Abstract
A total of 874 fecal specimens (446 diarrheal cases and 428 controls) from diarrheal children admitted in the Infectious Diseases Hospital, Kolkata and age and sex matched asymptomatic subjects from an urban community were assessed for the prevalence of enterotoxigenic Bacteroides fragilis (ETBF). Isolates of B. fragilis were tested for the presence of enterotoxin gene (bft) by PCR. The detection rate of ETBF was 7.2% (63 of 874 specimens) that prevailed equally in diarrheal cases and controls (7.2% each; 32 of 446 cases and 31 of 428 controls). Male children up to one year age group was significantly (p<0.05) associated with ETBF infection as compared to children > 2 years of age in cases and controls. In 25 ETBF isolates, the bft gene was genotyped using PCR-RFLP and only two alleles were identified with prevalence rate of 40% and 60% for bft-1 and bft-3, respectively. All the ETBF isolates were susceptible for chloramphenicol and imipenem but resistant to clindamycin (48%), moxifloxacin (44%) and metronidazole (32%). Resistance of ETBF to moxifloxacin (44%) and metronidazole is an emerging trend. Pulsed-field gel electrophoresis (PFGE) revealed that majority of the ETBF isolates are genetically diverse. In the dendrogram analysis, two clusters were identified, one with ETBF resistant to 5–8 antimicrobials and the other cluster with metronidazole and moxifloxacin susceptible isolates from diarrheal cases. To our knowledge, this is the first detailed report on ETBF from India indicating its clinical importance and molecular characteristics.
Introduction
The Bacteroides species are a group of Gram-negative anaerobes, which generally represent as a major constituent of the human gut microbiota. Although Bacteroides species play an important role in mediating mucosal and systemic immunity, this group sometimes cause opportunistic infections. Bacteroides fragilis is the only known species of the Bacteroides that can cause diarrhea and frequently isolated from abscesses, soft tissue infections and bacteremia [1]. B. fragilis does not have other known niches except the gut of mammals [2]. A study conducted in southern India showed that B. fragilis was frequently detected in humans with and without diarrhea [3]. In India, this pathogen has been identified in different clinical specimens and also in healthy persons but its virulence factors are not confirmed [4]. B. fragilis is categorized into two subgroups: non-enterotoxigenic B. fragilis (NTBF) and enterotoxigenic B. fragilis (ETBF). In developing counties, ETBF is an emerging pathogen associated with diarrhea in children and travelers [5]–[7]. In children, ETBF is associated with secretory diarrhea with mild severity and hence much attention has not been paid to this pathogen. The other syndromes of ETBF associated infection include extraintestianl infections, abdominal pain, tenesmus, inflammatory diarrhea, antibiotic associated diarrhea and chronic inflammation that lead to colon cancer [8]–[11].
ETBF produces a specific virulence factor known as fragilysin, which is a heat-labile enterotoxin responsible for mucosal inflammation. Based on the sequence variation in the B. fragilis enterotoxin encoding gene bft, three subtypes namely bft-1, bft-2, and bft-3 have been identified and these are predominantly found in specific geographical locations [10]. Several methods have been reported for the diagnosis of ETBF including conventional culture technique, cell culture assay, enzyme immunoassays, immunomagnetic separation followed by PCR (IM-PCR), and nested PCR [10]. Among these methods, nested PCR has been considered as a most simple and sensitive method [10], [11]. As this pathogen is associated with a wide variety of infections, information regarding its prevalence and characterization are important for the successful clinical management.
In India, only few studies have been made to detect ETBF associated with diarrhea or among non-diarrhea patients [3], [4], [6], [12]. This study was undertaken to detect the prevalence of ETBF among children with diarrhea admitted in the Infectious Diseases Hospital (IDH), Kolkata and without diarrhea from an urban community. In addition, phenotypic and genotypic characteristics of the ETBF were also investigated.
Materials and Methods
Study populations and sample collection
From February to August 2012, 874 fecal specimens collected from children below five years of age were processed. Of these, 446 fecal specimens were obtained from diarrheal children treated in the IDH and 428 samples of age and sex matched asymptomatic controls from an urban community. Before the initiation of antimicrobial therapy, the stool specimens were collected from these children. The children admitted in the hospital were treated with intravenous fluid (IVF) or oral rehydration solution (ORS) depends on the nature of dehydration and oral ciprofloxacin (6–10 mg per Kg of the body weight) and metronidazole (35–50 mg per Kg) was given in divided doses. After microbial screening, aliquots of the fecal specimens were stored at−80°C for subsequent use.
Extraction of total nucleic acid from fecal specimens
Fecal specimens (∼100 mg semi solid or 200 µl if liquid) were suspended in nuclease free water (final concentration ∼10%) with equal volume of vortel XF (Miller-Stephenson Chemical Co, Inc, Danbury, CT) and vortexed for 2 min followed by centrifugation at 4500 rpm for 10 min. Two hundred micro liters of the supernatant was used for extraction of the total nucleic acid using an automated system (NucliSens EasyMAG; bioMérieux, Marcy l'Etoile, France).
