to Tammy Phung for technical assistance with flow cytometry.
Footnotes
Abbreviations:
- IL
- interleukin
- iNOS
- inducible NO synthase
- KO
- knockout
- LDLR
- LDL receptor
- LPS
- lipopolysaccharide
- LXR
- liver X receptor
- STAT6
- signal transducer and activator of transcription 6
- TG
- triglyceride
- TNFα
- tumor necrosis factor alpha
- WBC
- white blood cell
- WT
- wild-type
This work is supported by National Institutes of Health Grants HL-066088 (P.T.), HL-030568 (P.T), DK-057978 (R.M. E), HL-088093 (R.M. E), HL-105278 (R.M. E), the Glenn Foundation (R.M. E), the Helmsley Charitable Trust (R.M. E), and the Cancer Center (R.M. E). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables and six figures.
REFERENCES
- 1.Tontonoz P. 2011. Transcriptional and posttranscriptional control of cholesterol homeostasis by liver X receptors. Cold Spring Harb. Symp. Quant. Biol. 76: 129–137 [DOI] [PubMed] [Google Scholar]
- 2.Li A. C., Binder C. J., Gutierrez A., Brown K. K., Plotkin C. R., Pattison J. W., Valledor A. F., Davis R. A., Willson T. M., Witztum J. L., et al. 2004. Differential inhibition of macrophage foam-cell formation and atherosclerosis in mice by PPARalpha, beta/delta, and gamma. J. Clin. Invest. 114: 1564–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamers A. A., Vos M., Rassam F., Marinković G., Kurakula K., van Gorp P. J., de Winther M. P., Gijbels M. J., de Waard V., de Vries C. J. 2012. Bone marrow-specific deficiency of nuclear receptor Nur77 enhances atherosclerosis. Circ. Res. 110: 428–438 [DOI] [PubMed] [Google Scholar]
- 4.Hanna R. N., Shaked I., Hubbeling H. G., Punt J. A., Wu R., Herrley E., Zaugg C., Pei H., Geissmann F., Ley K., et al. 2012. NR4A1 (Nur77) deletion polarizes macrophages toward an inflammatory phenotype and increases atherosclerosis. Circ. Res. 110: 416–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y., Howatt D. A., Gizard F., Nomiyama T., Findeisen H. M., Heywood E. B., Jones K. L., Conneely O. M., Daugherty A., Bruemmer D. 2010. Deficiency of the NR4A orphan nuclear receptor NOR1 decreases monocyte adhesion and atherosclerosis. Circ. Res. 107: 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maxwell M. A., Muscat G. E. 2006. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl. Recept. Signal. 4: e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao L. C., Wroblewski K., Zhang Z., Pei L., Vergnes L., Ilkayeva O. R., Ding S. Y., Reue K., Watt M. J., Newgard C. B., et al. 2009. Insulin resistance and altered systemic glucose metabolism in mice lacking Nur77. Diabetes. 58: 2788–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao L. C., Zhang Z., Pei L., Saito T., Tontonoz P., Pilch P. F. 2007. Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Mol. Endocrinol. 21: 2152–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng L. E., Chan F. K., Cado D., Winoto A. 1997. Functional redundancy of the Nur77 and Nor-1 orphan steroid receptors in T-cell apoptosis. EMBO J. 16: 1865–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pei L., Waki H., Vaitheesvaran B., Wilpitz D. C., Kurland I. J., Tontonoz P. 2006. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat. Med. 12: 1048–1055 [DOI] [PubMed] [Google Scholar]
- 11.Woronicz J. D., Calnan B., Ngo V., Winoto A. 1994. Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas. Nature. 367: 277–281 [DOI] [PubMed] [Google Scholar]
- 12.Zetterström R. H., Solomin L., Jansson L., Hoffer B. J., Olson L., Perlmann T. 1997. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 276: 248–250 [DOI] [PubMed] [Google Scholar]
