Abstract
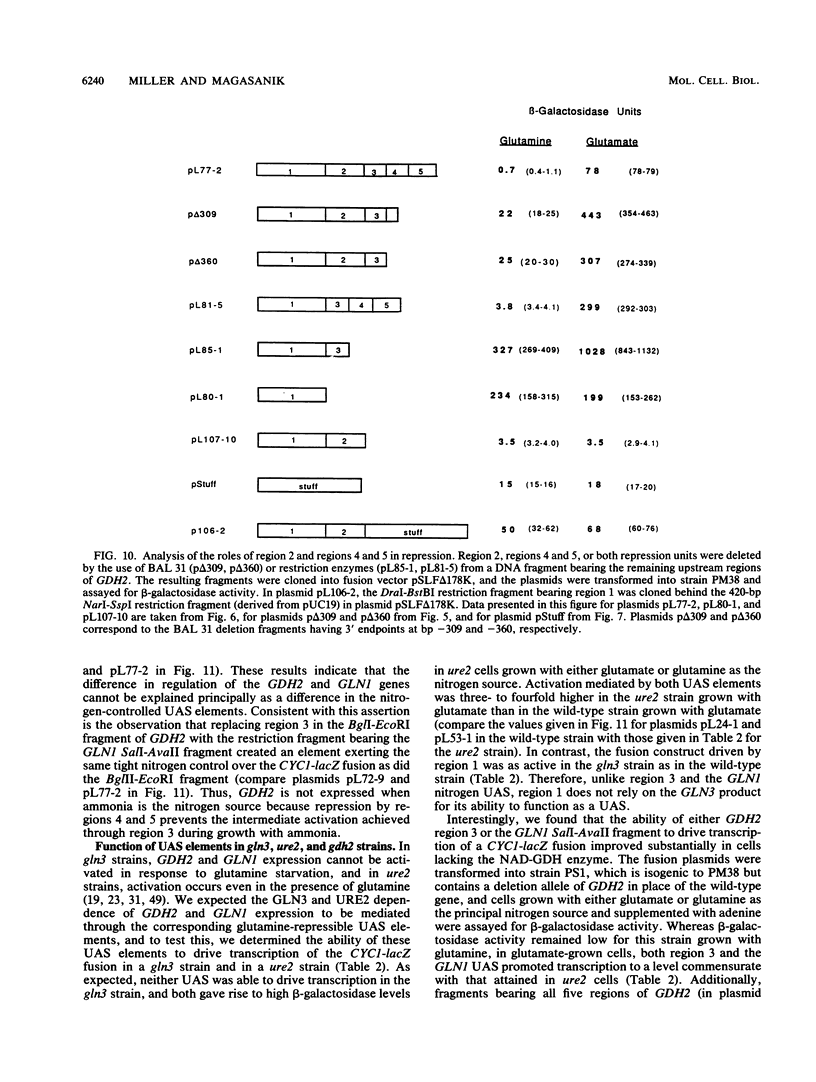
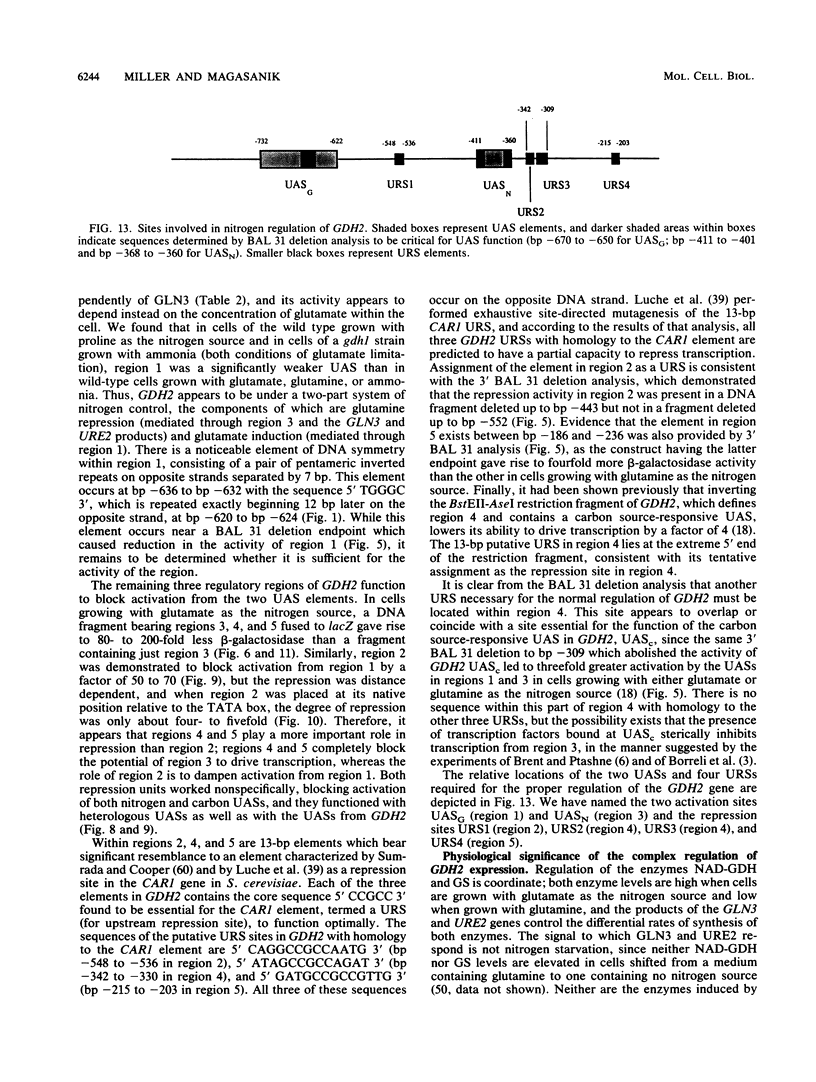
We analyzed the upstream region of the GDH2 gene, which encodes the NAD-linked glutamate dehydrogenase in Saccharomyces cerevisiae, for elements important for the regulation of the gene by the nitrogen source. The levels of this enzyme are high in cells grown with glutamate as the sole source of nitrogen and low in cells grown with glutamine or ammonium. We found that this regulation occurs at the level of transcription and that a total of six sites are required to cause a CYC1-lacZ fusion to the GDH2 gene to be regulated in the same manner as the NAD-linked glutamate dehydrogenase. Two sites behaved as upstream activation sites (UASs). The remaining four sites were found to block the effects of the two UASs in such a way that the GDH2-CYC1-lacZ fusion was not expressed unless the cells containing it were grown under conditions favorable for the activity of both UASs. This complex regulatory system appears to account for the fact that GDH2 expression is exquisitely sensitive to glutamine, whereas the expression of GLN1, coding for glutamine synthetase, is not nearly as sensitive.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benjamin P. M., Wu J. I., Mitchell A. P., Magasanik B. Three regulatory systems control expression of glutamine synthetase in Saccharomyces cerevisiae at the level of transcription. Mol Gen Genet. 1989 Jun;217(2-3):370–377. doi: 10.1007/BF02464906. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Borrelli E., Hen R., Chambon P. Adenovirus-2 E1A products repress enhancer-induced stimulation of transcription. Nature. 1984 Dec 13;312(5995):608–612. doi: 10.1038/312608a0. [DOI] [PubMed] [Google Scholar]
- Bossinger J., Cooper T. Possible failure of NADP-glutamate dehydrogenase to participate directly in nitrogen repression of the allantoin degradative enzymes in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1975 Oct 6;66(3):889–892. doi: 10.1016/0006-291x(75)90723-8. [DOI] [PubMed] [Google Scholar]
- Bossinger J., Lawther R. P., Cooper T. G. Nitrogen repression of the allantoin degradative enzymes in Saccharomyces cerevisiae. J Bacteriol. 1974 Jun;118(3):821–829. doi: 10.1128/jb.118.3.821-829.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature. 1984 Dec 13;312(5995):612–615. doi: 10.1038/312612a0. [DOI] [PubMed] [Google Scholar]
