Abstract
The hydroxy-carboxylic acid receptor (HCA1) is a G protein-coupled receptor that is highly expressed on adipocytes and considered a potential target for the treatment of dyslipidemia. In the current study, we investigated the pharmacological properties of naturally occurring variants in this receptor (H43Q, A110V, S172L, and D253H). After transient expression of these receptors into human embryonic kidney 293 cells, basal and ligand-induced signaling were assessed using luciferase reporter gene assays. The A110V, S172L, and D253 variants showed reduced basal activity; the S172L mutant displayed a decrease in potency to the endogenous ligand l-lactate. Both the S172L and D253H variants also showed impaired cell surface expression, which may in part explain the reduced activity of these receptors. The impact of a loss in HCA1 function on lipid accumulation was investigated in the adipocyte cell line, OP9. In these cells, endogenous HCA1 transcript levels rapidly increased and reached maximal levels 3 days after the addition of differentiation media. Knockdown of HCA1 using siRNA resulted in an increase in lipid accumulation as assessed by quantification of Nile Red staining and TLC analysis. Our data suggest that lipid homeostasis may be altered in carriers of selected HCA1 missense variants.
Keywords: polymorphism, OP9 cell, G protein-coupled receptor, dyslipidemia, constitutive activity, hydroxy-carboxylic acid receptor
The hydroxy-carboxylic acid receptor 1 (HCA1), formerly known as orphan receptor GPR81, is a G protein-coupled receptor (GPCR) that is expressed almost exclusively in adipocytes (1–3). Based on its expression profile and relation to HCA2 (also known as GPR109a), which is the target of the lipid-lowering drug niacin, HCA1 is considered a potential target for the treatment of dyslipidemia (4–6). Activation of this receptor by the endogenous ligand l-lactate triggers a Gαi-mediated inhibition of cAMP accumulation in receptor-expressing cells (2). Notably, HCA1 also shows ligand- independent basal activity (i.e., it is a constitutively active GPCR) (1, 2). When studied in mature isolated adipocytes from human, rat, or mouse, activation of HCA1 by l-lactate inhibits lipolysis (2, 7). HCA1 has also been shown to modulate the anti-lipolytic effects of insulin in vivo (8). In contrast to the role of HCA1 in mature adipocytes, limited information is available regarding the function of this receptor during fat cell development and differentiation.
Given the localization of HCA1 in fat tissue and its role in modulating lipolysis, we were interested in determining how naturally occurring variants influence receptor function. Using both the NCBI dbSNP database and the NHLBI ESP Exome Variant Server, we identified four missense mutants of interest. H43Q, annotated in NCBI dbSNP, was exclusively reported in a cohort of Sub-Saharan Africans. From the NHLBI ESP Exome Variant Server, a database of single-nucleotide polymorphisms (SNPs) assembled based on whole-exome sequencing, we selected three additional variants to be included in our study: A110V, S172L, and D253H. These missense mutations had the highest prevalence among HCA1 SNPs and have been identified most frequently in individuals of either African American (A110V and D253H) or European American (S172L) descent. We found that each of the variants examined, with the exception of the H43Q isoform, showed some degree of loss of function in assays assessing basal activity, ligand induced signaling, and/or surface expression. As a predictive model of how loss-of-function variants may alter lipid accumulation in vivo, we explored how HCA1 RNAi in the preadipocyte cell line OP9 influences lipid accumulation during experimentally induced differentiation. Taken together, our results suggest that reduced HCA1-mediated signaling may alter lipid deposition during terminal differentiation of adipocytes. Based on these findings, it will be of interest to assess potential phenotypic manifestations of individuals within targeted populations (e.g., obese, thin cohorts) who harbor these loss-of-function missense mutations.
EXPERIMENTAL PROCEDURES
Materials
DMEM, MEMalpha, OptiMEM, l-glutamine, FBS, optifect, lipofectamine, and Nile Red were purchased from Invitrogen (Chicago, IL). FBS for the culture of human embryonic kidney (HEK) 293 cells was purchased from Atlanta Biologicals (Lawrenceville, GA), and premium FBS for the culture of OP9 cells from Lonza (Chicago, IL). Polyethylenimine (PEI), 2-nitrophenyl β-d-galactopyranoside, l-lactate, Trizol reagent, and 4’,6-diamidino-2-phenylindole (DAPI) were obtained from Sigma (Atlanta, GA). Forskolin was purchased from Calbiochem/EMD Serono (Rockland, MA), SteadyLite reagent from Perkin Elmer (Chicago, IL), and peroxidase substrate BM-blue (3.3′-5.5′-tetramethylbenzidine) from Roche (Indianapolis, IN). All plates for tissue culture experiments were purchased from Corning (Lowell, MA). Silencer select siRNA and starter kit, DNase treatment and removal kits, reverse transcription reagents, and Sybr Green Master Mix were obtained from Applied Biosystems (Carlsbad, CA). Primers for RT-PCR were designed using NIH primer design software. Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). The plasmids encoding a cAMP response element (CRE6×) or a serum-response element (SRE5×) ligated upstream of a luciferase reporter gene have been described previously (9, 10).
Cell culture
All cells were maintained at 37°C in a humidified environment containing 5% CO2. HEK293 cells were grown in DMEM containing 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. Parental OP9 cells were generously provided by Dr. Coleen McNamara (University of Virginia, Charlottesville) (11). Dilution cloning was used to identify clone K, an OP9 cell line that was particularly responsive to experimentally induced differentiation. After exposure to differentiation media for 3 days, a large proportion of these cells (>50%) showed features of mature adipocytes (i.e., formation of lipid droplets). All subsequent experiments were performed using clonal cell line K. Clone K OP9 cells were maintained in OP9 propagation media: MEMalpha with 20% premium FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin. OP9 cells were differentiated using the following media: MEMalpha with 0.2% premium FBS, 175 nM insulin, 900 µM oleate bound to albumin (5.5:1 molar ratio), 100 U/ml penicillin, and 100 µg/ml streptomycin, as previously described (12).
Recombinant HCA1 constructs
Plasmid encoding HCA1 cDNA was purchased from the Missouri S and T cDNA Resource Center (Rolla, MO) and subcloned into pcDNA1.1. Single amino acid substitutions were introduced using oligonucleotide-directed, site-specific mutagenesis (13, 14). PCR was used to introduce a hemagglutinin (HA) epitope tag (YPYDYPDYA) after the initiator methionine of each receptor construct. The nucleotide sequences of all receptor constructs were confirmed by automated DNA sequencing.
