Abstract
The discovery of methods to convert somatic cells into induced pluripotent stem cells (iPSCs) through expression of a small combination of transcription factors has raised the possibility of producing custom-tailored cells for the study and treatment of numerous diseases. Indeed, iPSCs have already been derived from patients suffering from a large variety of disorders. Here we review recent progress that has been made in establishing iPSC-based disease models, discuss associated technical and biological challenges, and highlight possible solutions to overcome these barriers. We believe that a better understanding of the molecular basis of pluripotency, cellular reprogramming and lineage-specific differentiation of iPSCs is necessary for progress in regenerative medicine.
Medical advances within the past century, such as the discovery of antibiotics and the development of vaccines, have led to remarkable breakthroughs in our ability to treat and even cure some of the most challenging ailments. The recent finding that pluripotency can be induced in somatic cells may represent yet another key discovery in the area of drug discovery and cell-based therapy.
The search for a method to induce developmental reprogramming of a somatic cell into an embryonic state stems from seminal frog studies that demonstrated that differentiated cell nuclei introduced into enucleated oocytes support the development of genetically identical animals or clones1–3. Cloned animals were also later produced in mammalian species4–9. However, the identity of the cocktail of factors from the oocyte cytoplasm that was reverting the differentiated nucleus to its primitive state remained elusive. By systematically examining the effect of pluripotency-specific transcription factors on fibroblasts, Takahashi and Yamanaka discovered in 2006 that retroviral expression of a set of four genes (Oct4, Sox2, Klf4 and c-Myc) converted somatic cells into a pluripotent state, albeit at an extremely low efficiency10. These iPSCs exhibited transcriptional and epigenetic features that were highly similar to those of embryonic stem cells (ESCs)11–13. Different groups subsequently repeated these findings with human cells14–16.
Remarkable progress made in reprogramming technology over the past few years has facilitated the generation of virus-free and/or vector-free iPSCs, eliminating the potential risk of virally-induced tumour formation17–23. iPSCs have been derived at increased efficiencies from several easily accessible human cell types, including blood cells, keratinocytes and dermal fibroblasts23–27. These and other advances now allow basic and translational scientists to develop strategies for the use of iPSC technology in disease modelling and drug screening, and could enable autologous cell transplantation in clinical therapy in the future. As our understanding of the inherent similarities and differences between ESCs and iPSCs improve3, we will be better equipped to tackle challenges that have hampered the use of ESCs in clinical and translational applications thus far (see Box 1).
Here, we explore the growing interest in using disease-specific iPSCs as an in vitro platform for drug screening and disease pathway discovery. Given the potential for iPSCs to serve as a source of cell replacement in degenerative diseases, we will also discuss recent preclinical animal studies using iPSC derivatives in cell-based therapy and outline the challenges to be overcome before the full potential of iPSC technology can be realized in pharmaceutical and clinical applications.
iPSC-based disease modelling
The ability to generate pluripotent cell lines from patients afflicted with diseases of known and suspected aetiologies should allows us to obtain, in theory, genetically matched cell types from all major organs of interest in unlimited quantity. Indeed, recent studies have described the generation of iPSC lines from patients with a full range of genetically inherited as well as sporadic diseases (Table 1). In most cases, in vitro differentiation of iPSCs to the cell type relevant to the disorder has been reported, and there are now many studies that suggest that patient-specific iPSCs exhibit certain disease features. For example, a progressive loss of motor neurons was observed during in vitro differentiation of iPSCs derived from spinal muscular atrophy (SMA) patients, which may reflect the developmental loss of motor neurons seen during this disease28. Similarly, cardiomyocytes derived from iPSCs from patients with LEOPARD syndrome were found to be enlarged, possibly reflecting the hypertrophic cardiomyopathy associated with this disease29. Patients suffering from Long QT and Timothy syndrome exhibit increased QT intervals on electrocardiography, and differentiated cardiomyocytes produced from iPSCs from such patients had prolongation of action potentials in single-cell electrophysiological assays30,31. iPSCs derived from methyl CpG binding protein 2 (MeCP2)-deficient female patients with RETT syndrome give rise to glutamatergic neurons with fewer synapses and decreased calcium transients when compared with controls, as is usually seen in RETT patients32. Familial dysautonomia-derived iPSCs exhibit decreased neurogenic differentiation and migration behaviours, compared with control iPSCs33. Two recent studies investigated the disease phenotypes of iPSCs derived from Hutchinson-Gilford progeria patients and found that the differentiated smooth muscle cells had premature senescence, demonstrating that vascular defects seen in patients could also be observed in vitro34,35. Because some of these studies were performed with rather low numbers of iPSC lines or used ESC lines instead of iPSC lines from unaffected individuals as controls (Table 1), it remains an open question how reproducible the observed phenotypes are when larger sets of genetically matched patient and control cell lines are being compared with each other. Nevertheless, these in vitro studies provide the first proof-of-principle that disease modelling using iPSC technology may indeed be feasible.
Table 1.
