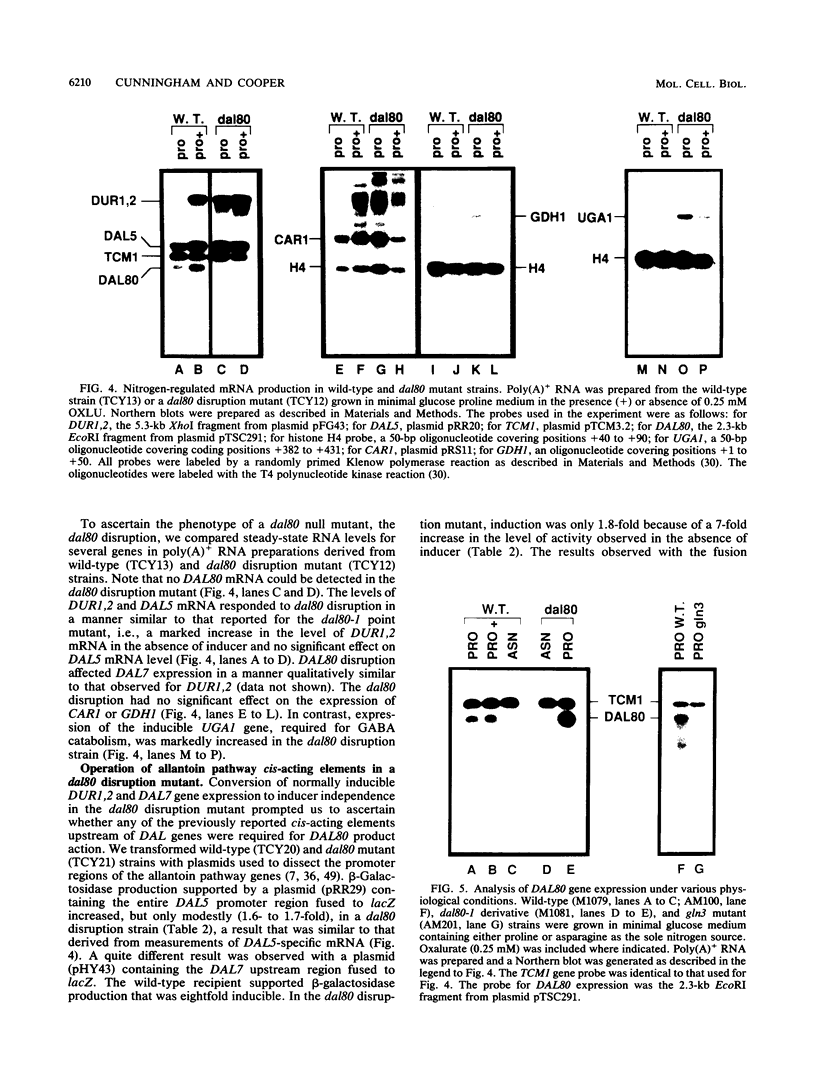
Abstract
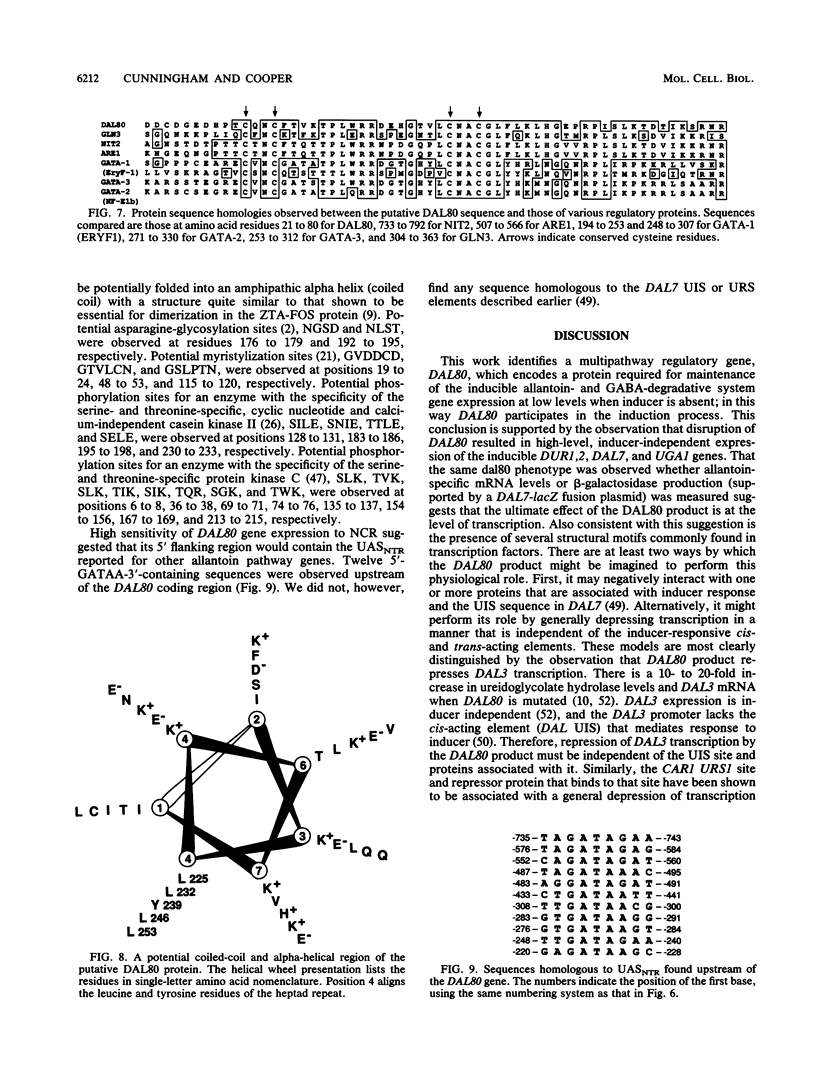
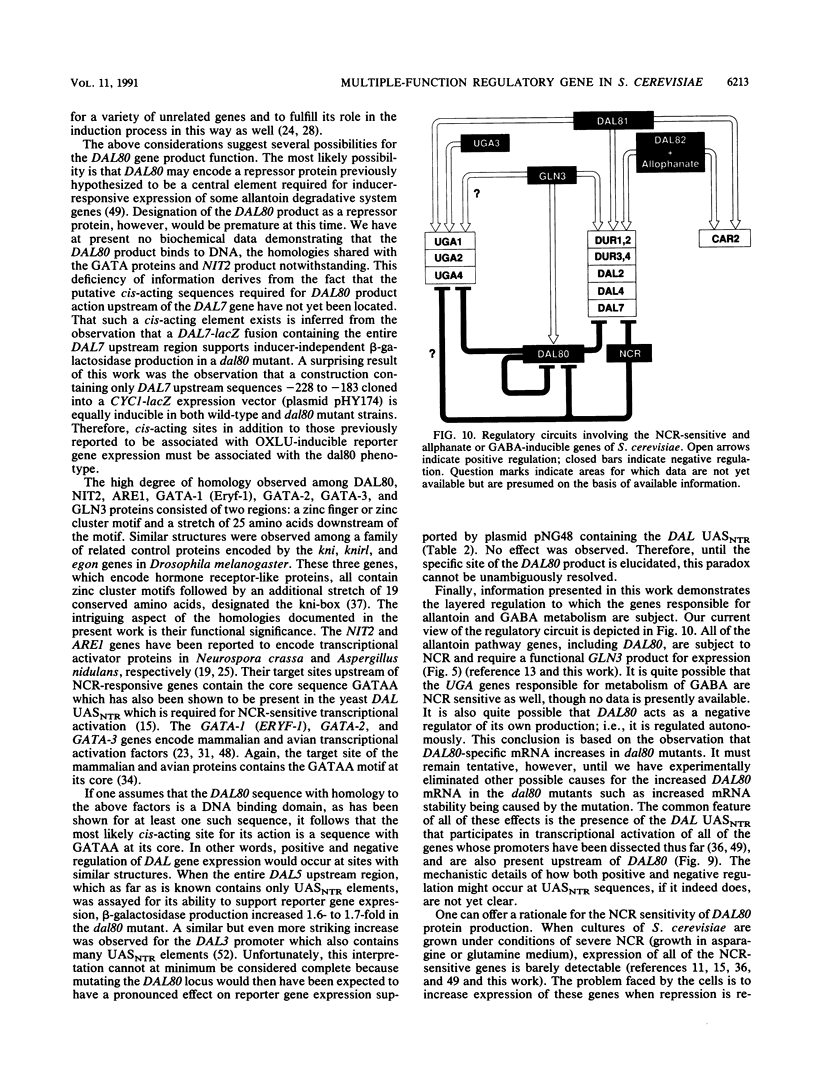
We have cloned the negative regulatory gene (DAL80) of the allantoin catabolic pathway, characterized its structure, and determined the physiological conditions that control DAL80 expression and its influence on the expression of nitrogen catabolic genes. Disruption of the DAL80 gene demonstrated that it regulates multiple nitrogen catabolic pathways. Inducer-independent expression was observed for the allantoin pathway genes DAL7 and DUR1,2, as well as the UGA1 gene required for gamma-aminobutyrate catabolism in the disruption mutant. DAL80 transcription was itself highly sensitive to nitrogen catabolite repression (NCR), and its promoter contained 12 sequences homologous to the NCR-sensitive UASNTR. The deduced DAL80 protein structure contains zinc finger and coiled-coil motifs. The DAL80 zinc finger motif possessed high homology to the transcriptional activator proteins required for expression of NCR-sensitive genes in fungi and the yeast GLN3 gene product required for functioning of the NCR-sensitive DAL UASNTR. It was also homologous to the three GATAA-binding proteins reported to be transcriptional activators in avian and mammalian tissues. The latter correlations raise the possibility that both positive and negative regulators of allantoin pathway transcription may bind to similar sequences.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
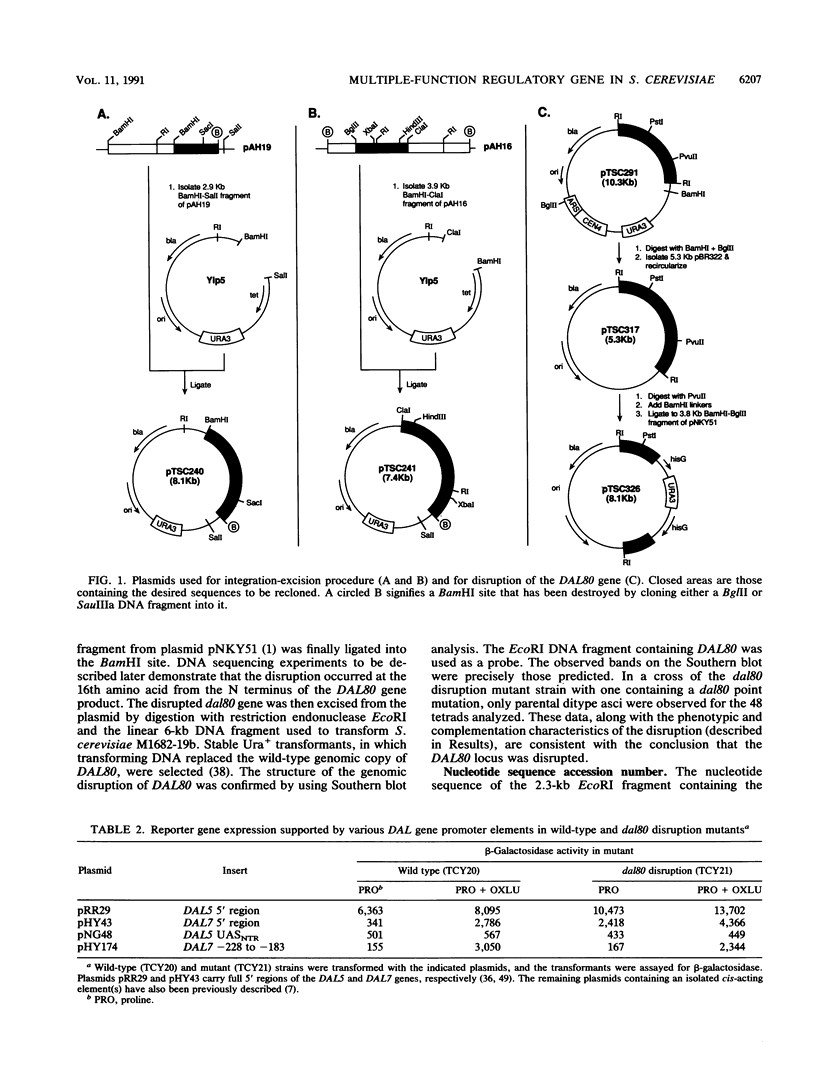
- Alani E., Cao L., Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987 Aug;116(4):541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bause E. Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes. Biochem J. 1983 Feb 1;209(2):331–336. doi: 10.1042/bj2090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossinger J., Lawther R. P., Cooper T. G. Nitrogen repression of the allantoin degradative enzymes in Saccharomyces cerevisiae. J Bacteriol. 1974 Jun;118(3):821–829. doi: 10.1128/jb.118.3.821-829.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricmont P. A., Cooper T. G. A gene product needed for induction of allantoin system genes in Saccharomyces cerevisiae but not for their transcriptional activation. Mol Cell Biol. 1989 Sep;9(9):3869–3877. doi: 10.1128/mcb.9.