Luciferase reporter gene assays
Receptor-mediated signaling was assessed using a previously described method with slight modifications (9). Briefly, HEK293 cells were plated at a density of 6,000 cells/well onto clear-bottom, white 96-well plates (Corning 3903) and grown to ∼80% confluence. Cells were transiently transfected in serum-free media using PEI (0.1 µl/well of a 1 mg/ml solution) (15, 16) with cDNAs encoding i) wild-type or mutant HCA1 (or pcDNA1.1, the empty expression vector), ii) an SRE-luciferase reporter gene (SRE5×-luc), iii) Gq5i66V, a chimeric Gαq protein with five carboxyl-terminal amino acids corresponding to Gαi and valine substituted for glycine at amino acid position 66, and iv) a β-galactosidase control to enable correction of interwell variability. In this experimental design, expression of Gαq5i66V directs signaling of a Gαi-coupled receptor to stimulation of a Gαq-dependent SRE5×-luc reporter gene (17–20), thereby providing a robust positive readout that is particularly useful for quantifying basal receptor activity. In these experiments, cells were transfected for 24 h, followed by aspiration of medium and determination of luciferase activity using SteadyLite reagent.
A complementary approach for measuring Gαi-coupled receptor signaling was also utilized. Cells were first stimulated with forskolin to trigger cAMP production; inhibition of this response was used as a quantifiable index of Gαi-mediated signaling. For these studies, cells were transfected with i) wild-type or mutant HCA1 (or pcDNA1.1, the empty expression vector), ii) a CRE-luciferase reporter gene (CRE6×-luc), and iii) a β-galactosidase control to enable correction of interwell variability. The cells were transfected for 24 h, followed by stimulation for 4 h with serum-free media containing 0.5 µM forskolin in either the presence or absence of the HCA1 agonist, l-lactate. The media were then aspirated, and luciferase activity was measured using SteadyLite reagent.
The luciferase assays that were utilized had the advantage of not being sensitive to high concentrations of l-lactate [i.e., no effect on receptor signaling was observed in cells transfected with either the vector control (pcDNA1.1) or with an unrelated Gαi- coupled receptor (mu opioid receptor)]. In contrast, we examined the effects of l-lactate on cAMP signaling utilizing two different cAMP assay kits [Lance Ultra cAMP kit (Perkin Elmer) and HitHunter cAMP XS+ Assay kit (DiscoveRx)]. With either kit, l-lactate nonspecifically decreased the forskolin-stimulated readout when applied to cells expressing pcDNA1.1 (the vector control). This precluded the use of these kits to directly assess cAMP levels in HEK293 cells expressing HCA1 polymorphisms.
In all experiments using luciferase readouts, a β-galactosidase assay was performed following assessment of luminescence. After incubation of cell lysates with 2-nitrophenyl β-d-galactopyranoside at 37°C for 30 to 60 min, cleavage of this substrate was quantified by measuring the optical density at 420 nm using a SpectraMax microplate reader (Molecular Devices, Sunnyvale, CA). Luciferase reporter gene activities in each well were then normalized to corresponding β-galactosidase activity controls.
ELISA assessment of HCA1 expression
Receptor expression levels were determined as previously described (13, 17). In brief, HEK293 cells were grown in clear 96-well plates (BD Biosciences; Bedford, MA) and transiently transfected with a plasmid encoding wild-type or variant receptors with an HA epitope tag. At either 24 or 48 h after transfection (as indicated), the medium was aspirated and cells were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. Cells were then permeabilized with 0.1% Triton X-100 in PBS for 2 min to enable measurement of total receptor expression. To assess HCA1 cell surface expression, parallel assays were performed without permeabilization. After washing with 100 mM glycine in PBS, cells were incubated for 30 min in blocking solution (PBS containing 20% FBS). An HRP-conjugated antibody (Roche; clone 3F10) directed against the HA epitope tag was then added (1:500 dilution in blocking solution). After 2 h, cells were washed five times with PBS. The peroxidase substrate BM-blue was then added at 50 µl per well. After incubation for 30–60 min at room temperature, conversion of this substrate by HRP conjugated to the HA antibody was terminated by addition of 2.0 M sulfuric acid, 50 µl/well. Converted substrate was quantified by measuring absorbance at 450 nm on a SpectraMax microplate reader (Molecular Devices). Absorbance values obtained for HA-tagged receptors were corrected for background by subtracting the absorbance measured in parallel wells transfected with the pcDNA1.1 expression plasmid.
OP9 cell transfection with siRNA
Initial control experiments were carried out to optimize the effectiveness of siRNA knockdown in OP9 cells. OP9 cells were seeded on 6-well plates (Corning 3506) at a density of 62,500 cells/well. The following day, cells were transfected with siRNA using Optifect reagent. In brief, a 30 nM final concentration of RNAi directed against the housekeeping gene GAPDH was mixed with Optifect reagent in OptiMEM media as per the manufacturer's protocol. After incubation, the mix was added to OP9 cells in antibiotic-free propagation media. Cells were transfected overnight. The following day, cells were trypsinized and replated onto 96-well plates at a density of ∼3,500 cells/well in OP9 propagation media. After 24 h, differentiation media was added, and cells were incubated for an additional 3 days. Using quantification of transcript levels by RT-PCR as described below with predesigned primers for GAPDH (Silencer siRNA Starter Kit AM1640), we observed an ∼70–80% knockdown of GAPDH expression levels in cells transfected with GAPDH RNAi. These experiments established that OP9 cells were readily transfectable using this method.
A parallel approach was then applied to induce knockdown of HCA1 expression in OP9 cells. Silencer Select predesigned siRNAs for HCA1 (s109951, s109953, s211558) and Silencer Select negative control siRNA (#439084) were tested. siRNAs were reconstituted in nuclease-free water and maintained as 50 µM stocks. Each of the three siRNAs for HCA1 gave comparable knockdown of gene expression as assessed by RT-PCR (described below); s109951 was used for all subsequent experiments.
For studies involving RNA collection, cells were seeded onto clear-bottom plates (Corning 258600), whereas for cell imaging, black 96-well plates were used (Corning 3606). On experiment day 0 (one day after replating, two days after transfection), RNA was collected in a subset of cells. In the remaining plates, propagation media was aspirated, and differentiation media was added. RNA was collected at multiple subsequent time points. Quantification of Nile Red staining and TLC analysis of lipids were assessed as outlined below.