Summary of published human iPSC disease models
| Disease type | Disease name | Genetic cause | Number of lines | Cell type | Control line | Phenotype | Drug test | Reference |
|---|---|---|---|---|---|---|---|---|
| Neurological | Parkinson's disease | Polygenic | 23 | Dopaminergic neurons | hiPSC | No obvious defect | ND | 20 |
| Polygenic (with LRRK2 mutation) | 4 | Dopaminergic neurons | hiPSC | Neuronal death with chemicals | Yes | 51 | ||
| Amyotrophic lateral sclerosis | Polygenic | 3 | Motor neurons | hESC | ND | ND | 107 | |
| Spinal muscular atrophy | Monogenic | 2 | Motor neurons | hiPSC | Loss of neuron formation, loss of SMN gene expression | Yes | 28 | |
| Familial dysautonomia | Monogenic | 2 | Neural crest cells | hiPSC, hESC | Loss of neural crest cells | Yes | 33 | |
| RETT syndrome | Monogenic | 4 | Neurons | hiPSC | Loss of synapses, reduced spine density, smaller soma size | Yes | 32 | |
| Huntington's disease | Monogenic | 2 | ND | hiPSC, hESC | ND | ND | 108 | |
| Friedreich ataxia | Monogenic | 6+ | ND | hESC | Changes GAA-TTC repeat | ND | 109 | |
| Blood | Fanconi anaemia | Monogenic | 19 | Blood cell | hiPSC, hESC | Corrected loss of FANCA function | ND | 110 |
| Fragile X syndrome | Monogenic | 11 | ND | hiPSC, hESC | Loss of FMR1 expression | ND | 111 | |
| Cardiac and vascular | Long QT 1 syndrome | Monogenic | 6 | Cardiomyocytes | hiPSC | Increased cardiomyocyte depolarization | Yes | 30 |
| Long QT 2 syndrome | Monogenic | Not reported | Cardiomyocytes | hiPSC | Increased cardiomyocyte depolarization | Yes | 112 | |
| LEOPARD syndrome | Monogenic | 6 | Cardiomyocytes | hiPSC, hESC | Increased cardiomyocyte size, decreased MAPK signalling | ND | 29 | |
| Timothy syndrome | Monogenic | 16 | cardiomyocytes | hiPSC | Increased cardiomyocyte depolarization | Yes | 31 | |
| Hutchinson Gilford Progeria | Monogenic | 4 | Smooth muscle cells, mesenchymal stem cells | hiPSC, hESC | Smooth muscle and mesenchymal cell apoptosis | ND | 34 | |
| Monogenic | 6 | Smooth muscle cells | hiPSC | Smooth muscle cell nuclear morphology and ageing phenotype | ND | 35 | ||
| Duchenne muscular dystrophy | Monogenic | 2 | ND | hiPSC, hESC | ND | ND | 108 | |
| Pancreatic | Type 1 diabetes | Polygenic | 4 | Insulin- and glucagon-producing cells | hESC | ND | ND | 113 |
| Hepatic | A1-antitrypsin deficiency | Monogenic | 19 | Hepatocytes | hiPSC | Loss of A1-antitrypsin expression | Yes | 114 |
| Others | Prader-Willi syndrome | Monogenic | 4 | Neurons | hiPSC, hESC | Imprint disorder | ND | 115 |
| Angelman and Prader-Willi syndrome | Monogenic | 13 | Neurons | hiPSC, hESC | Loss of paternal UBE3A expression | ND | 116 | |
| Down syndrome | Monogenic | 2 | ND | hiPSC, hESC | ND | ND | 108 | |
ND – not determined
Although production of disease phenotypes from differentiated iPSCs in vitro is the necessary first step towards disease modelling, the identification of novel pathways or drugs that could affect the disease process is the ultimate goal of this approach (Fig. 1). The loss of neurons associated with in vitro differentiation of iPSCs derived from SMA patients was ameliorated by treatment with small-molecule candidates that reverse disease features in other neuronal culture assays28. Likewise, the sensory neuron defect of familial-dysautonomia-iPSCs and the synapse defect of RETT-iPSCs were partially restored on exposure of cells to previously reported candidate drugs32,33. Thus, iPSCs from human patients suffering from various diseases can be used to bridge the gap between small animal models, which may not always reflect the true human disease phenotype, and clinical testing, which is expensive and time consuming. However, it would be premature to assume that small molecules active in iPSC-based assays in vitro would be immediately ready for clinical studies in humans. Further evaluation of their ADME (absorption, distribution, metabolism and excretion) properties and of their toxicity and efficacy will need to be done in animal models if they have not been tested previously in human patients.
Figure 1.
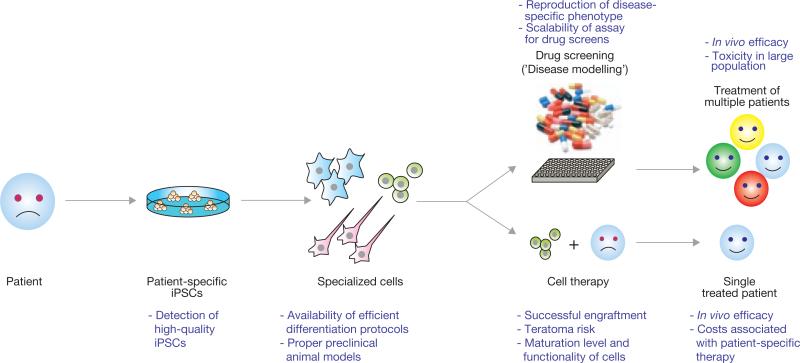
Schematic representation of the potential utility of iPSC technology in regenerative medicine. Introduction of reprogramming factors, such as Oct4, Sox2, Klf4 and c-Myc, into somatic cells of patients (for example, skin cells, keratinocytes or blood cells) gives rise to iPSCs. These patient-specific iPSCs can then be differentiated into a variety of specialized cell types for a potential use in disease modelling (top) or cell therapy (bottom). The concept behind disease modelling is to reproduce a cellular phenotype in cultured iPSC-derived cells as it occurs in the patient. Such a phenotype could be employed to model this disease for mechanistic studies as well as for large-scale drug screening efforts to identify compounds that could be used to treat any patient suffering from the same disease. The idea behind cell therapy is to generate autologous specialized cells from iPSCs for transplantation into individual patients. Shown in purple are the current limitations in using iPSC technology in regenerative medicine.