- Buchanan J. M. The amidotransferases. Adv Enzymol Relat Areas Mol Biol. 1973;39:91–183. doi: 10.1002/9780470122846.ch2. [DOI] [PubMed] [Google Scholar]
- Carey M., Lin Y. S., Green M. R., Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990 May 24;345(6273):361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Carlson M., Osmond B. C., Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981 May;98(1):25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman D. I., Leatherwood J., Carey M., Ptashne M., Kornberg R. D. Activation of yeast polymerase II transcription by herpesvirus VP16 and GAL4 derivatives in vitro. Mol Cell Biol. 1989 Nov;9(11):4746–4749. doi: 10.1128/mcb.9.11.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm G., Cooper T. G. Isolation and characterization of mutants that produce the allantoin-degrading enzymes constitutively in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Sep;2(9):1088–1095. doi: 10.1128/mcb.2.9.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie-Martin F., Magasanik B. Role of integration host factor in the regulation of the glnHp2 promoter of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1631–1635. doi: 10.1073/pnas.88.5.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Ferguson D., Rai R., Bysani N. The GLN3 gene product is required for transcriptional activation of allantoin system gene expression in Saccharomyces cerevisiae. J Bacteriol. 1990 Feb;172(2):1014–1018. doi: 10.1128/jb.172.2.1014-1018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Rai R., Yoo H. S. Requirement of upstream activation sequences for nitrogen catabolite repression of the allantoin system genes in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Dec;9(12):5440–5444. doi: 10.1128/mcb.9.12.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschigano P. W., Magasanik B. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione s-transferases. Mol Cell Biol. 1991 Feb;11(2):822–832. doi: 10.1128/mcb.11.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschigano P. W., Miller S. M., Magasanik B. Physiological and genetic analysis of the carbon regulation of the NAD-dependent glutamate dehydrogenase of Saccharomyces cerevisiae. Mol Cell Biol. 1991 Sep;11(9):4455–4465. doi: 10.1128/mcb.11.9.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne W. E., Magasanik B. Ammonia regulation of amino acid permeases in Saccharomyces cerevisiae. Mol Cell Biol. 1983 Apr;3(4):672–683. doi: 10.1128/mcb.3.