RNA isolation and qRT-PCR
To isolate RNA, Trizol reagent was used as per the manufacturer's protocol. Following isolation, RNA was DNase treated using a commercially available kit from Ambion/Applied Biosystems. cDNA was synthesized from the DNase-treated RNA using MuLV Reverse Transcriptase and accompanying Applied Biosystems reagents. qRT-PCR assays were performed using an ABI PRISM 7900HT instrument (Applied Biosystems). Primers for HCA1 and 36B4 were designed using NIH primer design software and were synthesized by Integrated DNA Technologies. The forward and reverse primers were as follows: HCA1 5′-ACAGCCCAACAACGAGGCAGAG-3′ and 5′-AGCCATGCAAAGCGACACGG-3′ 36B4 5′-CTTCCAGGCTTTGGGCATCAC-3′ and 5′-CGAAGGAGAAGGGGGAGATGTTC-3′. Standard curves were generated using 0–500 ng of total cDNA. Reactions were performed in triplicate using Sybr Green Master Mix and 0.75 µM of each primer. Thermocycling parameters were as follows: 95°C for 10 min followed by 40 cycles, alternating between 95°C for 15 s and 60°C for 1 min. Melting curves were generated for each sample on all runs to ensure that a single amplicon was generated. The amplicon size was also confirmed by gel electrophoresis. The relative amount of HCA1 mRNA was normalized to the amount of 36B4 mRNA (internal control).
Nile Red imaging and quantification
Stock solutions of 700 µM DAPI and 1 mg/ml Nile Red were prepared in water and acetone, respectively. For Nile Red imaging, the medium was gently removed and the cells were fixed in 4% paraformaldehyde for 20 min at room temperature. Cells were then incubated with DAPI (20 µl/ml of stock in 1:1 glycerol-PBS) for 5 min at room temperature. DAPI was subsequently removed and Nile Red added (1:10,000 dilution in PBS) for a minimum of 25 min. During incubation with Nile Red, plates were stored in foil to avoid exposure to light. An ImageXpress MICRO imaging system (Molecular Devices) was used to image both DAPI and Nile Red staining. Thirty-six grids (898 × 671 µM area per grid) were imaged per well of the 96-well plate. Data from each grid were integrated and analyzed using MetaXpress software (Molecular Devices) with defined settings for cell morphology and Nile Red quantification. Threshold values for Nile Red detection were set to exclude staining of cell membrane lipid. The following parameters were measured and/or calculated for each well: the number of DAPI-positive cells, the percentage of Nile Red-positive cells, the total area covered by Nile Red staining, the overall intensity of Nile Red staining per well, and the intensity of Nile Red staining divided by the number of Nile Red-positive cells (a surrogate marker of cellular lipid content). Data from each experiment were expressed relative to corresponding values in cells transfected with negative-control siRNA. Averages across three independent experiments are reported.
TLC analysis of triglycerides
TLC was performed as previously described with slight modifications (21, 22). The medium was gently removed from the cells, and lipids were extracted with hexane-isopropyl alcohol (3:2). Extracts were evaporated to dryness and resuspended in chloroform-methanol (2:1). Cellular lipids were separated by TLC on silica gel 60 plates and visualized using iodine vapor. The triglyceride band was identified by comparison with a known standard and quantified by densitometry using ImageJ analysis software.
Statistical analysis
EC50 values were determined by nonlinear curve fitting using Graph Pad Prism 5.0 software. Statistical comparisons were made using one-way ANOVA with Tukey's posthoc test. For Nile Red quantification and TLC analysis, a one-sample t-test versus a hypothetical value of 100 was utilized. Data were considered to be statistically significant at P < 0.05.
RESULTS
Human HCA1 variants
Receptor constructs encoding naturally occurring HCA1 missense mutations (H43Q, A110V, S172L, and D253H) were generated. A cartoon representation of corresponding amino acid substitutions in HCA1 is shown in Fig. 1. Genotype frequency for the H43Q variant, as listed in Table 1, was based on the PGA_YORUB-PANEL of a Sub-Saharan African population. Frequencies of the A110V, S172L, and D253H variants were obtained from the NHLBI ESP Exome Variant Server [Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/)]. The amino acid sequences flanking each missense variant were compared between human HCA1 and several species homologs of this receptor using Vector NTI software (Invitrogen). As illustrated in Table 2, the adjacent amino acids were highly conserved.
Fig. 1.
A diagram of HCA1 illustrating the position of missense mutations within the receptor. Respective residues in the wild-type protein are indicated by the corresponding single-letter amino acid code.
TABLE 1.
HCA1 variants with corresponding amino acid alteration, reference identification, nucleotide substitution, frequency, ethnicity, origin of study cohort, and data source
| SNP | Reference | Nucleotide Substitution | Frequency | Ethnicity | Cohort/Panel | Source |
| H43Q | rs35292336 | C/G | 0.053 | Sub-Saharan African | PGA_YORUB-PANEL | dbSNP |
| A110V | rs61747158 | C/T | 32/3706 | African American | ESP_Cohort_Populations | NHLBI Exome Variant Server |
| S172L | rs61742326 | C/T | 22/6998 | European American | ESP_Cohort_Populations | NHLBI Exome Variant Server |
| D253H | rs36124671 | G/C | 71/3667 | African American | ESP_Cohort_Populations | NHLBI Exome Variant Server |
TABLE 2.
Interspecies comparison of amino acid sequences that flank residues affected by HCA1 mutants
| Species | RefSeq | H43Q | A110V | S172L | D253H |
| Homo sapiens | NP_115943 | CGFCFHMKTWK | FLTVVAADRYFK | SFIMESANGWH | PSSACDPSVHG |
| Mus musculus | NP_780729 | CGFCFHMKTWK | FLTVVAV DRYFK | SFIMESANGWH | P T SACDPSVH T |
| Rattus norvegicus | NP_001138806 | CGFCFHMKTWK | FLTVVAV DRYFK | SFIMESANGWH | PSSACDPSVH I |
| Bos taurus | NP_001138706 | CGFCFHMKTWK | FLTVVAV DRYFK | SFIM VSANGWH | PSSACNPSVH V |
Amino acid substitutions resulting from the variant of interest are highlighted in bold. Residues that differ from homo sapiens are underlined.