It is worth noting that there have been no published studies that use diseased iPSCs in a high-throughput screening platform to discover novel small molecules that can potentially reverse a disease phenotype. However, given the converging interests of chemical and stem cell biology, successful applications of such therapeutic screens using diseased iPSCs are expected in the near future. Besides providing a powerful tool for drug discovery, iPSC technology may allow researchers to model pre-symptomatic abnormalities in patient-derived cells that could yield valuable insights into disease mechanisms and may lead to the development of diagnostic tools and drugs for early intervention.
iPSCs in cell therapy
One of the most exciting aspects of iPSC technology is the possibility of generating autologous cells for cell-replacement therapy (Fig. 1). The somatic origin of iPSCs has minimized but not eliminated some of the challenges that have hampered the development of human ESC-based therapies. As cell transplantation for tissue repair outside of the haematopoietic and skin systems is a relatively nascent area of investigation, the regulatory requirements for the approval of pluripotent stem cell-derived cells in clinical studies remain extremely high. A recent trial by the biopharmaceutical company Geron provides insights into the challenges that lie ahead (see Box 1).
Ultimately, our ability to bring pluripotent stem cell biology into cell-based therapy will depend on the efficiency of cell-lineage-specific differentiation, efficiency of cell purification to eliminate the risk of teratoma, and development of novel cell delivery methods to introduce cells of interest into relevant organs (Fig. 1). Despite a decade of research on the mechanisms driving human pluripotent stem cell differentiation, it remains challenging to reliably generate large quantities of well-differentiated and functional cells from human ESCs or iPSCs.
With regards to transplantation of iPSCs for therapeutic regeneration, the most compelling study so far showed that haematopoietic cells derived from iPSCs can reduce the blood cell phenotype in a humanized mouse model of sickle cell anemia36. iPSCs were derived from a transgenic mouse carrying a mutation in the human haemoglobin sequence and then genetically corrected through homologous recombination. In vitro differentiation of the ‘corrected’ iPSCs into haematopoietic progenitors and subsequent transplantation into the original transgenic mice resulted in restoration of normal haemoglobin levels and an improved phenotype. Although this rescue is remarkable, the fact that retroviral HoxB4-transduced haematopoietic progenitor cells were used is a caveat, and it remains to be seen whether similar rescue effects can be obtained using non-HoxB4 transduced cells. More importantly, a bona fide haematopoietic stem cell with the capacity for long-term multilineage reconstitution has yet to be generated from human iPSCs. Thus, the translational potential of this strategy for sickle cell anaemia patients remains unclear.
Similar transplantation-based approaches have been reported for other organs. For example, partially-purified dopaminergic neurons derived from mouse iPSCs improved the clinical symptoms of a rat model of Parkinson's disease37. Likewise, transplantation of human iPSC-derived cells into experimentally injured rodent heart showed some degree of short-term functional improvement in cardiac contractile function38,39.
These examples of successful transplantation of iPSC-derived mesodermal and ectodermal cells into animals involved the cell lineages that are more easily produced from pluripotent stem cells (for example, neurons, blood and cardiomyocytes)40. The generation of endodermal lineage cells with bona fide differentiated characteristics has been more difficult. However, researchers have recently succeeded in generating insulin-producing cells41, hepatocytes42, anterior foregut endoderm43 and intestinal cells44 from human pluripotent stem cells. As previously shown for the derivation of motor neurons from mouse ESCs45, the exposure of pluripotent cell lines to growth factors encountered by embryonic cells from that tissue during normal development improves differentiation towards the lineage of choice.
Generating chimaeric animals could also allow the production of endodermal tissue. In a recent study, wild-type iPSCs were injected into the blastocysts of mice unable to form a normal pancreas (Pdx1 mutants)46 resulting in chimaeric mice that harboured pancreases composed entirely of the introduced iPSCs. Mature β cells from these mice were then harvested and transplanted into syngeneic mice treated with streptozocin to induce diabetes, leading to recovery of their glucose regulatory capacity46. Wild-type rat β cells generated by injection of Pdx1–/– mouse blastocysts with rat iPSCs also rescued the glucose dysregulation in streptozocin-treated rats following their transplantation (Fig. 2). Although it is unclear if such interspecies chimeras would ever be ethically or technically feasible with human iPSCs and whether the resulting cells would ever be safe for human application given the potential for reactivation of endogenous host-derived viruses47, the use of such assays to generate mature, differentiated and functional derivatives from iPSCs may open a new approach for disease modelling in vivo.
Figure 2.
Xenogeneic rat-mouse chimaera to produce entirely iPSC-derived rat pancreas. The introduction of wild type rat iPSCs into Pdx1-deficient blastocyst-stage mouse embryos resulted in the generation of a chimaeric rat-mouse that harbours a rat iPSC-derived pancreas. This pancreas is expected to be composed entirely of rat iPSC-derived cells as the loss of Pdx1 in mouse embryos results in the complete absence of a developing pancreas.
Challenges to iPSC-based disease modelling and drug discovery
Several challenges must be overcome before successful implementation of iPSC-based drug screening and pathway discovery can be achieved (Fig. 1). The most critical issues are whether the relevant disease phenotypes can be faithfully reproduced in vitro and, if so, whether they can accurately predict disease behaviour in vivo. Despite promising studies suggesting that certain features of familial dysautonomia, SMA and RETT syndrome can be generated using iPSC-derived neural cells, other neurological disorders such as Parkinson's disease seem more difficult to model so far20. Three main factors may influence the amenability of diseases to in vitro modelling: the onset of disease in patients, the cell-autonomous nature of the disorder and the complexity of the underlying genetic defects. For example, evidence from animal models and clinical data indicate that familial dysautonomia, SMA and RETT syndrome manifest early in life, may have a strong cell-autonomous component and are caused by mutations in single genes, whereas Parkinson's disease generally occurs later in life and is caused by environmental and complex genetic factors. However, it is still unclear which of these three elements most strongly influences our ability to generate the relevant disease phenotype in vitro. It is possible that a disease such as autism, which involves complex genetics but manifests early in life, could still be modelled with an appropriate iPSC-derived cell type. Many diseases with the greatest societal impact are polygenic and highly influenced by environment (for example, congestive heart failure, Alzheimer's disease, diabetes, sudden cardiac death, emphysema and Parkinson's disease). It remains to be seen whether their key phenotypes can be reproduced in vitro using iPSCs.