Culture and confirmation of ETBF
Stool specimens collected from patients and controls were transported to the laboratory within 2 hrs of collection in a cold chamber maintained at 4°C. The fecal specimens were streaked on to respective selective agars for isolation of vibrios, Salmonella spp, Shigella spp, Campylobacter spp, and Aeromonas spp and identified these pathogens as described before [13]. Three different Escherichia coli colonies from MacConkey agar were tested for different pathogroups by multiplex PCR [13]. For the isolation of Bacteroides species, Bile-Esculin (BBE) agar (Becton, Dickinson and Company, Sparks, MD) plate was used and incubated at 37°C for 48 hrs in an anaerobic jar (BD GasPak EZ anaerobic systems). After incubation, several individual gray, raised circular colonies surrounded by esculin hydrolyses from each specimen was subcultured on Colombia blood agar (CBA) plate (bioMérieux) and incubated anaerobically for 48 hrs. A portion of pure culture from the blood agar was suspended in an anaerobic broth [Luria Broth supplemented with beef extract (0.3%), cysteine HCl (0.04%), glucose (0.1%) sodium hydrogen phosphate (0.4%) and glycerol (15%)] and preserved at −80°C. The remaining portion of the culture was suspended in TE buffer for the confirmation of ETBF by PCR. In addition to culture methods, ELISA was performed to detect rotavirus, adenovirus, and parasites such as Giardia lamblia, Cryptosporidium spp. and Entamoeba histolytica directly from the stool specimens [13]. Helicobacter pylori in the stools were detected using a commercial ELISA kit (Amplified IDEIA™ HPStAR, Oxoid, Basingstoke, Hants, UK). The DNA extracted from the stool specimens were used for the detection of Astro virus, Sapo virus, and Noro virus (Genotype I and Genotype II) by reverse transcriptase PCR [13].
Detection of B. fragilis and ETBF by PCR
The pure cultures of Bacteroides from CBA plates were suspended in a TE buffer and boiled for 15 min in the water bath, snap cooled on ice and centrifuged at 10, 000 rpm for 5 min. The resulting supernatants were screened for the 16S-rRNA gene-specific for B. fragilis group and bft by PCR [14], [15]. In addition, the total nucleic acid extracted from the fecal specimens was tested for the presence of bft by PCR.
Genotyping of bft
From the 63 ETBF positive stool specimens by PCR, 30 ETBF were isolated by culture method. Due to non-viability of 5 isolates, only 25 isolates harboring bft was amplified by PCR and restriction fragment length polymorphism technique (RFLP) was applied to detect the bft subtypes [16]. The PCR amplified products were purified using QIAquick PCR purification kit (QIGEN, GmBH, Hilden, Germany) and then digested with Sau3A1 (Thermo Scientific Inc., Waltham, MA) according to the manufacturer's recommendations. The digested DNA was separated in a 2% agarose gel, and visualized after ethidium bromide staining.
Antimicrobial susceptibility testing
ETBF isolates were grown on CBA and suspended in sterile saline and the cell density was determined using a densitometer (bioMérieux), which is equivalent to a 1.0 McFarland standard (∼3×108 CFU/mL). The cell suspension was spread uniformly on the Brucella blood agar (BBA, Oxoid, Basingstoke, UK) supplemented with 5% laked sheep blood, hemin, and vitamin K according to CLSI guidelines [17]. After the inoculation, E-test strips (AB Biodisk-bioMérieux) for each drug (amoxicillin-clavulanic acid, AMC; ampicillin, AMP; ampicillin-sulbactam, SAM; cefoxitin, FOX; chloramphenicol C; ciprofloxacin, CIP; clindamycin, CLI; imipenem, IPM; moxifloxacin, MXF; norfloxacin, NOR; and metronidazole, MTZ) was placed and incubated for 48 hrs at 37°C in an anaerobic atmosphere. Reference strain B. fragilis ATCC 25285 was used as control. Resistant and susceptibility of ETBF were estimated according to quality control ranges for B. fragilis assigned by the manufacturer's instruction and also using breakpoints information of CLSI and other reports [17]–[19].
Pulse Field Gel Electrophoresis (PFGE)
PFGE protocol described by Yamasaki et al. [20] was slightly modified and adopted in this study. Briefly, 25 ETBF isolates were anaerobically grown for 14–18 hrs on CBA plates at 37°C. Bacterial cultures were suspended in cell suspension buffer (CSB; 10 mM Tris-HCl (pH 7.2), (20 mM NaCl) (50 mM EDTA [pH 8.0]) using sterile cotton swabs. Cells were harvested by centrifugation, washed and resuspended in CSB and adjusted to an optical density of 1.0 to 1.2 at 610 nm. The cell suspension (500 µl) was mixed with (500 µl) molten low melting agarose (2%) at 50°C. The mixture was carefully dispensed into a sample mold (Bio-Rad, Hercules, CA). After solidification, the plugs were transferred to a 2.0-ml micro centrifuge tubes containing 1.0 ml of cell lysis buffer (1 mg/ml lysozyme, 10 mM Tris-HCl (pH 7.2) 50 mM NaCl, 0.2% sodium deoxycholate and 0.5% of sodium laurylsarcosine) and incubated at 37°C for 3 hrs. After incubation, plugs were washed twice with reagent grade water and treated with 1 ml of proteinase K solution (1 mg/ml proteinase K, 100 mM EDTA (pH 8.0) 0.2% sodium deoxycholate and 1% of sodium laurylsarcosine) at 50°C for overnight. The plugs were washed with washing buffer (10 mM Tris-HCl and 50 mM EDTA [pH 8.0]) two times for one hr, once with phenylmethylsulfonyl fluoride (1 mM) containing wash buffer and twice with diluted wash buffer (0.1X) with agitation at room temperature.