HCA1 variants exhibit decreased basal and agonist-induced signaling in HEK293 cells
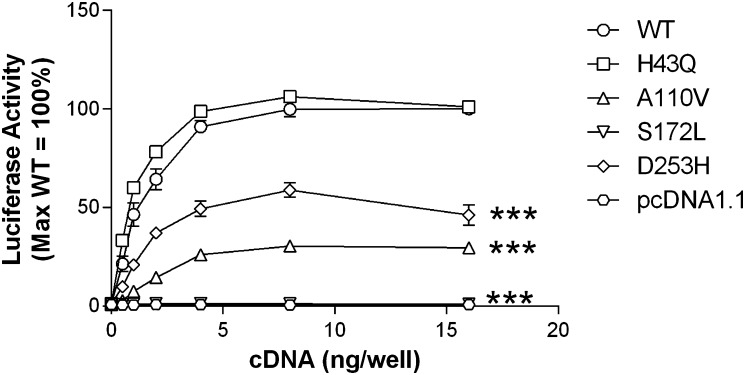
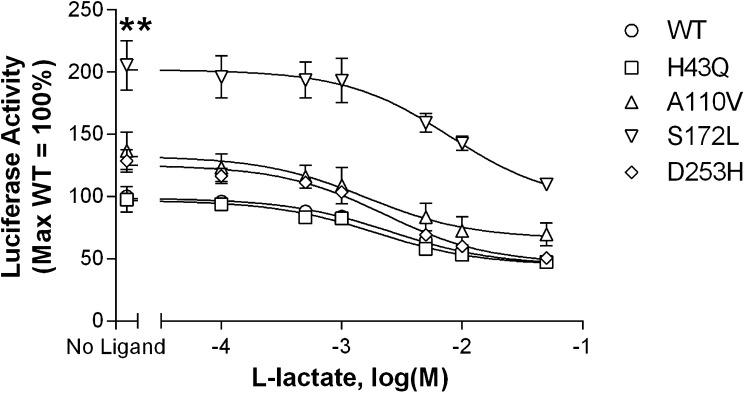
Basal signaling of wild-type versus variant receptors is illustrated in Fig. 2. In this assay, HEK293 cells were transfected with cDNAs encoding an SRE5×-Luc reporter gene construct, a Gαq5i66V chimera, and HCA1 receptors. HCA1 constitutive activity is reflected by an increase in luciferase output, with increasing amounts of transfected receptor cDNA. The wild-type receptor, as well as the H43Q, A110V, and D253H variants, exhibited constitutive activity. A significant decrease in basal activity versus the wild-type receptor was observed with the A110V, S172L, and D253H variants. Notably, constitutive activity was abolished with the S172L substitution. As a complementary measure of receptor function, basal and ligand-stimulated signaling were also assessed using a forskolin inhibition assay with a CRE6×-Luc reporter gene construct. This assay further confirmed the observed decrease in basal activity at the S172L variant. Although not quite reaching significance, the A110V and D253H mutants also showed reduced constitutive activity. Despite the loss of basal activity observed with HCA1 variants, each receptor could be further stimulated by the endogenous ligand l-lactate (Fig. 3). In contrast to the other variants, the S172L isoform showed a significant shift in the l-lactate concentration response curve compared with the wild-type receptor. This is reflected by a 3-fold higher EC50 value, indicating decreased agonist potency (Table 3). In cells transfected with pcDNA1.1 vector control, l-lactate had no effect on forskolin-mediated signaling (data not shown).
Fig. 2.
HCA1 is constitutively active with three receptor variants showing decreased basal activity. HEK293 cells were transiently transfected with cDNAs encoding i) indicated receptors, ii) a SRE5×-Luc reporter gene, iii) a Gαq5i66V chimera, and iv) a β-galactosidase control. Twenty-four hours after transfection, luciferase activity was determined as described in Materials and Methods. Luciferase activity values are expressed as a percentage of wild-type when 16 ng/well of receptor cDNA was transfected (=100%). Data points represent the mean ± SEM from at least three independent experiments, each performed in triplicate. Comparison of wild-type versus variant receptor activity with transfection of 16 ng/well cDNA: ***P < 0.001.
Fig. 3.
All HCA1 variants signal in response to l-lactate stimulation. The S172L isoform shows a significant decrease in basal activity (reflected by the ‘no ligand’ data point). The other variants (A110V, D253H) illustrate a trend toward decreased constitutive activity. HEK293 cells were transiently transfected with 1 ng/well of receptor-encoding cDNA together with a CRE6×-Luc reporter gene and a β-galactosidase control construct. Twenty-four hours after transfection, cells were incubated with 0.5 µM forskolin, with or without increasing concentrations of l-lactate. Luciferase activity was determined as described in Materials and Methods. Activities were normalized relative to the value observed at the wild-type receptor stimulated with forskolin in the absence of l-lactate (no ligand = 100%). Data points represent the mean ± SEM from at least three independent experiments, each performed in triplicate. Comparison of wild-type versus variant receptor activity with no ligand stimulation: **P < 0.01.
TABLE 3.
Agonist potency of l-lactate is decreased at the S172L variant versus the wild-type receptor
| Receptor | EC50 | pEC50 |
| mM | ||
| WT | 2.54 | 2.60 ± 0.10 |
| H43Q | 1.90 | 2.74 ± 0.15 |
| A110V | 1.57 | 2.81 ± 0.14 |
| S172L | 7.88 | 2.08 ± 0.11** |
| D253H | 2.27 | 2.65 ± 0.05 |
Half-maximal effective concentration (EC50 values) for each receptor isoform was calculated and the corresponding pEC50 value compared with the wild-type control: **P < 0.01.
Surface expression of the S172L and D253H variants is decreased
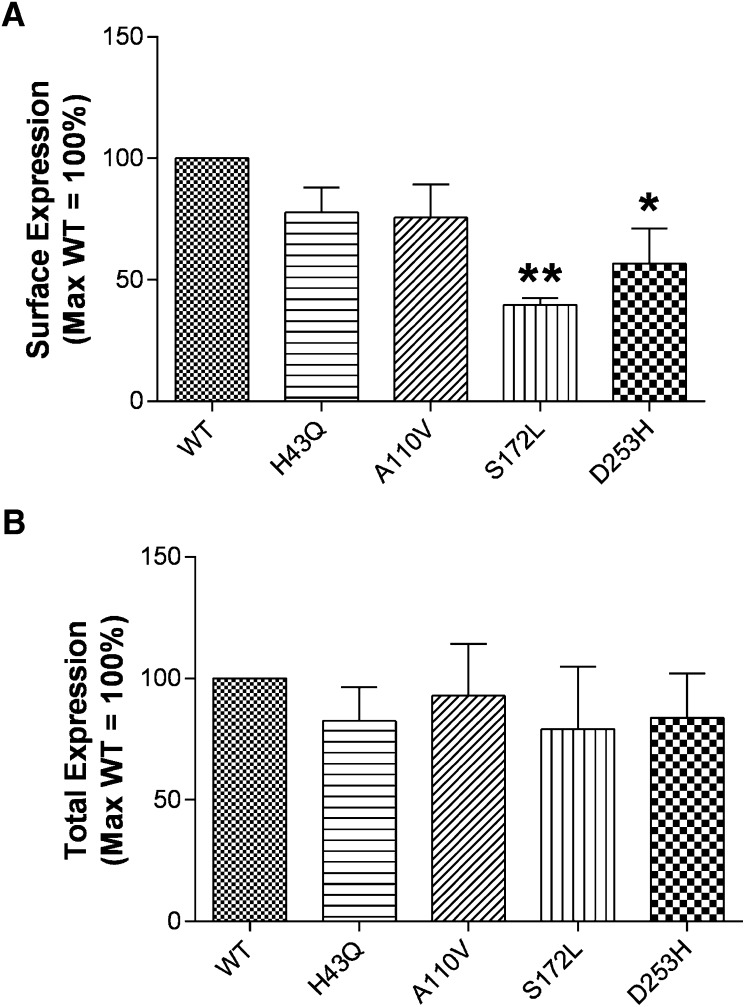
To determine whether the decrease in basal signaling observed with HCA1 variants is attributable to reduced receptor expression, ELISAs were performed on cells transfected with cDNAs encoding HA-tagged receptors. These assays were run with or without cell permeabilization to measure total and surface receptor expression, respectively (Fig. 4). Although there were no changes in total expression, a significant decrease in surface expression was observed with the S172L and D253H variants versus wild-type HCA1 when assessed 24 h after transfection. At 48 h, a significant decrease in S172L surface expression was observed; a similar trend was seen with the D253H variant. It is of note that despite the observed decrease in basal activity of the A110V variant, this receptor showed a level of surface expression comparable to wild-type.