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricmont P. A., Daugherty J. R., Cooper T. G. The DAL81 gene product is required for induced expression of two differently regulated nitrogen catabolic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Feb;11(2):1161–1166. doi: 10.1128/mcb.11.2.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bysani N., Daugherty J. R., Cooper T. G. Saturation mutagenesis of the UASNTR (GATAA) responsible for nitrogen catabolite repression-sensitive transcriptional activation of the allantoin pathway genes in Saccharomyces cerevisiae. J Bacteriol. 1991 Aug;173(16):4977–4982. doi: 10.1128/jb.173.16.4977-4982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Chang Y. N., Dong D. L., Hayward G. S., Hayward S. D. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J Virol. 1990 Jul;64(7):3358–3369. doi: 10.1128/jvi.64.7.3358-3369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm G., Cooper T. G. Isolation and characterization of mutants that produce the allantoin-degrading enzymes constitutively in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Sep;2(9):1088–1095. doi: 10.1128/mcb.2.9.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Chisholm V. T., Cho H. J., Yoo H. S. Allantoin transport in Saccharomyces cerevisiae is regulated by two induction systems. J Bacteriol. 1987 Oct;169(10):4660–4667. doi: 10.1128/jb.169.10.4660-4667.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Ferguson D., Rai R., Bysani N. The GLN3 gene product is required for transcriptional activation of allantoin system gene expression in Saccharomyces cerevisiae. J Bacteriol. 1990 Feb;172(2):1014–1018. doi: 10.1128/jb.172.2.1014-1018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Lawther R. P. Induction of the allantoin degradative enzymes in Saccharomyces cerevisiae by the last intermediate of the pathway. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2340–2344. doi: 10.1073/pnas.70.8.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Rai R., Yoo H. S. Requirement of upstream activation sequences for nitrogen catabolite repression of the allantoin system genes in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Dec;9(12):5440–5444. doi: 10.1128/mcb.9.12.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert D., Vissers S., André B. The pleiotropic UGA35(DURL) regulatory gene of Saccharomyces cerevisiae: cloning, sequence and identity with the DAL81 gene. Gene. 1991 Jan 15;97(2):163–171. doi: 10.1016/0378-1119(91)90048-g. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. nit-2, the major nitrogen regulatory gene of Neurospora crassa, encodes a protein with a putative zinc finger DNA-binding domain. Mol Cell Biol. 1990 Mar;10(3):1056–1065. doi: 10.1128/mcb.10.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genbauffe F. S., Cooper T. G. Induction and repression of the urea amidolyase gene in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Nov;6(11):3954–3964. doi: 10.1128/mcb.6.11.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grand R. J. Acylation of viral and eukaryotic proteins. Biochem J. 1989 Mar 15;258(3):625–638. doi: 10.1042/bj2580625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko L. J., Yamamoto M., Leonard M. W., George K. M., Ting P., Engel J. D. Murine and human T-lymphocyte GATA-3 factors mediate transcription through a cis-regulatory element within the human T-cell receptor delta gene enhancer. Mol Cell Biol. 1991 May;11(5):2778–2784. doi: 10.1128/mcb.11.5.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovari L., Sumrada R., Kovari I., Cooper T. G. Multiple positive and negative cis-acting elements mediate induced arginase (CAR1) gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Oct;10(10):5087–5097. doi: 10.1128/mcb.10.10.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla B., Caddick M. X., Langdon T., Martinez-Rossi N. M., Bennett C. F., Sibley S., Davies R. W., Arst H. N., Jr The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990 May;9(5):1355–1364. doi: 10.1002/j.1460-2075.1990.tb08250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenzel E. A., Mulligan J. A., Sommercorn J., Krebs E. G. Substrate specificity determinants for casein kinase II as deduced from studies with synthetic peptides. J Biol Chem. 1987 Jul 5;262(19):9136–9140. [PubMed] [Google Scholar]
- Lawther R. P., Cooper T. G. Kinetics of induced and repressed enzyme synthesis in Saccharomyces cerevisiae. J Bacteriol. 1975 Mar;121(3):1064–1073. doi: 10.1128/jb.121.3.1064-1073.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche R. M., Sumrada R., Cooper T. G. A cis-acting element present in multiple genes serves as a repressor protein binding site for the yeast CAR1 gene. Mol Cell Biol. 1990 Aug;10(8):3884–3895. doi: 10.1128/mcb.10.8.