The agarose-embedded ETBF DNA plugs were digested with 50 U of NotI enzyme (New England Biolabs Inc., Ipswich, MA) and Salmonella enterica serovar Braenderup (H9812) was digested by XbaI and its DNA fragments were used as molecular size markers. The digested DNA fragments were resolved with 1% PFGE-grade agarose (SeaKem Gold agarose, Lonza, Rockland, ME) in 0.5X trisborate EDTA buffer at 6 V/cm for 16 hrs at 14°C. Run conditions were generated by the autoalgorithm mode of the CHEF Mapper PFGE system (Bio-Rad) with a size range of 30–600 kb. After electrophoresis, the ethidium bromide (Sigma, St. Louis, MO) stained agarose gel was visualized and the captured images were digitized for computer-aided analysis (Gel Doc system, Bio-Rad). PFGE profiles were analyzed using the BioNumerics version 4.0 software (Applied Maths, Sint Martens Latem, Belgium). The tagged image file formats were normalized by using the universal Salmonella enterica serotype Braenderup (H9812) size standard on each gel against the reference in the database. PFGE profiles were matched using the Dice coefficient and unweighted pair group method using arithmetic averages (UPGMA) clustering with a 1.5% band position tolerance window and 1.5% optimization. The clustering of the PFGE patterns and band assignments were verified visually.
Statistical Analysis
The inferential age groups were evaluated for ETBF positive specimens from under five years children by Multinomial Logistic Regression (MLR)(1,2) analysis using SPSS software (Version 19.0, SPSS Inc., Chicago, Illinois). The age groups were classified into 3 categories: ≤1 year, >1–2 years and >2 years and coded as 1–3, respectively. The relationship between the risk dependent variable and each of the categorical explanatory variables are shown in Table 1. Infection caused by a ETBF was classified in number as ‘1’ for organism present and ‘2’ for its absence. The extreme values of the classified age group was fixed as reference category (>2 yrs).
Table 1. Multinomial Logistic Regression Models exploring significant risk age group of ETBF infection.
| Age | B-value | OR 95% CI | P value | |
| Cases | ||||
| Up to 1 year | 0.944 | 2.57 (1.07–6.16) | 0.034* | |
| Male | 1.83 | 6.57 (1.08–39.88) | 0.041* | |
| Female | 1.33 | 3.80 (0.64–22.4) | 0.141 | |
| 1–2 years | 0 | 1.00 (0.35–2.85) | 1 | |
| Male | 0 | 1.00 (0.12–8.54) | 1 | |
| Female | 0 | 1.00 (0.12–8.28) | 1 | |
| >2 years | Reference category | |||
| Controls | ||||
| Up to 1 year | 1.041 | 2.83 (1.12–7.19) | 0.028* | |
| Male | 2.06 | 7.87 (1.1951.97) | 0.032* | |
| Female | 1.77 | 5.91 (0.88–39.43) | 0.067 | |
| 1–2 years | 0.288 | 1.33 (0.46–3.84) | 0.594 | |
| Male | 0.71 | 2.03 (0.24–17.31) | 0.516 | |
| Female | 0.778 | 2.18 (0.25–18.71) | 0.478 | |
| >2 years | Reference category | |||
Statistically significant
Ethics Statement
Ethical approval was obtained from the National Institute of Cholera and Enteric Diseases Ethics Committee (Ref.C-48/2012-T&E), and parents of the children gave written informed consent.
Results
Prevalence of ETBF
A total of 874 fecal specimens were analyzed in this study including 446 from diarrheal children and 428 from controls. The overall detection rate of ETBF was 7.2% (63 of 874) that prevailed evenly in cases and controls (7.2% each; 32 of 446 cases and 31 of 428 controls). ETBF was detected as the sole pathogen in 14 of 32 (44%) cases and 12 of 31(39%) controls. However, these results were not statistically significant. Thirty-five ETBF positive samples were associated with different enteric pathogens and equally found in cases and controls (57% each). Details of co-pathogens associated with ETBF are presented in Table 2.