Fig. 4.
Cell surface expression (A) of the S172L and D253H variants is decreased versus wild-type HCA1 with no change in total expression (B). HEK293 cells were transfected with 16 ng/well of cDNA encoding wild-type or mutant HA-tagged receptors. After 24 h, expression levels were measured by ELISA as described in Materials and Methods. Expression data are graphed as a percentage of the corresponding wild-type receptor value. Each bar represents the mean ± SEM from at least three independent experiments, each performed with n ≥ 3 replicates. Comparison of wild-type versus variant receptor expression: *P < 0.05, **P < 0.01.
HCA1 mRNA levels are increased during the differentiation of OP9 cells
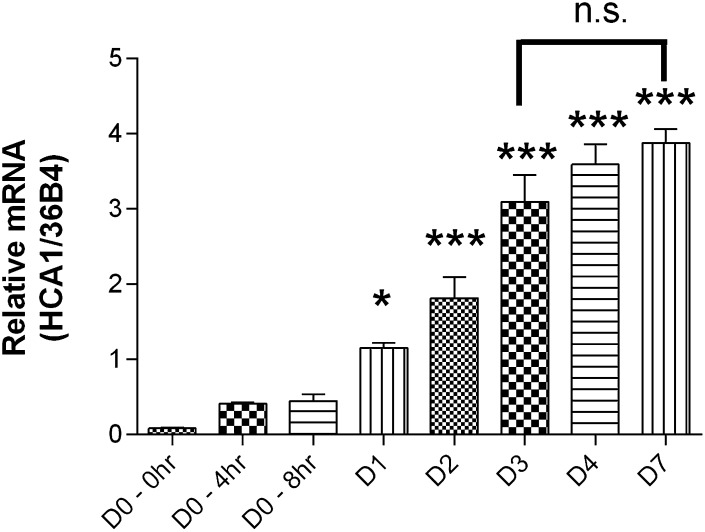
OP9 cells are a preadipocyte cell line that can be experimentally differentiated in vitro to mature adipocytes (12). We observed that this process is accompanied by a marked increase in HCA1 mRNA expression. As shown in Fig. 5, the HCA1 transcript begins to rise significantly at 24 h (day 1) after the initiation of differentiation. The mRNA levels approach a plateau at day 3 and show no significant further increase during the subsequent 4 days. By day 3, insulin-oleate-differentiated OP9 cells are lipid laden and show other markers of fully differentiated adipocytes (12). These features were independently confirmed for OP9 clone K cells (J.L., data not shown). All subsequent experiments were therefore performed using the day 3 time point.
Fig. 5.
HCA1 mRNA levels increase during differentiation of OP9 cells. Over a 7 day period, cells were differentiated using insulin-oleate media as described in Materials and Methods. RNA was harvested on specified days and analyzed using real-time RT-PCR with primers specific for HCA1. The relative amount of HCA1 mRNA in each sample was normalized to the relative amount of 36B4 mRNA (internal control). HCA1 mRNA levels were significantly increased 24 h (D1) after the addition of differentiation media. No significant difference in mRNA levels was observed among days 3, 4, and 7. Bars represent the mean ± SEM, n = 3. Comparison of mRNA levels versus day 0: *P < 0.05, ***P < 0.001.
Knockdown of HCA1 using siRNA during differentiation of OP9 cells increases lipid
The OP9 model was used to determine whether HCA1 knockdown during differentiation altered the adipocyte phenotype. Using the transfection protocol described above, we observed on average a 70% knockdown of HCA1 mRNA levels. Figure 6 illustrates that knockdown of HCA1 during differentiation increased the amount of lipid accumulated on day 3, as assessed by Nile Red staining. To quantify the amount of lipid, we utilized high-throughput imaging software to assess both DAPI staining of nuclei and Nile Red staining of lipid. As illustrated in Table 4, no significant changes in DAPI-positive cells were observed with HCA1 knockdown. In contrast, the percentage of cells that were Nile Red positive (i.e., lipid was detected above threshold) was increased compared with the control group. The overall intensity of Nile Red staining per well was also significantly enhanced after HCA1 knockdown, as was the amount of lipid deposited per cell (i.e., the intensity of Nile Red divided by the number of Nile Red-positive cells). A trend toward an increase in the total area of Nile Red staining (reflecting the combined area of lipid droplets) was also observed (P = 0.098). Comparing the images of negative control versus HCA1 siRNA-treated cells demonstrates that knockdown of HCA1 during differentiation enhances lipid accumulation. Both an increase in the number of cells depositing lipid and an increase in the amount of lipid per cell contribute to this phenotype.
Fig. 6.
Transfection of OP9 cells with HCA1 silencing RNAi increases differentiation-induced lipid accumulation. Representative images of Nile Red and DAPI staining in OP9 cells after 3 days of incubation in differentiation media.
TABLE 4.