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Minehart P. L., Magasanik B. Sequence and expression of GLN3, a positive nitrogen regulatory gene of Saccharomyces cerevisiae encoding a protein with a putative zinc finger DNA-binding domain. Mol Cell Biol. 1991 Dec;11(12):6216–6228. doi: 10.1128/mcb.11.12.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2758–2766. doi: 10.1128/mcb.4.12.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive M. G., Daugherty J. R., Cooper T. G. DAL82, a second gene required for induction of allantoin system gene transcription in Saccharomyces cerevisiae. J Bacteriol. 1991 Jan;173(1):255–261. doi: 10.1128/jb.173.1.255-261.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumb M., Frampton J., Wainwright H., Walker M., Macleod K., Goodwin G., Harrison P. GATAAG; a cis-control region binding an erythroid-specific nuclear factor with a role in globin and non-globin gene expression. Nucleic Acids Res. 1989 Jan 11;17(1):73–92. doi: 10.1093/nar/17.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Genbauffe F. S., Cooper T. G. Structure and transcription of the allantoate permease gene (DAL5) from Saccharomyces cerevisiae. J Bacteriol. 1988 Jan;170(1):266–271. doi: 10.1128/jb.170.1.266-271.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Genbauffe F. S., Sumrada R. A., Cooper T. G. Identification of sequences responsible for transcriptional activation of the allantoate permease gene in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):602–608. doi: 10.1128/mcb.9.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe M., Nauber U., Jäckle H. Three hormone receptor-like Drosophila genes encode an identical DNA-binding finger. EMBO J. 1989 Oct;8(10):3087–3094. doi: 10.1002/j.1460-2075.1989.tb08460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles J. I. Use of integrative transformation of yeast in the cloning of mutant genes and large segments of contiguous chromosomal sequences. Methods Enzymol. 1983;101:290–300. doi: 10.1016/0076-6879(83)01022-8. [DOI] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Isolation of the CAR1 gene from Saccharomyces cerevisiae and analysis of its expression. Mol Cell Biol. 1982 Dec;2(12):1514–1523. doi: 10.1128/mcb.2.12.