Table 2. Prevalence of ETBF as sole and with other pathogens in children with diarrhea and controls.
| Pathogen | Case (n = 446) | Control (n = 428) |
| ETBF as a sole pathogen | 14 | 12 |
| Adenovirus | 1 | 1 |
| C. difficile | 2 | 1 |
| Campylobacter jejuni and Adenovirus | 1 | |
| C. jejuni and EPEC | 1 | |
| C. jejuni and Norovirus GII | 1 | |
| C. coli, Adenoviurs and Giardia | 1 | |
| EAEC | 1 | 2 |
| EAEC and Adenovirus | 1 | |
| EPEC | 1 | |
| EPEC, Adeno virus and Giardia | 1 | |
| EPEC, H. pylori | 1 | |
| Giardia | 3 | 6 |
| Giardia and Adenovirus | 1 | |
| Giardia, Cryptosporidium | 1 | |
| Giardia, H. pylori | 3 | |
| Giardia and Norovirus GII | 1 | |
| H. pylori | 2 | 2 |
| Norovirus GII | 1 | |
| Shigella spp | 1 |
Comparative analysis revealed that the detection rate of ETBF among the three age groups in both case and control were 18(9.3%), 7 (5.6%), 7 (5.6%) and 17 (10.1%), 8 (6.6%), 6 (4.3%), respectively for up to 1 years, >1–2 years and >2 years. There was no difference between cases and controls in the prevalence of ETBF. However, ETBF detection rate in male children under 1year of age group was significant (p<0.05) in cases as well as controls as compared to>2 years of children (Table 1). By culture method, 30 (47.6%) ETBF isolates (16 and 14 from cases and controls, respectively) were identified from 63 stool DNA-direct PCR positive specimens and all the ETBF isolates were also positive in the species-specific 16S-rRNA PCR. Five ETBF isolates lost their viability during storage.
ETBF subtypes
In the PCR-RFLP analysis, bft-1 and bft-3 alleles of the toxin encoding genes were identified among 25 ETBF isolates. There were no differences in the distribution of these alleles among ETBF isolates from cases and controls (bft-1 40% and bft-3 60% in cases and controls, respectively).
Antimicrobial susceptibility testing
Based on the MIC cut-off values, the antimicrobial testing results were categorized as resistant and susceptible. The ETBF isolates were uniformly susceptible to imipenem and chloramphenicol. The resistance frequencies to ampicillin, ampicillin/sulbactum, amoxicillin/potassium clavulanate, cefoxitin, clindamycin, ciprofloxacin, metronidazole, moxifloxacin, and norfloxacin remained 92, 48, 60, 8, 48, 88, 44, 32 and 92%, respectively.
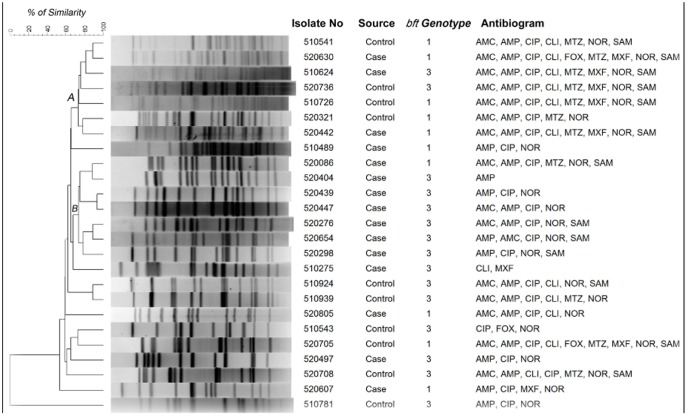
PFGE
PFGE was performed with ETBF isolated from 15 cases and 10 controls. The UPGMA based dendrogram displayed two major clades (A and B) containing 14 isolates with 75% homology (Fig. 1). Except two isolates in the clade B, majority of the ETBF remained genetically heterogeneous and there is no clear demarcation of ETBF isolated from cases and controls. However, majority of ETBF in clade B were isolated from diarrheal cases. In the clade A, of the 7 ETBF, 4 were from controls and resistant to 5–8 antimicrobials including metronidazole and moxifloxacin. ETBF isolates in the clade B were resistant for 1–5 antimicrobials but susceptible for metronidazole and moxifloxacin. In addition, all the isolates in clade B harbored bft-3 (Fig.1).
Figure 1. Not1 restriction patterns of genomic DNA of enterotoxigenic B. fragilis isolates.
The dendrogram was generated by using UPGAMA method.
Discussion
In this study, bft-PCR assay was performed using the DNA extracted from the stools. The designed primers for bft amplification cover all the three toxin genotypes. PCR assay for the detection of ETBF is more useful than culture method as the later needs prolonged anaerobic incubation followed by confirmation of the isolates. Of the 63 PCR positive specimens, culture results yielded 47.6% positivity for ETBF, with an overall isolation rate of 3.4%. In a previous study based on culture and bft-PCR from Goa, India showed that the detection rate of ETBF among the travelers with diarrhea was 13% [6]. In a study conducted in Kolkata by bacterial culture and toxin assay using tissue culture revealed that isolation rate of ETBF among acute diarrheal cases was 2.6% [12]. Similar to our results, low prevalence of ETBF was documented in Bangladesh (3–4%) and Brazil (2%) [21], [22]. In the HT29/C1 cell assay and the PCR based detection method showed that the association of ETBF is case and controls were not significant (Table 3).