Quantification of OP9 Nile Red staining and TLC analysis of triglycerides on differentiation day 3
| Parameter | Negative-Control siRNA | HCA1 siRNA | P |
| DAPI-positive cells | 100 | 90.7 ± 9.09 | 0.415 |
| % Nile Red-positive cells | 100 | 115 ± 3.11* | 0.040 |
| Total area of Nile Red staining | 100 | 130 ± 10.2 | 0.098 |
| Integrated intensity of Nile Red | 100 | 140 ± 8.95* | 0.046 |
| Integrated intensity of Nile Red/number of Nile Red-positive cells | 100 | 136 ± 4.20* | 0.014 |
| TLC analysis | 100 | 118 ± 5.24* | 0.04 |
Data reflect analysis of images analogous to those shown in Fig. 6 and are presented as a percentage of negative-control siRNA values. Each value represents the mean ± SEM from three to four independent experiments, each performed with n = 2–3 replicates. Comparison of negative control versus HCA1 siRNA: *P < 0.05.
As a complementary approach to quantifying lipid by Nile Red staining, we also assessed the amount of triglyceride on day 3 using TLC. Lipids were measured in cells where HCA1 was knocked down during differentiation. Quantification of bands on the TLC plates revealed a significant increase in triglyceride between negative control versus HCA1 siRNA-treated cells (P = 0.04, Table 4).
DISCUSSION
In the current study, we examined the pharmacological consequences resulting from four naturally occurring missense mutations in HCA1. Initial assessment focused on changes in constitutive activity. A comparison with the wild-type receptor revealed that three naturally occurring receptor isoforms (A110V, S172L, and D253H) cause a significant decrease in basal signaling. The reduced level of constitutive activity observed in cells expressing the S172L and D253H mutants may in part be explained by the observed decrease in cell surface expression. It is possible that these missense mutations lead to misfolding or structural instability of the receptor with consequent intracellular retention (23). It is unlikely that these receptors are degraded intracellularly, inasmuch as total expression of these variants is unchanged. In contrast to the S172L and D253H missense mutations, the A110V variant displayed a decrease in basal activity with no accompanying change in surface expression. Given the location of this variant within the second intracellular loop, a well-established site of GPCR-G protein interaction (24, 25), the reduced level of constitutive signaling with the A110V variant may be related to altered G protein coupling.
In addition to the changes in basal activity and expression observed for the S172L variant, this receptor showed a significant decrease in potency when stimulated with l-lactate, the endogenous agonist. The observed shift in EC50 may be explained by a modification of the ligand binding site and/or defective transitioning from the inactive to the active receptor state (26, 27). Involvement of the latter mechanism is suggested by the concurrent loss of constitutive activity. According to the extended ternary model of receptor function, diminished basal activity is often paralleled by reduced ligand potency (26). Alternatively, given that the affected residue is located in the second extracellular loop of HCA1, it is plausible that the S172L substitution directly or indirectly affects the binding of l-lactate and subsequent receptor activation. Supporting this possibility, S172L is in close proximity to a highly conserved amino acid motif (residues 165–168) that is implicated in ligand binding and HCA1 function (28).
Given the interest in developing HCA1 agonists for the treatment of dyslipidemia (29), it will be of interest to determine whether the efficacy and/or potency of novel ligands are affected by the variants highlighted in this report. The activity of new drugs may potentially be compromised by HCA1 mutants (as observed with l-lactate at the S172L substitution). Alternatively, compounds may rescue the loss-of-function (reduced basal signaling and receptor expression, diminished responsiveness to endogenous agonist) resulting from selected naturally occurring variations in this receptor. Studying the impact of receptor polymorphisms on the activity of synthetic HCA1 agonists may be a factor in selecting which molecules to develop and/or identify patient populations that could potentially benefit most from treatment with HCA1-targeted therapeutics.
Our observation that several HCA1 variants compromise receptor function raised the question of how reduced receptor signaling may modify lipid accumulation in adipocytes. As a model system to explore this potential link, we assessed the consequences of knocking down endogenous HCA1 expression during the differentiation of OP9 cells. With this model system, an insulin-oleate cocktail is sufficient to initiate the transition from premature to mature adipocytes. The addition of compounds that may affect cell and/or HCA1 function by unrelated pharmacological mechanisms may therefore be avoided. Such reagents include thiazolidinediones and 1-methyl-3-isobutylxanthine (IBMX), which are commonly used in other models of adipocyte differentiation (e.g., 3T3-L1 cells, primary human adipocytes, human multipotent adipose-derived stem cells, and human Simpson-Golabi Behmel syndrome cells) (30–32). It is known that thiazolidinediones like rosiglitazone pharmacologically upregulate HCA1 through activation of a PPAR-retinoid X receptor binding site in the proximal promoter of the HCA1 gene (33). IBMX enhances cAMP accumulation via inhibition of phosphodiesterase, thus potentially interfering with HCA1-induced signaling through the same second-messenger pathway. Use of either reagent could thus confound the interpretation of our studies, which were designed to explore how HCA1 expression influences lipid deposition in adipocytes during the differentiation process.
We have shown that the expression of HCA1 robustly increases as OP9 cells acquire an adipocyte phenotype. Receptor transcript levels plateau after 3 days of incubation in insulin-oleate-containing media. During this process, terminal adipocyte differentiation occurs and can be morphologically quantified using this model system. We also demonstrate a feasible approach for efficient knockdown of genes prior to OP9 differentiation and a parallel assay to examine lipid accumulation. Application of these technologies enabled us to demonstrate that reduced expression of HCA1 during adipocyte differentiation results in an increase in lipid deposition. Both the number of cells that accumulate lipid as well as the amount of lipid per cell were enhanced, reflecting hyperplasia and hypertrophy, respectively (34). Adipocyte hyperplasia can be protective against both lipid and insulin dysregulation, whereas adipocyte hypertrophy is a characteristic of obesity and can lead to the development of insulin resistance (35–38). In light of our findings, it is of interest to note that genetically obese mice (ob/ob) have a dramatic decrease in HCA1 mRNA levels (39). It remains to be determined, however, whether the predominant effect of HCA1 signaling is beneficial or detrimental to human health.
We have shown that taken together, selected missense variants in HCA1 result in loss of function and that reduced signaling of this receptor promotes enhanced lipid accumulation in a cellular model of preadipocyte differentiation. There is increased interest in understanding the phenotypic consequences of naturally occurring polymorphisms and missense mutations in GPCRs (40–42). As the clinical correlates of rare variants across the genome are defined, it will be of interest to examine how loss-of-function mutations in HCA1 modify lipid deposition, dyslipidemia, and insulin resistance. These data may help to guide the development of clinically useful HCA1-targeted drugs and to better understand the role of HCA1 in human physiology and disease.