- 13.Mullican S. E., Zhang S., Konopleva M., Ruvolo V., Andreeff M., Milbrandt J., Conneely O. M. 2007. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat. Med. 13: 730–735 [DOI] [PubMed] [Google Scholar]
- 14.Ramirez-Herrick A. M., Mullican S. E., Sheehan A. M., Conneely O. M. 2011. Reduced NR4A gene dosage leads to mixed myelodysplastic/myeloproliferative neoplasms in mice. Blood. 117: 2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pei L., Castrillo A., Chen M., Hoffmann A., Tontonoz P. 2005. Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J. Biol. Chem. 280: 29256–29262 [DOI] [PubMed] [Google Scholar]
- 16.Pei L., Castrillo A., Tontonoz P. 2006. Regulation of macrophage inflammatory gene expression by the orphan nuclear receptor Nur77. Mol. Endocrinol. 20: 786–794 [DOI] [PubMed] [Google Scholar]
- 17.Nomiyama T., Zhao Y., Gizard F., Findeisen H. M., Heywood E. B., Jones K. L., Conneely O. M., Bruemmer D. 2009. Deficiency of the NR4A neuron-derived orphan receptor-1 attenuates neointima formation after vascular injury. Circulation. 119: 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonta P. I., van Tiel C. M., Vos M., Pols T. W., van Thienen J. V., Ferreira V., Arkenbout E. K., Seppen J., Spek C. A., van der Poll T., et al. 2006. Nuclear receptors Nur77, Nurr1, and NOR-1 expressed in atherosclerotic lesion macrophages reduce lipid loading and inflammatory responses. Arterioscler. Thromb. Vasc. Biol. 26: 2288–2294 [DOI] [PubMed] [Google Scholar]
- 19.Waki H., Park K. W., Mitro N., Pei L., Damoiseaux R., Wilpitz D. C., Reue K., Saez E., Tontonoz P. 2007. The small molecule harmine is an antidiabetic cell-type-specific regulator of PPARgamma expression. Cell Metab. 5: 357–370 [DOI] [PubMed] [Google Scholar]
- 20.Lee S. L., Wesselschmidt R. L., Linette G. P., Kanagawa O., Russell J. H., Milbrandt J. 1995. Unimpaired thymic and peripheral T cell death in mice lacking the nuclear receptor NGFI-B (Nur77). Science. 269: 532–535 [DOI] [PubMed] [Google Scholar]
- 21.Ponnio T., Burton Q., Pereira F. A., Wu D. K., Conneely O. M. 2002. The nuclear receptor Nor-1 is essential for proliferation of the semicircular canals of the mouse inner ear. Mol. Cell. Biol. 22: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong C., Bradley M. N., Rong X., Wang X., Wagner A., Grijalva V., Castellani L. W., Salazar J., Realegeno S., Boyadjian R., et al. 2012. LXRalpha is uniquely required for maximal reverse cholesterol transport and atheroprotection in ApoE-deficient mice. J. Lipid Res. 53: 1126–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tangirala R. K., Rubin E. M., Palinski W. 1995. Quantitation of atherosclerosis in murine models: correlation between lesions in the aortic origin and in the entire aorta, and differences in the extent of lesions between sexes in LDL receptor-deficient and apolipoprotein E-deficient mice. J. Lipid Res. 36: 2320–2328 [PubMed] [Google Scholar]
- 24.Bradley M. N., Hong C., Chen M., Joseph S. B., Wilpitz D. C., Wang X., Lusis A. J., Collins A., Hseuh W. A., Collins J. L., et al. 2007. Ligand activation of LXR beta reverses atherosclerosis and cellular cholesterol overload in mice lacking LXR alpha and apoE. J. Clin. Invest. 117: 2337–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tangirala R. K., Bischoff E. D., Joseph S. B., Wagner B. L., Walczak R., Laffitte B. A., Daige C. L., Thomas D., Heyman R. A., Mangelsdorf D. J., et al. 2002. Identification of macrophage liver X receptors as inhibitors of atherosclerosis. Proc. Natl. Acad. Sci. USA. 99: 11896–11901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joseph S. B., Castrillo A., Laffitte B. A., Mangelsdorf D. J., Tontonoz P. 2003. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat. Med. 9: 213–219 [DOI] [PubMed] [Google Scholar]