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R., Cooper T. G. Oxaluric acid: a non-metabolizable inducer of the allantoin degradative enzymes in Saccharomyces cerevisiae. J Bacteriol. 1974 Mar;117(3):1240–1247. doi: 10.1128/jb.117.3.1240-1247.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoscy V., Cooper T. G. Pleiotropic control of five eucaryotic genes by multiple regulatory elements. J Bacteriol. 1982 Sep;151(3):1237–1246. doi: 10.1128/jb.151.3.1237-1246.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers S., Andre B., Muyldermans F., Grenson M. Induction of the 4-aminobutyrate and urea-catabolic pathways in Saccharomyces cerevisiae. Specific and common transcriptional regulators. Eur J Biochem. 1990 Feb 14;187(3):611–616. doi: 10.1111/j.1432-1033.1990.tb15344.x. [DOI] [PubMed] [Google Scholar]
- Winston F., Chumley F., Fink G. R. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 1983;101:211–228. doi: 10.1016/0076-6879(83)01016-2. [DOI] [PubMed] [Google Scholar]
- Woodgett J. R., Gould K. L., Hunter T. Substrate specificity of protein kinase C. Use of synthetic peptides corresponding to physiological sites as probes for substrate recognition requirements. Eur J Biochem. 1986 Nov 17;161(1):177–184. doi: 10.1111/j.1432-1033.1986.tb10139.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Ko L. J., Leonard M. W., Beug H., Orkin S. H., Engel J. D. Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev. 1990 Oct;4(10):1650–1662. doi: 10.1101/gad.4.10.1650. [DOI] [PubMed] [Google Scholar]
- Yoo H. S., Cooper T. G. The DAL7 promoter consists of multiple elements that cooperatively mediate regulation of the gene's expression. Mol Cell Biol. 1989 Aug;9(8):3231–3243. doi: 10.1128/mcb.9.8.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. S., Genbauffe F. S., Cooper T. G. Identification of the ureidoglycolate hydrolase gene in the DAL gene cluster of Saccharomyces cerevisiae. Mol Cell Biol. 1985 Sep;5(9):2279–2288. doi: 10.1128/mcb.5.9.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vuuren H. J., Daugherty J. R., Rai R., Cooper T. G. Upstream induction sequence, the cis-acting element required for response to the allantoin pathway inducer and enhancement of operation of the nitrogen-regulated upstream activation sequence in Saccharomyces cerevisiae. J Bacteriol. 1991 Nov;173(22):7186–7195. doi: 10.1128/jb.173.22.7186-7195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]