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne W. E., Magasanik B. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: roles of the URE2 and GLN3 genes. J Bacteriol. 1988 Feb;170(2):708–713. doi: 10.1128/jb.170.2.708-713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse G. F., Frischauf A., Lehrach H. An integrated and simplified approach to cloning into plasmids and single-stranded phages. Methods Enzymol. 1983;101:78–89. doi: 10.1016/0076-6879(83)01006-x. [DOI] [PubMed] [Google Scholar]
- Drillien R., Aigle M., Lacroute F. Yeast mutants pleiotropically impaired in the regulation of the two glutamate dehydrogenases. Biochem Biophys Res Commun. 1973 Jul 17;53(2):367–372. doi: 10.1016/0006-291x(73)90671-2. [DOI] [PubMed] [Google Scholar]
- Dubois E., Vissers S., Grenson M., Wiame J. M. Glutamine and ammonia in nitrogen catabolite repression of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1977 Mar 21;75(2):233–239. doi: 10.1016/0006-291x(77)91033-6. [DOI] [PubMed] [Google Scholar]
- Dunlop P. C., Meyer G. M., Roon R. J. Nitrogen catabolite repression of asparaginase II in Saccharomyces cerevisiae. J Bacteriol. 1980 Jul;143(1):422–426. doi: 10.1128/jb.143.1.422-426.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T., Felsenfeld G. The erythroid-specific transcription factor Eryf1: a new finger protein. Cell. 1989 Sep 8;58(5):877–885. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Mutational analysis of upstream activation sequence 2 of the CYC1 gene of Saccharomyces cerevisiae: a HAP2-HAP3-responsive site. Mol Cell Biol. 1988 Feb;8(2):647–654. doi: 10.1128/mcb.8.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. Site-directed mutagenesis of the 'zinc finger' DNA-binding domain of the nitrogen-regulatory protein NIT2 of Neurospora. Mol Microbiol. 1990 Nov;4(11):1847–1852. doi: 10.1111/j.1365-2958.1990.tb02033.x. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. nit-2, the major positive-acting nitrogen regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5331–5335. doi: 10.1073/pnas.87.14.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Grenson M., Dubois E., Piotrowska M., Drillien R., Aigle M. Ammonia assimilation in Saccharomyces cerevisiae as mediated by the two glutamate dehydrogenases. Evidence for the gdhA locus being a structural gene for the NADP-dependent glutamate dehydrogenase. Mol Gen Genet. 