Acknowledgments
The authors would like to acknowledge the National Institutes of Health National Heart, Lung, and Blood Institute GO Exome Sequencing Project for providing an online database that includes sequence variations in HCA1. This consortium includes: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926), and the Heart GO Sequencing Project (HL-103010). The authors would like to thank Ci Chen (Molecular Cardiology Research Institute, Tufts Medical Center) and Albert Tai (Study Center on the Immunogenetics of Infectious Disease core facility, Tufts University) for excellent technical assistance, as well as Colleen McNamara and Amanda Doran for advice on OP9 cell maintenance and differentiation. The authors would also like to thank Laura Liscum (Departments of Physiology and Neuroscience, Tufts University) for help with thin-layer chromatography analysis of triglycerides.
Footnotes
Abbreviations:
- CRE
- cAMP response element
- DAPI
- 4’,6-diamidino-2-phenylindole
- GPCR
- G protein-coupled receptor
- HA
- hemagglutinin
- HCA1
- hydroxy-carboxylic acid receptor 1
- HEK
- human embryonic kidney
- IBMX
- 1-methyl-3-isobutylxanthine
- PEI
- polyethylenimine
- SNP
- single-nucleotide polymorphism
- SRE
- serum-response element
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases [Grant R01-DK072497] and the National Heart Lung and Blood Institute [Grant T32 HL069770]. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
REFERENCES
- 1.Ge H., Weiszmann J., Reagan J. D., Gupte J., Baribault H., Gyuris T., Chen J. L., Tian H., Li Y. 2008. Elucidation of signaling and functional activities of an orphan GPCR, GPR81. J. Lipid Res. 49: 797–803 [DOI] [PubMed] [Google Scholar]
- 2.Liu C., Wu J., Zhu J., Kuei C., Yu J., Shelton J., Sutton S. W., Li X., Yun S. J., Mirzadegan T., et al. 2009. Lactate inhibits lipolysis in fat cells through activation of an orphan G-protein-coupled receptor, GPR81. J. Biol. Chem. 284: 2811–2822 [DOI] [PubMed] [Google Scholar]
- 3.Offermanns S., Colletti S. L., Lovenberg T. W., Semple G., Wise A., IJzerman A. P. 2011. International Union of Basic and Clinical Pharmacology. LXXXII: nomenclature and classification of hydroxy-carboxylic acid receptors (GPR81, GPR109A, and GPR109B). Pharmacol. Rev. 63: 269–290 [DOI] [PubMed] [Google Scholar]
- 4.Ahmed K., Tunaru S., Offermanns S. 2009. GPR109A, GPR109B and GPR81, a family of hydroxy-carboxylic acid receptors. Trends Pharmacol. Sci. 30: 557–562 [DOI] [PubMed] [Google Scholar]
- 5.Offermanns S. 2006. The nicotinic acid receptor GPR109A (HM74A or PUMA-G) as a new therapeutic target. Trends Pharmacol. Sci. 27: 384–390 [DOI] [PubMed] [Google Scholar]
- 6.Wise A., Foord S. M., Fraser N. J., Barnes A. A., Elshourbagy N., Eilert M., Ignar D. M., Murdock P. R., Steplewski K., Green A., et al. 2003. Molecular identification of high and low affinity receptors for nicotinic acid. J. Biol. Chem. 278: 9869–9874 [DOI] [PubMed] [Google Scholar]
- 7.Cai T. Q., Ren N., Jin L., Cheng K., Kash S., Chen R., Wright S. D., Taggart A. K., Waters M. G. 2008. Role of GPR81 in lactate-mediated reduction of adipose lipolysis. Biochem. Biophys. Res. Commun. 377: 987–991 [DOI] [PubMed] [Google Scholar]
- 8.Ahmed K., Tunaru S., Tang C., Muller M., Gille A., Sassmann A., Hanson J., Offermanns S. 2010. An autocrine lactate loop mediates insulin-dependent inhibition of lipolysis through GPR81. Cell Metab. 11: 311–319 [DOI] [PubMed] [Google Scholar]
- 9.Al-Fulaij M. A., Ren Y., Beinborn M., Kopin A. S. 2007. Identification of amino acid determinants of dopamine 2 receptor synthetic agonist function. J. Pharmacol. Exp. Ther. 321: 298–307 [DOI] [PubMed] [Google Scholar]
- 10.Fortin J. P., Schroeder J. C., Zhu Y., Beinborn M., Kopin A. S. 2010. Pharmacological characterization of human incretin receptor missense variants. J. Pharmacol. Exp. Ther. 332: 274–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doran A. C., Meller N., Cutchins A., Deliri H., Slayton R. P., Oldham S. N., Kim J. B., Keller S. R., McNamara C. A. 2008. The helix-loop-helix factors Id3 and E47 are novel regulators of adiponectin. Circ. Res. 103: 624–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolins N. E., Quaynor B. K., Skinner J. R., Tzekov A., Park C., Choi K., Bickel P. E. 2006. OP9 mouse stromal cells rapidly differentiate into adipocytes: characterization of a useful new model of adipogenesis. J. Lipid Res. 47: 450–460 [DOI] [PubMed] [Google Scholar]
- 13.Doyle J. R., Fortin J. P., Beinborn M., Kopin A. S. 2012. Selected melanocortin 1 receptor single-nucleotide polymorphisms differentially alter multiple signaling pathways. J. Pharmacol. Exp. Ther. 342: 318–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortin J. P., Zhu Y., Choi C., Beinborn M., Nitabach M. N., Kopin A. S. 2009. Membrane-tethered ligands are effective probes for exploring class B1 G protein-coupled receptor function. Proc. Natl. Acad. Sci. USA. 106: 8049–8054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrhardt C., Schmolke M., Matzke A., Knoblauch A., Will C., Wixler V., Ludwig S. 2006. Polyethylenimine, a cost-effective transfection agent. Signal Transduct. 6: 179–184 [Google Scholar]
- 16.Boussif O., Lezoualc'h F., Zanta M. A., Mergny M. D., Scherman D., Demeneix B., Behr J. P. 1995. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. USA. 92: 7297–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Fulaij M. A., Ren Y., Beinborn M., Kopin A. S. 2008. Pharmacological analysis of human D1 and D2 dopamine receptor missense variants. J. Mol. Neurosci. 34: 211–223 [DOI] [PubMed] [Google Scholar]
- 18.Conklin B. R., Farfel Z., Lustig K. D., Julius D., Bourne H. R. 1993. Substitution of three amino acids switches receptor specificity of Gq alpha to that of Gi alpha. Nature. 363: 274–276 [DOI] [PubMed] [Google Scholar]