- 27.Odegaard J. I., Chawla A. 2011. Alternative macrophage activation and metabolism. Annu. Rev. Pathol. 6: 275–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon S. 2003. Alternative activation of macrophages. Nat. Rev. Immunol. 3: 23–35 [DOI] [PubMed] [Google Scholar]
- 29.Shirey K. A., Cole L. E., Keegan A. D., Vogel S. N. 2008. Francisella tularensis live vaccine strain induces macrophage alternative activation as a survival mechanism. J. Immunol. 181: 4159–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szanto A., Balint B. L., Nagy Z. S., Barta E., Dezso B., Pap A., Szeles L., Poliska S., Oros M., Evans R. M., et al. 2010. STAT6 transcription factor is a facilitator of the nuclear receptor PPARgamma-regulated gene expression in macrophages and dendritic cells. Immunity. 33: 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng M., Huntley D., Huang I. F., Foye-Jackson O., Wang L., Sarkissian A., Zhou Q., Walker W. A., Cherayil B. J., Shi H. N. 2007. Alternatively activated macrophages in intestinal helminth infection: effects on concurrent bacterial colitis. J. Immunol. 179: 4721–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanna R. N., Carlin L. M., Hubbeling H. G., Nackiewicz D., Green A. M., Punt J. A., Geissmann F., Hedrick C. C. 2011. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat. Immunol. 12: 778–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMorrow J. P., Murphy E. P. 2011. Inflammation: a role for NR4A orphan nuclear receptors? Biochem. Soc. Trans. 39: 688–693 [DOI] [PubMed] [Google Scholar]
- 34.Mix K. S., McMahon K., McMorrow J. P., Walkenhorst D. E., Smyth A. M., Petrella B. L., Gogarty M., Fearon U., Veale D., Attur M. G., et al. 2012. Orphan nuclear receptor NR4A2 induces synoviocyte proliferation, invasion, and matrix metalloproteinase 13 transcription. Arthritis Rheum. 64: 2126–2136 [DOI] [PubMed] [Google Scholar]
- 35.Mix K. S., Attur M. G., Al-Mussawir H., Abramson S. B., Brinckerhoff C. E., Murphy E. P. 2007. Transcriptional repression of matrix metalloproteinase gene expression by the orphan nuclear receptor NURR1 in cartilage. J. Biol. Chem. 282: 9492–9504 [DOI] [PubMed] [Google Scholar]
- 36.Saijo K., Winner B., Carson C. T., Collier J. G., Boyer L., Rosenfeld M. G., Gage F. H., Glass C. K. 2009. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 137: 47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caesar R., Fak F., Backhed F. 2010. Effects of gut microbiota on obesity and atherosclerosis via modulation of inflammation and lipid metabolism. J. Intern. Med. 268: 320–328 [DOI] [PubMed] [Google Scholar]
- 38.Mencarelli A., Cipriani S., Renga B., Bruno A., D'Amore C., Distrutti E., Fiorucci S. 2012. VSL#3 resets insulin signaling and protects against NASH and atherosclerosis in a model of genetic dyslipidemia and intestinal inflammation. PLoS ONE. 7: e45425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geissmann F., Jung S., Littman D. R. 2003. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 19: 71–82 [DOI] [PubMed] [Google Scholar]
- 40.Geissmann F., Manz M. G., Jung S., Sieweke M. H., Merad M., Ley K. 2010. Development of monocytes, macrophages, and dendritic cells. Science. 327: 656–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gautier E. L., Chow A., Spanbroek R., Marcelin G., Greter M., Jakubzick C., Bogunovic M., Leboeuf M., van Rooijen N., Habenicht A. J., et al. 2012. Systemic analysis of PPARgamma in mouse macrophage populations reveals marked diversity in expression with critical roles in resolution of inflammation and airway immunity. J. Immunol. 189: 2614–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.