Table 3. Prevalence of enterotoxigenic Bacteroides fragilis in diarrheal cases and controls in different studies.
| Place | Prevalence of ETBE (%) | Detection method | Remark | bft genotype (%) | Reference | |||
| Case | Control | Case | Control | |||||
| Bangladesh | 22 (6.1) n = 358 | 5 (1.2) n = 425 | HT29/C1 assay | p = 0.0001 | ND | ND | [7] | |
| Apache and Bangladesh | 44 (4.4) n = 991 | 18 (3.1) n = 581 | HT29/C1 assay | NS | ND | ND | [48] | |
| India | 6 (2.6) n = 226 | 3 (1.7) n = 172 | HT29/C1 assay | NS | ND | ND | [12] | |
| Italy | 14 (21.5) n = 65 | 9 (6.9) n = 129 | HT29Cl assay | NS | ND | ND | [24] | |
| Bangladesh | 28 (3.5) n = 814 | 12 (1.5) n = 814 | HT29/C1 assay | p = 0.01 | ND | ND | [21] | |
| Sweden | 195 (26.8) n = 728 | 24 (12.4) n = 194 | HT29/C1 assay | NS | ND | ND | [28] | |
| Brazil | 2 (2.1) n = 96 | 0 n = 74 | HT29/C1 assay | NS | ND | ND | [22] | |
| Bangladesh | 40 (2.3) n = 1750 | 15 (0.3) n = 5679 | HT29/C1 assay | p = 0.001 | ND | ND | [30] | |
| Vietnam | 43 (7.3) n = 587 | 6 (2.4)n = 249 | Immuno and PCR | p = 0.01 | Bft-1 (67.4), Bft-2 (18.6) Bft-3 (16.0) | Bft-2 (83.0), Bft-2 (17.0) | [27] | |
| Turkey | 13 (11.0) n = 117 | 8 (7.8)n = 102 | PCR | p = 0.05 | ND | ND | [25] | |
| Turkey | 28 (38.0) n = 73 | 7 (12.0) n = 59 | PCR | p = 0.009 | ND | ND | [34]* | |
| Turkey | 29 (15.0) n = 200 | 27 (14.0) n = 200 | PCR | NS | Bft-1 (82.7), Bft-2 (17.3) | Bft-1 (88.9), Bft-2 (11.1) | [23] | |
| Brazil | 9 (8.2) n = 110 | 7 (4.7) n = 150 | Real time PCR | NS | Bft-1 (8.2), Bft-3 (0.9) | Bft-1 (4.7) | [46] | |
Abbreviations: ND, not done; NS, not significant. * Study with colorectal cancer patients
In almost half the number of PCR-positive stools, we could not isolate ETBF, though we tested several typical B. fragilis colonies from each specimen. The recovery rate of B. fragilis would have been better if we used strict anaerobic conditions at the time of stool collection, transport and during processing. However, with the use of DNA based PCR assay, we could detect the ETBF almost two times more (7.2%) than the culture method. For the detection of ETBF, molecular based detection methods are very useful as the assays are sensitive, rapid and easier to perform. A real-time PCR approach may also be helpful for the rapid diagnosis of ETBF.
This study is the first of its kind as we examined the ETBF burden among young children in India. The prevalence rate of ETBF among diarrheal patients and asymptomatic controls from different countries are shown in Table 3. In accordance with studies conducted in Turkey and Italy, the prevalence of ETBF in Kolkata was almost equal in children with diarrhea and controls [23], [24]. However, reports from Vietnam, Turkey, Apache Indians in USA and Bangladesh, showed that the prevalence of ETBF was significantly high in cases and controls [25]–[27]. The highest prevalence of ETBF has been documented in countries such as Turkey (25%), Sweden (23%), Italy (17%), and in apache region of Arizona-USA (12%).
Overall, the prevalence of ETBF was the same in both cases and controls. However, considering the sole infection status, ETBF was comparatively identified more in cases (44%; 14/32) than in controls (39%; 12/31). In the investigations carried out in Bangladesh, Sweden, Turkey, Japan and Nicaragua, ETBF was detected as the only pathogen from 39 to 88% of the diarrheal cases [9], [25], [28], [29]. These findings support the view that there must be specific geographical difference in the prevalence of ETBF. In addition, findings from several countries show that ETBF significantly high in children with older age group [7], [9], [30]. Our findings show that ETBF was more frequently found in children less than 1 year age group (Table 1).
Polymicrobial etiology in diarrheal cases is a common trend in many endemic regions [31], [32]. We observed that the co-infection rate of ETBF with other pathogens was 4% in children with diarrhea, which is almost similar to the findings from Vietnam [27] or with higher age group patients from Bangladesh and Turkey [7], [23]. The significantly associated pathogens found with ETBF include enteropathogenic E. coli (EPEC), Shigella spp, Campylobacter spp., Salmonella spp., Clostridium difficile, Entameoeba histolytica, Cryptosporidium, Giardia spp. Rota virus and Adeno virus [7], [25], [27], [28]. Although we screened for all these pathogens, we found no significant association between ETBF and other pathogens.
Three different genotypes of bft have been documented in the ETBF and detection of these genetic signatures is useful in assessing the severity of the infection. Although the BFT has similar biological activity, their toxicity seems to differ based on its genotype. The purified BFT-2 elucidated higher biological activity than the other two genotypes [9], [10]. In addition, the bft-2 allele harboring ETBF colonize well in the intestines of children than in adults [33] and exhibit antibiotic associated diarrhea [9]. In this study, majority of the ETBF isolates harbored the bft-3 allele than bft-1 and none had the bft-2 allele. ETBF harboring the bft-1 allele has been reported from many countries (Table 3) [9], [23], [34]. In Turkey, in addition to bft-1, bft-2 allele was also identified in ETBF from diarrheal children and adults [23]. In Japan and Korea, prevalence of ETBF harboring bft-3 was reported in septicemia and diarrheal cases [9], [16] but this allele is rarely found in European countries [10], [33], [35]. We identified bft-3 predominantly in diarrheal cases and controls in Kolkata and perhaps this is the first report on the prevalence of bft-3 in Southeast Asia region.