If the aetiology for disease development is known or suspected, there may be ways to introduce the causal agent into purified iPSC-derived cells to induce or accelerate the manifestation of disease phenotypes (see Table 2). For example, in amyotrophic lateral sclerosis (ALS), superoxide dismutase (SOD) mutations affect the function of glial cells surrounding motor neurons. Studies have shown that co-culture of human ESC-derived motor neurons with glial cells carrying the mutation induces neuronal death48,49. An in vitro disease model could therefore potentially be generated that uses glial cells and motor neurons derived from iPSCs from an ALS patient in a similar co-culture system. Another example is Duchenne muscular dystrophy (DMD); the skeletal muscle phenotype of this disease is thought to be due to both the presence of dystrophin mutations and cumulative mechanical stretch injury from muscle use50. Thus, mechanical stress (or catecholamine stimulation) may need to be applied to iPSC-derived skeletal muscle to appropriately model this disease in vitro. For other complex diseases, exposure of relevant chemical agents or toxins to iPSC-derived cells may reveal phenotypes that would otherwise remain undetectable. For example, in one study that generated dopaminergic neurons from iPSCs derived from patients with sporadic cases of Parkinson's disease, no obvious abnormalities could be detected20. However, in a subsequent study, dopaminergic neurons derived from iPSCs obtained from a single Parkinson's disease patient harbouring a mutation in the leucine-rich repeat kinase 2 gene (LRRK2) were exposed to oxidative stress and demonstrated increased susceptibility to cell death51.
Table 2.
Potential human iPSC models of complex diseases involving environmental factors
| Possible pertubations to induce iPSC disease phenotype in vitro |
|||||
|---|---|---|---|---|---|
| Disease Name | Toxin | Genes | Chemical | Physical | Cells |
| Amyotrophic lateral sclerosis | Selenium | SOD, TDP-43, FUS/TLS | Reactive oxygen | – | Astrocytes |
| Duchenne muscular dystrophy | Cardiotoxin | Dystrophin/delta-sarcoglycan gene knockdown | Catecholamine | Mechanical stretch | – |
| Hypertrophic or dilated cardiomyopathy | Ethanol | Introduce sarcomeric gene mutations | Doxorubicin, herceptin | Mechanical stretch or electrical pacing | – |
| Hepatocellular carcinoma | Aflatoxin B1, ethanol, arsenic | Hepatitis B and C virus, p53, K-ras | Vinyl chloride, Polycyclic aromatic hydrocarbon | – | – |
| Chronic obstructive pulmonary disease | Tobacco | Elastase, α1 anti-trypsin | Nicotine, reactive oxygen | Mechanical stretch | Neutrophils |
Assuming that disease features can be reproduced in vitro, it is still unclear whether the phenotypes can be used for high throughput small-molecule screening. A major limitation is the lack of robust lineage-specific differentiation protocols that enable researchers to generate sufficient quantities of purified cells of a specific type for large-scale screening applications. Although significant advances have been made to direct the differentiation of ESCs or iPSCs into certain types of neurons52,53, cardiomyocytes54–57, blood58–61 and pancreatic cells41,62, none of these protocols generates the cell types of interest with > 95% purity. Sorting of these cells from the heterogeneous iPSC mixture to reproduce the disease phenotypes for high-throughput small molecule screening remains a challenge. Improvements in cell-purification strategies (for example, fluorescence activated cell sorting, drug selection, gradient centrifugation and functional marker isolation) may eventually allow us to overcome this barrier. The use of small-molecule screens to identify compounds that can enrich for a cell type of interest has also proved valuable51,63–67. Despite these challenges, some companies already offer human iPSC (hiPSC)-derived cardiomyocytes in quantities that are suitable for drug discovery and toxicology testing.
The heterogeneity of the maturation stage of the differentiated iPSCs is also a potential limitation. A high-throughput screen aimed at identifying small molecules that improve cardiomyocyte contractility may have a high rate of false-positive and -negative hits if there are well-to-well differences in differentiation state, as mature cardiomyocytes exhibit greater contractility than their immature counterparts. Similar issues may apply to small-molecule screening using hepatocytes or pancreatic β cells if the end-point of analysis is the secretion of specific enzymes or hormones, which strictly depend on the cells’ maturation stages.
Once these barriers to the development of robust in vitro disease models using iPSCs are overcome and small molecules that can reverse the disease phenotype in vitro have been identified, an appropriate animal model will be needed to validate the in vitro screen ‘hits’ in vivo. For candidates that are Food and Drug Administration (FDA)-approved drugs, with known pharmacokinetic and toxicity profiles, no additional animal studies might be needed. For small molecules that have not been previously tested for their pharmacokinetic, toxicity and efficacy profiles, a standard pre-clinical evaluation of these molecules in vivo will still be required. It is likely that a number of small-molecule candidates identified from such screens might show efficacy only in the artificial conditions of an in vitro assay. Thus, large-animal models of disease would be essential to help eliminate these candidates with insufficient biological efficacy or enhanced toxicity in vivo. Investment of research resources to create reliable animal disease models should thus be a significant priority if we are to realize the full potential of therapeutic drug screening efforts using disease-specific human iPSCs.
Challenges to iPSC-based therapy
Major hurdles remain before iPSC-derived cells can be safely introduced into human patients. First, as for any pluripotent stem-cell-based therapy, the risk of teratoma formation can be substantial. As most pre-clinical human ESC/iPSC-derived cell transplantation studies have been performed in immunosuppressed animals, it is unclear whether the risk for teratoma formation will be similar or greater with patient-matched iPSCs than that observed in immunosuppressed animals. The frequency of teratoma formation following human ESC transplantation into animal hosts is directly related to the degree of immunosuppression68, so transplantation of genetically matched iPSC derivatives into patients, which are expected to elicit no immune reactivity against the transplanted cells, may result in an even greater rate of teratoma formation than the rate observed in animal studies. Alternatively, incomplete reprogramming or genetic aberrations accrued during the iPSC derivation process (see below) may render even genetically matched iPSC lines immunogenic. So far, no study has evaluated the immunogenicity of genetically matched iPSCs on transplantation into syngeneic hosts.