- 19.Fortin J. P., Ci L., Schroeder J., Goldstein C., Montefusco M. C., Peter I., Reis S. E., Huggins G. S., Beinborn M., Kopin A. S. 2010. The mu-opioid receptor variant N190K is unresponsive to peptide agonists yet can be rescued by small-molecule drugs. Mol. Pharmacol. 78: 837–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kostenis E., Martini L., Ellis J., Waldhoer M., Heydorn A., Rosenkilde M. M., Norregaard P. K., Jorgensen R., Whistler J. L., Milligan G. 2005. A highly conserved glycine within linker I and the extreme C terminus of G protein alpha subunits interact cooperatively in switching G protein-coupled receptor-to-effector specificity. J. Pharmacol. Exp. Ther. 313: 78–87 [DOI] [PubMed] [Google Scholar]
- 21.Liscum L., Faust J. R. 1987. Low density lipoprotein (LDL)-mediated suppression of cholesterol synthesis and LDL uptake is defective in Niemann-Pick type C fibroblasts. J. Biol. Chem. 262: 17002–17008 [PubMed] [Google Scholar]
- 22.Goldstein J. L., Basu S. K., Brown M. S. 1983. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 98: 241–260 [DOI] [PubMed] [Google Scholar]
- 23.Conn P. M., Ulloa-Aguirre A. 2010. Trafficking of G-protein-coupled receptors to the plasma membrane: insights for pharmacoperone drugs. Trends Endocrinol. Metab. 21: 190–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J., Wang Y., Zhang X., Lloyd J. R., Li J. H., Karpiak J., Costanzi S., Wess J. 2010. Structural basis of G protein-coupled receptor-G protein interactions. Nat. Chem. Biol. 6: 541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wess J. 1998. Molecular basis of receptor/G-protein-coupling selectivity. Pharmacol. Ther. 80: 231–264 [DOI] [PubMed] [Google Scholar]
- 26.Lefkowitz R. J., Cotecchia S., Samama P., Costa T. 1993. Constitutive activity of receptors coupled to guanine nucleotide regulatory proteins. Trends Pharmacol. Sci. 14: 303–307 [DOI] [PubMed] [Google Scholar]
- 27.Beinborn M., Ren Y., Blaker M., Chen C., Kopin A. S. 2004. Ligand function at constitutively active receptor mutants is affected by two distinct yet interacting mechanisms. Mol. Pharmacol. 65: 753–760 [DOI] [PubMed] [Google Scholar]
- 28.Kuei C., Yu J., Zhu J., Wu J., Zhang L., Shih A., Mirzadegan T., Lovenberg T., Liu C. 2011. Study of GPR81, the lactate receptor, from distant species identifies residues and motifs critical for GPR81 functions. Mol. Pharmacol. 80: 848–858 [DOI] [PubMed] [Google Scholar]
- 29.Liu C., Kuei C., Zhu J., Yu J., Zhang L., Shih A., Mirzadegan T., Shelton J., Sutton S., Connelly M. A. 2012. 3,5-Dihydroxybenzoic acid, a specific agonist for HCA1, inhibits lipolysis in adipocytes. J. Pharmacol. Exp. Ther. 341: 794–801 [DOI] [PubMed] [Google Scholar]
- 30.Ntambi J. M., Young-Cheul K. 2000. Adipocyte differentiation and gene expression. J. Nutr. 130: 3122S–3126S [DOI] [PubMed] [Google Scholar]
- 31.Wabitsch M., Brenner R. E., Melzner I., Braun M., Moller P., Heinze E., Debatin K. M., Hauner H. 2001. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int. J. Obes. Relat. Metab. Disord. 25: 8–15 [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez A. M., Elabd C., Delteil F., Astier J., Vernochet C., Saint-Marc P., Guesnet J., Guezennec A., Amri E. Z., Dani C., et al. 2004. Adipocyte differentiation of multipotent cells established from human adipose tissue. Biochem. Biophys. Res. Commun. 315: 255–263 [DOI] [PubMed] [Google Scholar]
- 33.Jeninga E. H., Bugge A., Nielsen R., Kersten S., Hamers N., Dani C., Wabitsch M., Berger R., Stunnenberg H. G., Mandrup S., et al. 2009. Peroxisome proliferator-activated receptor gamma regulates expression of the anti-lipolytic G-protein-coupled receptor 81 (GPR81/Gpr81). J. Biol. Chem. 284: 26385–26393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jo J., Gavrilova O., Pack S., Jou W., Mullen S., Sumner A. E., Cushman S. W., Periwal V. 2009. Hypertrophy and/or hyperplasia: dynamics of adipose tissue growth. PLOS Comput. Biol. 5: e1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arner E., Westermark P. O., Spalding K. L., Britton T., Ryden M., Frisen J., Bernard S., Arner P. 2010. Adipocyte turnover: relevance to human adipose tissue morphology. Diabetes. 59: 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arner P., Bernard S., Salehpour M., Possnert G., Liebl J., Steier P., Buchholz B. A., Eriksson M., Arner E., Hauner H., et al. 2011. Dynamics of human adipose lipid turnover in health and metabolic disease. Nature. 478: 110–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffstedt J., Arner E., Wahrenberg H., Andersson D. P., Qvisth V., Lofgren P., Ryden M., Thorne A., Wiren M., Palmer M., et al. 2010. Regional impact of adipose tissue morphology on the metabolic profile in morbid obesity. Diabetologia. 53: 2496–2503 [DOI] [PubMed] [Google Scholar]
- 38.Meissburger B., Stachorski L., Roder E., Rudofsky G., Wolfrum C. 2011. Tissue inhibitor of matrix metalloproteinase 1 (TIMP1) controls adipogenesis in obesity in mice and in humans. Diabetologia. 54: 1468–1479 [DOI] [PubMed] [Google Scholar]
- 39.Feingold K. R., Moser A., Shigenaga J. K., Grunfeld C. 2011. Inflammation inhibits GPR81 expression in adipose tissue. Inflam. Res. 60: 991–995 [DOI] [PubMed] [Google Scholar]
- 40.Farooqi I. S., O'Rahilly S. 2007. Genetic factors in human obesity. Obes. Rev. 8 (Suppl.): 37–40 [DOI] [PubMed] [Google Scholar]
- 41.Gibson G. 2011. Rare and common variants: twenty arguments. Nat. Rev. Genet. 13: 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seifert R., Wenzel-Seifert K. 2002. Constitutive activity of G-protein-coupled receptors: cause of disease and common property of wild-type receptors. Naunyn Schmiedebergs Arch. Pharmacol. 366: 381–416 [DOI] [PubMed] [Google Scholar]