Although ETBF causes self-limiting diarrhea, antimicrobial therapy is recommended to reduce the possibility of imminent extraintestinal complications. Despite antibiotic therapy, intestinal inflammation caused by ETBF may persist for about 3 weeks [15]. Several antimicrobial susceptibility studies have been documented with clinically isolated B. fragilis [36]–[38] but only few reports exist on ETBF [34], [39], [40]. Moxifloxacin alone or in combination with metronidazole is advocated for the empirical treatment of infections caused by Gram-negative anaerobes [41]. In addition, cefoxitin, clindamycin, and carbapenems are recommended for anaerobic infections. Recently, acquisition of resistance by B. fragilis to many of these antimicrobials has been documented [42]. To generate basic information on resistance nature of ETBF, we have used E-test method in this study, as this assay has been adopted for many anaerobes [19], [43]. We found that the ETBF are susceptible for chloramphenicol, and imipenem, but resistant to moxifloxacin and clindamycin. In addition, only 8 and 44% of the isolates are resistant to cefoxitin and metronidazole, respectively. Studies from in Brazil and Poland have documented that majority of the ETBF produced beta-lactamase, but susceptible for amoxicillin/clavulanic acid, imipenem and metronidazole [34], [44]. Rarely, some of the B. fragilis isolates from human stools were resistant for clindamycin and cefoxitin [39].
In the PFGE, the metronidazole and moxifloxacin susceptible and resistant ETBF isolates are clustered into two distinct groups. Overall, the PFGE results with ETBF isolates showed that they are genetically distinct. This trend seems to be common in many countries [39], [45]. Though we have identified many ETBF harboring bft-1 and bft-3 in this study, they are genetically different as evidenced from the PFGE. It is well known that in some bacterial species such as Campylobacter jejuni, diarrheagenic E. coli etc., the genetic constitution is largely diverse and hence the clonality of isolates in any given area may vary extensively.
Conclusion
This study highlights the prevalence of ETBF in children without any significant association with diarrheal cases or in controls. However, ETBF was significantly detected in male children younger than one year of age group compared to>2 years group. Overall, ETBF was predominantly detected as a co-pathogen along with enteric parasites and viruses. The bft-3 genotype was mostly seen than bft-1, without any specific age groups or the specimen category. Antimicrobial susceptibility results showed that all the ETBF isolates were susceptible to chloramphenicol, imipenem. Resistance of ETBF for clindamycin, moxifloxacin and metronidazole seems to be an emerging trend. Majority of the isolates are genetically heterogeneous as detected in the PFGE. More controlled long-term studies are required to prove the role of ETBF as an etiological agent for diarrhea.
Funding Statement
This study was supported by grants from Global Enteric Multicentric Study, University of Maryland, Baltimore, USA, and Indian Council of Medical Research (ICMR), India. AS is a recipient of Junior Research Fellowship from the National Academy of Sciences, Allahabad, India. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Claros MC, Claros ZC, Hecht DW, Citron DM, Goldstein EJ, et al. (2006) Characterization of the Bacteroides fragilis pathogenicity island in human blood culture isolates. Anaerobe 12: 17–22. [DOI] [PubMed] [Google Scholar]
- 2. Pamer EG (2007) Immune responses to commensal and environmental microbes. Nature Immunology 8: 1173–1178. [DOI] [PubMed] [Google Scholar]
- 3. Balamurugan R, Janardhan HP, George S, Raghava MV, Muliyil J, et al. (2008) Molecular studies of fecal anaerobic commensal bacteria in acute diarrhea in children. Journal of Pediatric Gastroenterology and Nutrition 46: 514–519. [DOI] [PubMed] [Google Scholar]
- 4. Lalitha MK, Koshi G (1980) Bacteroides in clinical infections. Indian Journal of Medical Research 71: 701–707. [PubMed] [Google Scholar]
- 5. Hill DR, Beeching NJ (2010) Travelers' diarrhea. Current Opinion in Infectious Diseases 23: 481–487. [DOI] [PubMed] [Google Scholar]
- 6. Jiang ZD, Dupont HL, Brown EL, Nandy RK, Ramamurthy T, et al. (2010) Microbial etiology of travelers' diarrhea in Mexico, Guatemala, and India: importance of enterotoxigenic Bacteroides fragilis and Arcobacter species. Jounral of Clinical Microbiology 48: 1417–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sack RB, Albert MJ, Alam K, Neogi PK, Akbar MS (1994) Isolation of enterotoxigenic Bacteroides fragilis from Bangladeshi children with diarrhea: a controlled study. Journal of Clinical Microbiology 32: 960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cohen SH, Shetab R, Tang–Feldman YJ, Sarma P, Silva J Jr, et al. (2006) Prevalence of enterotoxigenic Bacteroides fragilis in hospital-acquired diarrhea. Diagnostic Microbiology and Infectious Disease 55: 251–254. [DOI] [PubMed] [Google Scholar]