At present, it is unclear whether any of the currently available strategies to generate differentiated cells from iPSCs and to separate them from residual pluripotent cells is able to eliminate the risk of teratoma formation. Although it is encouraging that lineage selective survival or engraftment has been observed on transplantation of human ESC-derived cardiomyocytes into immunosuppressed rodents55,56 or murine iPSC-derived neurons into Parkinsonian rats37, it is unknown whether such effects would persist when autologous human iPSC-derived cells are transplanted. The transdifferentiation of one adult cell type into another cell type would circumvent the teratoma risk associated with pluripotent cells and may provide an alternative approach to produce clinically relevant autologous cell types. Indeed, recent data suggest that the introduction of either pluripotency genes or lineage-specific transcription factors into fibroblasts can give rise to cells resembling haematopoietic progenitors69, cardiomyocytes70,71, myogenic cells72 and neurons73. It remains to be seen, however, if transdifferentiated cell types are as functional as ESC-/iPSC-derived cells and whether lineage switching into other cell types can be achieved with this strategy. A serious disadvantage of transdifferentiation over directed differentiation from pluripotent cells is that somatic cells in general have a limited lifespan and are therefore not expandable, whereas ESCs/iPSCs have limitless growth and can hence be repeatedly coaxed into the desired cell types.
Beyond the issue of teratoma formation, there is now a growing recognition that differentiated cells derived from ESCs/iPSCs are mostly immature. These cells mimic embryonic development and adopt phenotypes that resemble fetal or neonatal cells74,75. Whether this immaturity will influence their clinical applicability may vary between diseases and cell types. For the treatment of degenerative diseases, such as Parkinson's disease, Alzheimer's disease or congestive heart failure, the transplanted cells would need to be sufficiently mature to replace the lost cells of similar type (for example, dopaminergic neurons and ventricular cardiomyocytes) to ensure proper function. Likewise, cell maturity may also be critical for diseases that require the transplanted cells to correct lost secretory function or cell number (for example, pancreatic islet β cells, hepatocytes or haematopoietic cells). For instance, human ESC (hESC)-derived erythroid progenitors express mostly embryonic and fetal haemoglobin but have none76 or only limited77 ability to activate mature β-globin expression, and this could affect their functionality. Whether transplanted cells can undergo further maturation over time within their site of engraftment remains to be determined. Thus, future developments within this area should aim at enhancing the maturation of pluripotent stem-cell-derived cells in vitro before their therapeutic application78–80. Notably, a recent study suggested that direct conversion of human fibroblasts into haematopoietic cells entails activation of adult globin genes rather than fetal globins as is generally seen following ESC/iPSC differentiation, suggesting a possible alternative to producing certain mature cell types69.
A final consideration for the successful application of iPSC-derived cells in regenerative medicine is their ability to integrate with existing cells in the tissue. Most solid organs harbour an intrinsic architecture that shows an appropriate balance between the number of each cell type and their geometrical arrangement to reflect their developmental relationships. It remains to be seen whether transplantation of isolated single cells in suspension (mostly of one cell type) could auto-regulate the number of each of the cell types to produce the endogenous tissue architecture. Furthermore, the engrafted cells will need to function in concert with the existing cells. This is particularly important for organs such as the heart, lung, kidney and liver where individual functional units (for example, ventricular muscles, nephrons, alveolar sacs and the hepatobiliary network) are interconnected with other functional units and with the vasculature. Although the successes of haematopoietic stem cell transplantation therapy over the past four decades have spurred the interest and development of cell transplantation strategies in solid organs, it should be cautioned that observations made in the haematopoietic system may not necessarily apply to solid organs. The recent disappointing clinical data from the transplantation of skeletal myoblasts81 and bone marrow mononuclear cells82,83 for the treatment of myocardial injury should remind us that the route to a durable clinical therapy using stem cells remains largely obscure and much greater understanding in cell lineage specification, differentiation and function will be needed to advance this field.
Recent progress in tissue engineering raises the possibility that some of the structural limitations associated with cell transplantation may in fact be surmountable. Several reports have provided exciting proof-of-principle evidence that the seeding of decellularized tissue scaffolds with endothelial and epithelial cells grown in bioreactors can produce bioartificial lungs84,85, livers86 and hearts87 that engraft in animals and exhibit normal tissue function for up to several days.
Are hiPSCs and hESCs equivalent?
To what extent pluripotent cell lines exhibit biological variability among one another and whether hiPSCs have the same properties as hESCs are two additional important questions in the field. These issues may affect the functionality and safety of hiPSC-derived mature cells and answers to these questions may determine how many cell lines need to be derived to observe the desired phenotypes in a reliable fashion. These two topics are actively debated and we will attempt to summarize recent findings that address these issues.
Although initial studies concluded that hiPSCs are highly similar or even indistinguishable from hESCs15,16,88, a number of laboratories have subsequently documented substantial differences in gene expression89, DNA methylation90,91, in vitro differentiation potential92,93 and teratoma-forming propensity94. However, it remains unclear which of these differences are due to inherent differences between hESCs and hiPSCs rather than to differences associated with the generation of hiPSCs3. Indeed, recent studies showed that genetic background95, the use of viral integration20, lab-to-lab variation96 and passage number89,97 can have profound effects on gene expression and function in pluripotent cells.