- 9. Kato N, Liu CX, Kato H, Watanabe K, Tanaka Y, et al. (2000) A new subtype of the metalloprotease toxin gene and the incidence of the three bft subtypes among Bacteroides fragilis isolates in Japan. FEMS Microbiology Letters 182: 171–176. [DOI] [PubMed] [Google Scholar]
- 10. Sears CL (2009) Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clinical Microbiology Reviews 22: 349–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wick EC, Sears CL (2010) Bacteroides spp. and diarrhea. Current Opinion in Infectious Diseases 23: 470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Niyogi SK, Dutta P, Mitra U, Pal DK (1997) Association of enterotoxigenic Bacteroides fragilis with childhood diarrhoea. Indian Journal of Medical Research 105: 167–169. [PubMed] [Google Scholar]
- 13. Panchalingam S, Antonio M, Hossain A, Mandomando I, Ochieng B, et al. (2012) Diagnostic Microbiologic Methods in the GEMS-1 Case/Control Study. Clinical Infectious Diseases 4: S294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsuki T, Watanabe K, Fujimoto J, Miyamoto Y, Takada T, et al. (2002) Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Applied Environmental Microbiology 68: 5445–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sears CL, Islam S, Saha A, Arjumand M, Alam NH, et al. (2008) Association of enterotoxigenic Bacteroides fragilis infection with inflammatory diarrhea. Clinical Infectious Diseases 47: 797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chung GT, Franco AA, Wu S, Rhie GE, Cheng R, et al. (1999) Identification of a third metalloprotease toxin gene in extraintestinal isolates of Bacteroides fragilis . Infection and Immunity 67: 4945–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CLSI (2009) Performance standards for antimicrobial susceptibility testing of anaerobic bacteria; informational supplement M11–S1.
- 18. Rennie RP, Turnbull L, Brosnikoff C, Cloke J (2012) First comprehensive evaluation of the M.I.C. evaluator device compared to Etest and CLSI reference dilution methods for antimicrobial susceptibility testing of clinical strains of anaerobes and other fastidious bacterial species. Journal of Clinical Microbiology 50: 1153–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rosenblatt JE, Gustafson DR (1995) Evaluation of the Etest for susceptibility testing of anaerobic bacteria. Diagnostic Microbiology and Infectious Disease 22: 279–284. [DOI] [PubMed] [Google Scholar]
- 20. Yamasaki S, Nair GB, Bhattacharya SK, Yamamoto S, Kurazono H, et al. (1997) Cryptic appearance of a new clone of Vibrio cholerae serogroup O1 biotype El Tor in Calcutta, India. Microbiology and Immunology 41: 1–6. [DOI] [PubMed] [Google Scholar]
- 21. Albert MJ, Faruque AS, Faruque SM, Sack RB, Mahalanabis D (1999) Case-control study of enteropathogens associated with childhood diarrhea in Dhaka, Bangladesh. Journal of Clinical Microbiology 37: 3458–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krzyzanowsky F, Avila-Campos MJ (2003) Detection of non-enterotoxigenic and enterotoxigenic Bacteroides fragilis in stool samples from children in São Paulo, Brazil. Revista do Instituto de Medicina Tropical de São Paulo 45: 225–227. [DOI] [PubMed] [Google Scholar]
- 23. Akpınar M, Aktaş E, Cömert F, Külah C, Sümbüloğlu V (2010) Evaluation of the prevalence of enterotoxigenic Bacteroides fragilis and the distribution bft gene subtypes in patients with diarrhea. Anaerobe 6: 505–509. [DOI] [PubMed] [Google Scholar]
- 24. Pantosti A, Menozzi MG, Frate A, Sanfilippo L, D'Ambrosio F, et al. (1997) Detection of enterotoxigenic Bacteroides fragilis and its toxin in stool samples from adults and children in Italy. Clinical Infectious Diseases 24: 12–16. [DOI] [PubMed] [Google Scholar]
- 25. Durmaz B, Dalgalar M, Durmaz R (2005) Prevalence of enterotoxigenic Bacteroides fragilis in patients with diarrhea: a controlled study. Anaerobe 11: 318–321. [DOI] [PubMed] [Google Scholar]
- 26. Sack RB, Myers LL, Almeido-Hill J, Shoop DS, Bradbury WC, et al. (1992) Enterotoxigenic Bacteroides fragilis: epidemiologic studies of its role as a human diarrhoeal pathogen. Journal of Diarrhoeal Diseases Research. 1992 10: 4–9. [PubMed] [Google Scholar]
- 27. Vu Nguyen T, Le Van P, Le Huy C, Weintraub A (2005) Diarrhea caused by enterotoxigenic Bacteroides fragilis in children less than 5 years of age in Hanoi, Vietnam. Anaerobe 11: 109–114. [DOI] [PubMed] [Google Scholar]