In addition, three independent studies found that hiPSCs carry copy number variations (deletions and duplications)98–100 and point mutations101, as assessed by SNP (single nucleotide polymorphism) arrays and exon sequencing, respectively. Some of these alterations seem to be the result of culturing, as has been seen before for hESCs, whereas other mutations pre-existed in the somatic donor cells. Some may have arisen de novo during the reprogramming process. A limitation of these reports is that hESC lines, which probably originate from multiple embryonic founder cells, were compared with hiPSCs that are, per definition, clonal cell lines derived from a single fibroblast of unknown genomic integrity. It should, therefore, be informative to include clonal fibroblast and early-passage subcloned hESC lines in these analyses to evaluate the exact contributions of cell of origin, subcloning procedure and passage number on the mutational profile.
In any case, these studies clearly indicate that many hESCs and hiPSCs harbour subtle or severe chromosomal abnormalities, and thus may warrant careful examination before their potential use in therapy or disease modelling. Although large genomic amplifications, especially of areas comprising cancer-associated genes, would certainly be a reason for excluding such hiPSC lines in therapeutic applications, future work is needed to evaluate whether the observed heterozygous small deletions and point mutations result in functional consequences and thus pose a risk in a potential therapeutic setting.
Beyond genome integrity issues, there is now increasing recognition among investigators that human pluripotent stem cells are extremely variable in their propensity for lineage-specific differentiation102. In fact, some of the earlier differences, observed when smaller numbers of hESCs and hiPSCs were compared, may be explained by this variability. A recent study reported substantial variability between the global transcriptional and DNA methylation profiles of 20 different hESC and 12 hiPSC lines, indicating that a large sample size is critical to draw firm conclusions about potential differences103. Notably, this and a similar study104 also established a scoring algorithm that allowed these groups to prospectively identify pluripotent cell lines that efficiently give rise to neural lineage cells, based on gene expression data. The latter approaches depend on a simple differentiation assay combined with the analysis of several hundred transcripts and genome-wide expression profiling, respectively, to predict hiPSC differentiation potentials, which may be time- and cost-inefficient. In the future, however, it may become possible to rapidly pre-screen newly derived hiPSCs and hESCs for their potential to differentiate into desired cell lineages of all three germ layers using a smaller set of markers.
In conclusion, although there is evidence for subtle differences between ESCs and iPSCs at the transcriptional, epigenetic, genetic and functional levels, it remains unclear which of these are solely the result of biological variation or handling of the cells and which are a consequence of the reprogramming process itself. Further work is needed to examine this issue and to determine if the observed aberrations have any functional impact on their potential therapeutic utility.
Future perspectives
Although significant progress still needs to be made in understanding the molecular mechanisms of cellular pluripotency and reprogramming, the possibility that novel pathways and drugs may be discovered through the use of iPSC technology should sustain the great enthusiasm that basic and clinical/translational scientists have bestowed on this area of research. The idea that we have the knowledge and means to generate ‘spare parts’ for every failing organ may belong to the realm of science fiction for now. However, it is worth noting that the rat-mouse interspecies chimaerism study by Kobayashi et al. suggests one possible route to generate human tissues (if not organs) using iPSCs46.
The remarkable discovery of iPSCs by Takahashi and Yamanaka may be the molecular equivalent of the discovery of antibiotics or vaccines in the last century. Time will tell whether the efforts of stem cell biologists and translational scientists in this area today will be discussed in the same way. Our endeavour to overcome the barriers that prevent successful translation of stem cell biology into clinical therapy should help to improve our knowledge regarding disease pathogenesis itself and ways to prevent their onset or progression. In the end, this may prove to be the most important contribution of iPSC technology.
BOX 1 Challenges in pluripotent stem cell clinical trials.
The recent trial by Geron using human ESC-derived oligodendrocytes for patients with spinal cord injury provides a salient example of the challenges facing pluripotent stem cell therapy105. During the initial stage of FDA filing, graft-derived microscopic cysts were found in mice transplanted with cell preparations enriched for human ESC-derived oligodendrocytes. Subsequently, extensive analyses were performed to evaluate the lot-to-lot differences in cyst formation and to assess whether an in vitro assay could predict this potential. Furthermore, safety data from a 9-month follow-up of these cyst-containing grafts was demanded by the FDA. Geron has recently demonstrated that the microscopic cysts do not represent a manifestation of teratoma and are not harmful. Consequently, the clinical advisory panel of the FDA granted permission to Geron to resume enrolment of patients for the trial.
It will be interesting to see whether other human ESC-based clinical studies will encounter similar issues. For example, Advanced Cell Technology has recently received approval by the FDA to conduct a Phase I/II clinical trial using human ESC-derived retinal pigment epithelial cells to treat Stargardt disease and age-related macular regeneration. Likewise, Novocell will perform trials using human ESC-derived pancreatic progenitor cells for the treatment of type 1 diabetes. Although immunorejection of allogeneic grafts remains a serious challenge in hESC-based trials, a recent study found that short-term suppression of leukocyte co-stimulatory molecules could significantly improve engraftment efficiencies of both hESC- and hiPSC-derived cells in mice106.
ACKNOWLEDGMENTS
We thank J. Wu and G. Mostoslavsky for their insightful comments and critical reading of the manuscript and J. Gold for detailed discussion regarding the human ESC-derived oligodendrocyte clinical trial at Geron. This work was supported by grants from the NIH (OD003266 and HD058013 to K.H.; OD004411, HL081086, HL100408 to S.M.W.), the Harvard Stem Cell Institute (K.H. and S.M.W.), and the Howard Hughes Medical Institute (to K.H.). We apologize to colleagues whose work we could not cite in this brief review article.
Footnotes
COMPETING FINANCIAL INTERESTS
K.H. is an advisor to iPierian, Inc.