- 28. Zhang G, Svenungsson B, Kärnell A, Weintraub A (1999) Prevalence of enterotoxigenic Bacteroides fragilis in adult patients with diarrhea and healthy controls. Clinical Infectious Diseases 29: 590–594. [DOI] [PubMed] [Google Scholar]
- 29. Caceres M, Zhang G, Weintraub A, Nord CE (2000) Prevalence and antimicrobial susceptibility of enterotoxigenic Bacteroides fragilis in children with diarrhea in Nicaragua. Anaerobe 6: 143–148. [Google Scholar]
- 30. Pathela P, Hasan KZ, Roy E, Alam K, Huq F, et al. (2005) Enterotoxigenic Bacteroides fragilis -associated diarrhea in children 0–2 years of age in rural Bangladesh. Journal of Infectious Diseases 191: 1245–1252. [DOI] [PubMed] [Google Scholar]
- 31. Nair GB, Ramamurthy T, Bhattacharya MK, Krishnan T, Ganguly S, et al. (2010) Emerging trends in the etiology of enteric pathogens as evidenced from an active surveillance of hospitalized diarrhoeal patients in Kolkata, India. Gut Pathogens 2: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nimri LF, Elnasser Z, Batchoun R (2004) Polymicrobial infections in children with diarrhoea in a rural area of Jordan. FEMS Immunology and Medical Microbiology 42: 255–259. [DOI] [PubMed] [Google Scholar]
- 33. Scotto d'Abusco AS, Del Grosso M, Censini S, Covacci A, Pantosti A (2000) The alleles of the bft gene are distributed differently among enterotoxigenic Bacteroides fragilis strains from human sources and can be present in double copies. Journal of Clinical Microbiology 38: 607–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, et al. (2006) A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clinical Microbiology and Infection 12: 782–786. [DOI] [PubMed] [Google Scholar]
- 35. Ulger Toprak N, Rajendram D, Yagci A, Gharbia S, Shah HN, et al. (2006) The distribution of the bft alleles among enterotoxigenic Bacteroides fragilis strains from stool specimens and extraintestinal sites. Anaerobe 12: 71–74. [DOI] [PubMed] [Google Scholar]
- 36. Chaudhry R, Mathur P, Dhawan B, Kumar L (2001) Emergence of metronidazole-resistant Bacteroides fragilis, India. Emerging Infectious Diseases 7: 485–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee Y, Park Y, Kim MS, Yong D, Jeong SH, et al. (2010) Antimicrobial susceptibility patterns for recent clinical isolates of anaerobic bacteria in South Korea. Antimicrobial Agents and Chemotherapy 54: 3993–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paula GR, Falcão LS, Antunes EN, Avelar KE, Reis FN, et al. (2004) Determinants of resistance in Bacteroides fragilis strains according to recent Brazilian profiles of antimicrobial susceptibility. International Journal of Antimicrobial Agents 24: 53–58. [DOI] [PubMed] [Google Scholar]
- 39. Obuch-Woszczatyński P, Wintermans RG, Van Belkum A, Endtz H, Pituch H, et al. (2004) Enterotoxigenic Bacteroides fragilis (ETBF) strains isolated in The Netherlands and Poland are genetically diverse. Polish Journal of Microbiology 53: 35–39. [PubMed] [Google Scholar]
- 40. Rokosz A, Sawicka-Grzelak A, Kot K, Meszaros J, Łuczak M (2001) Use of the Etest method for antimicrobial susceptibility testing of obligate anaerobes. Medycyna Doswiadczalna i Mikrobiologia 53: 167–175. [PubMed] [Google Scholar]
- 41. Papaparaskevas J, Pantazatou A, Katsandri A, Houhoula DP, Legakis NJ, et al. (2008) Moxifloxacin resistance is prevalent among Bacteroides and Prevotella species in Greece. Journal of Antimicrobial Chemotherapy 62: 137–141. [DOI] [PubMed] [Google Scholar]
- 42. Wexler HM (2007) Bacteroides: the good, the bad, and the nitty-gritty. Clinical Microbiology Reviews 20: 593–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Galvão BP, Meggersee RL, Abratt VR (2011) Antibiotic resistance and adhesion potential of Bacteroides fragilis clinical isolates from Cape Town, South Africa. Anaerobe 17: 142–146. [DOI] [PubMed] [Google Scholar]
- 44. Nakano V, Padilla G, do Valle Marques M, Avila-Campos MJ (2004) Plasmid-related beta-lactamase production in Bacteroides fragilis strains. Research in Microbiology 155: 843–846. [DOI] [PubMed] [Google Scholar]
- 45. Antunes EN, Ferreira EO, Falcão LS, Paula GR, Avelar KE, et al. (2004) Non-toxigenic pattern II and III Bacteroides fragilis strains: coexistence in the same host. Research in Microbiology 155: 522–524. [DOI] [PubMed] [Google Scholar]
- 46. Merino VR, Nakano V, Liu C, Song Y, Finegold SM, et al. (2011) Quantitative detection of enterotoxigenic Bacteroides fragilis subtypes isolated from children with and without diarrhea. Journal of Clinical Microbiology 49: 416–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. San Joaquin VH, Griffis JC, Lee C, Sears CL (1995) Association of Bacteroides fragilis with childhood diarrhea. Scandinavian Journal of Infectious Diseases 27: 211–215. [DOI] [PubMed] [Google Scholar]