Contributor Information
Sean M. Wu, Cardiovascular Research Center, Division of Cardiology, Massachusetts General Hospital, Boston 02114, Massachusetts, USA and the Harvard Stem Cell Institute, Cambridge, Massachusetts 02138, USA. smwu@partners.org
Konrad Hochedlinger, Harvard Stem Cell Institute, Cambridge, Massachusetts 02138, USA; the Center for Regenerative Medicine and Cancer Center, Massachusetts General Hospital, Boston, Massachusetts 02114, USA; the Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, Massachusetts 02138, USA and the Howard Hughes Medical Institute, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, Massachusetts 02138, USA khochedlinger@helix.mgh.harvard.edu.
References
- 1.Briggs R, King TJ. Transplantation of living nuclei from blastula cells into enucleated frogs’ eggs. Proc. Natl Acad. Sci. USA. 1952;38:455–463. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J. Embryol. Exp. Morphol. 1962;10:622–640. [PubMed] [Google Scholar]
- 3.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rideout WM, III, Hochedlinger K, Kyba M, Daley GQ, Jaenisch R. Correction of a genetic defect by nuclear transplantation and combined cell and gene therapy. Cell. 2002;109:17–27. doi: 10.1016/s0092-8674(02)00681-5. [DOI] [PubMed] [Google Scholar]
- 5.Schnieke AE, et al. Human factor IX transgenic sheep produced by transfer of nuclei from transfected fetal fibroblasts. Science. 1997;278:2130–2133. doi: 10.1126/science.278.5346.2130. [DOI] [PubMed] [Google Scholar]
- 6.Hochedlinger K, Jaenisch R. Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature. 2002;415:1035–1038. doi: 10.1038/nature718. [DOI] [PubMed] [Google Scholar]
- 7.Lee BC, et al. Dogs cloned from adult somatic cells. Nature. 2005;436:641. doi: 10.1038/436641a. [DOI] [PubMed] [Google Scholar]
- 8.Yin XJ, et al. Cats cloned from fetal and adult somatic cells by nuclear transfer. Reproduction. 2005;129:245–249. doi: 10.1530/rep.1.00403. [DOI] [PubMed] [Google Scholar]
- 9.Li S, Chen X, Fang Z, Shi J, Sheng HZ. Rabbits generated from fibroblasts through nuclear transfer. Reproduction. 2006;131:1085–1090. doi: 10.1530/rep.1.01065. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 13.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:1–12. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 16.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 17.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 18.Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soldner F, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaji K, et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haase A, et al. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell. 2009;5:434–441. doi: 10.1016/j.stem.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 24.Lowry WE, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl Acad. Sci. USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loh YH, et al. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476–5479. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunisato A, et al. Direct generation of induced pluripotent stem cells from human non-mobilized blood. Stem Cells Dev. 2011;20:159–168. doi: 10.1089/scd.2010.0063. [DOI] [PubMed] [Google Scholar]
- 27.Aasen T, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 28.Ebert AD, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carvajal-Vergara X, et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moretti A, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl. J. Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 31.Yazawa M, et al. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchetto MC, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee G, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, et al. A human iPSC model of Hutchinson Gilford progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 2011;8:31–45. doi: 10.1016/j.stem.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Liu GH, et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. doi: 10.1038/nature09879. advance online publication, doi:10.1038/nature09879 (23 February 2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanna J, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 37.Wernig M, et al. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson's disease. Proc. Natl Acad. Sci. USA. 2008;105:5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ. Res. 2009;104:e30–e41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson TJ, et al. Repair of acute myocardial infarction by human stemness factors induced pluripotent stem cells. Circulation. 2009;120:408–416. doi: 10.1161/CIRCULATIONAHA.109.865154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 41.Kroon E, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 42.Song Z, et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 2009;19:1233–1242. doi: 10.1038/cr.2009.107. [DOI] [PubMed] [Google Scholar]
- 43.Green MD, et al. Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat. Biotechnol. 2011;29:267–272. doi: 10.1038/nbt.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi T, et al. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell. 2010;142:787–799. doi: 10.1016/j.cell.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 47.Coffin JM, Stoye JP. Virology. A new virus for old diseases? Science. 2009;326:530–531. doi: 10.1126/science.1181349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 49.Marchetto MC, et al. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 50.McNally EM. New approaches in the therapy of cardiomyopathy in muscular dystrophy. Annu. Rev. Med. 2007;58:75–88. doi: 10.1146/annurev.med.58.011706.144703. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen HN, et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8:267–280. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swistowski A, et al. Efficient generation of functional dopaminergic neurons from human induced pluripotent stem cells under defined conditions. Stem Cells. 2010;28:1893–1904. doi: 10.1002/stem.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nizzardo M, et al. Human motor neuron generation from embryonic stem cells and induced pluripotent stem cells. Cell Mol. Life Sci. 2010;67:3837–3847. doi: 10.1007/s00018-010-0463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang L, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 55.van Laake LW, et al. Human embryonic stem cell-derived cardiomyocytes survive and mature in the mouse heart and transiently improve function after myocardial infarction. Stem Cell Res. 2007;1:9–24. doi: 10.1016/j.scr.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Laflamme MA, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 57.Kattman SJ, et al. Expression of Flk-1/KDR and PDGFR-α marks the emergence of cardiac mesoderm from mouse and human pluripotent stem cells. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 58.Irion S, et al. Temporal specification of blood progenitors from mouse embryonic stem cells and induced pluripotent stem cells. Development. 2010;137:2829–2839. doi: 10.1242/dev.042119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi KD, et al. Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells. 2009;27:559–567. doi: 10.1634/stemcells.2008-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lengerke C, et al. Hematopoietic development from human induced pluripotent stem cells. Ann. N. Y. Acad. Sci. 2009;1176:219–227. doi: 10.1111/j.1749-6632.2009.04606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma YD, Lugus JJ, Park C, Choi K. Differentiation of mouse embryonic stem cells into blood. Curr. Protoc. Stem Cell Biol. 2008 doi: 10.1002/9780470151808.sc01f04s6. Chapter 1, Unit 1F.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D'Amour KA, et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 63.Borowiak M, et al. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu X, Ding S, Ding Q, Gray NS, Schultz PG. Small molecules that induce cardiomyogenesis in embryonic stem cells. J. Am. Chem. Soc. 2004;126:1590–1591. doi: 10.1021/ja038950i. [DOI] [PubMed] [Google Scholar]
- 65.Hao J, et al. Dorsomorphin, a selective small molecule inhibitor of BMP signaling, promotes cardiomyogenesis in embryonic stem cells. PLoS One. 2008;3:e2904. doi: 10.1371/journal.pone.0002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Desbordes SC, et al. High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell. 2008;2:602–612. doi: 10.1016/j.stem.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swijnenburg RJ, et al. Immunosuppressive therapy mitigates immunological rejection of human embryonic stem cell xenografts. Proc. Natl Acad. Sci. USA. 2008;105:12991–12996. doi: 10.1073/pnas.0805802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Szabo E, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 70.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Efe JA, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 72.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 73.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sachinidis A, et al. Cardiac specific differentiation of mouse embryonic stem cells. Cardiovasc. Res. 2003;58:278–291. doi: 10.1016/s0008-6363(03)00248-7. [DOI] [PubMed] [Google Scholar]
- 75.Boheler KR, et al. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ. Res. 2002;91:189–201. doi: 10.1161/01.res.0000027865.61704.32. [DOI] [PubMed] [Google Scholar]
- 76.Olivier EN, Qiu C, Velho M, Hirsch RE, Bouhassira EE. Large-scale production of embryonic red blood cells from human embryonic stem cells. Exp. Hematol. 2006;34:1635–1642. doi: 10.1016/j.exphem.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 77.Ma F, et al. Generation of functional erythrocytes from human embryonic stem cell-derived definitive hematopoiesis. Proc. Natl Acad. Sci. USA. 2008;105:13087–13092. doi: 10.1073/pnas.0802220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Domian IJ, et al. Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science. 2009;326:426–429. doi: 10.1126/science.1177350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Madden LR, et al. Proangiogenic scaffolds as functional templates for cardiac tissue engineering. Proc. Natl Acad. Sci. USA. 2010;107:15211–15216. doi: 10.1073/pnas.1006442107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zimmermann WH, Eschenhagen T. Embryonic stem cells for cardiac muscle engineering. Trends Cardiovasc. Med. 2007;17:134–140. doi: 10.1016/j.tcm.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 81.Menasche P, et al. The myoblast autologous grafting in ischemic cardiomyopathy (MAGIC) trial: first randomized placebo-controlled study of myoblast transplantation. Circulation. 2008;117:1189–1200. doi: 10.1161/CIRCULATIONAHA.107.734103. [DOI] [PubMed] [Google Scholar]
- 82.Lunde K, et al. Intracoronary injection of mononuclear bone marrow cells in acute myocardial infarction. N. Engl. J. Med. 2006;355:1199–1209. doi: 10.1056/NEJMoa055706. [DOI] [PubMed] [Google Scholar]
- 83.Janssens S, et al. Autologous bone marrow-derived stem-cell transfer in patients with ST-segment elevation myocardial infarction: double-blind, randomised controlled trial. Lancet. 2006;367:113–121. doi: 10.1016/S0140-6736(05)67861-0. [DOI] [PubMed] [Google Scholar]
- 84.Petersen TH, et al. Tissue-engineered lungs for in vivo implantation. Science. 2010;329:538–541. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ott HC, et al. Regeneration and orthotopic transplantation of a bioartificial lung. Nat. Med. 2010;16:927–933. doi: 10.1038/nm.2193. [DOI] [PubMed] [Google Scholar]
- 86.Uygun BE, et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat. Med. 2010;16:814–820. doi: 10.1038/nm.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ott HC, et al. Perfusion-decellularized matrix: using nature's platform to engineer a bioartificial heart. Nat. Med. 2008;14:213–221. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 88.Guenther MG, et al. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chin MH, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Doi A, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Deng J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu BY, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl Acad. Sci. USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Feng Q, et al. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;28:704–712. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- 94.Miura K, et al. Variation in the safety of induced pluripotent stem cell lines. Nat. Biotechnol. 2009;27:743–745. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]
- 95.Stadtfeld M, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Newman AM, Cooper JB. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell. 2010;7:258–262. doi: 10.1016/j.stem.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 97.Polo JM, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Laurent LC, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hussein SM, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 100.Mayshar Y, et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 101.Gore A, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Osafune K, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 103.Bock C, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Muller FJ, et al. A bioinformatic assay for pluripotency in human cells. Nat. Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Strauss S. Geron trial resumes, but standards for stem cell trials remain elusive. Nat. Biotechnol. 2010;28:989–990. doi: 10.1038/nbt1010-989. [DOI] [PubMed] [Google Scholar]
- 106.Pearl JI, et al. Short-term immunosuppression promotes engraftment of embryonic and induced pluripotent stem cells. Cell Stem Cell. 2011;8:309–317. doi: 10.1016/j.stem.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 108.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ku S, et al. Friedreich's ataxia induced pluripotent stem cells model intergenerational GAATTC triplet repeat instability. Cell Stem Cell. 2010;7:631–637. doi: 10.1016/j.stem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Raya A, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Itzhaki I, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 113.Maehr R, et al. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc. Natl Acad. Sci. USA. 2009;106:15768–15773. doi: 10.1073/pnas.0906894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rashid ST, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J. Clin. Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang J, et al. Induced pluripotent stem cells can be used to model the genomic imprinting disorder Prader-Willi syndrome. J. Biol. Chem. 2010;285:40303–40311. doi: 10.1074/jbc.M110.183392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chamberlain SJ, et al. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc. Natl Acad. Sci. USA. 2010;107:17668–17673. doi: 10.1073/pnas.1004487107. [DOI] [PMC free article] [PubMed] [Google Scholar]