1974;128(1):73–85. doi: 10.1007/BF00267295. [DOI] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain C., Vissers S., Dubois E., Legrain M., Wiame J. M. Regulation of glutamine synthetase from Saccharomyces cerevisiae by repression, inactivation and proteolysis. Eur J Biochem. 1982 Apr;123(3):611–616. doi: 10.1111/j.1432-1033.1982.tb06576.x. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Carey M., Ptashne M., Green M. R. How different eukaryotic transcriptional activators can cooperate promiscuously. Nature. 1990 May 24;345(6273):359–361. doi: 10.1038/345359a0. [DOI] [PubMed] [Google Scholar]
- Luche R. M., Sumrada R., Cooper T. G. A cis-acting element present in multiple genes serves as a repressor protein binding site for the yeast CAR1 gene. Mol Cell Biol. 1990 Aug;10(8):3884–3895. doi: 10.1128/mcb.10.8.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIDDELHOVEN W. J. THE PATHWAY OF ARGININE BREAKDOWN IN SACCHAROMYCES CEREVISIAE. Biochim Biophys Acta. 1964 Dec 9;93:650–652. doi: 10.1016/0304-4165(64)90349-6. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Colin D., ten Have J. P. Regulation of compartmentation of amino acid pools in Saccharomyces cerevisiae and its effects on metabolic control. Eur J Biochem. 1980 Jul;108(2):439–447. doi: 10.1111/j.1432-1033.1980.tb04740.x. [DOI] [PubMed] [Google Scholar]
- Middelhoven W. J. Induction and repression of arginase and ornithine transaminase in baker's yeast. Antonie Van Leeuwenhoek. 1970;36(1):1–19. doi: 10.1007/BF02069003. [DOI] [PubMed] [Google Scholar]
- Miller S. M., Magasanik B. Role of NAD-linked glutamate dehydrogenase in nitrogen metabolism in Saccharomyces cerevisiae. J Bacteriol. 1990 Sep;172(9):4927–4935. doi: 10.1128/jb.172.9.4927-4935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minehart P. L., Magasanik B. Sequence and expression of GLN3, a positive nitrogen regulatory gene of Saccharomyces cerevisiae encoding a protein with a putative zinc finger DNA-binding domain. Mol Cell Biol. 1991 Dec;11(12):6216–6228. doi: 10.1128/mcb.11.12.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2758–2766. doi: 10.1128/mcb.4.12.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P. The GLN1 locus of Saccharomyces cerevisiae encodes glutamine synthetase. Genetics. 1985 Oct;111(2):243–258. doi: 10.1093/genetics/111.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Genbauffe F. S., Sumrada R. A., Cooper T. G. Identification of sequences responsible for transcriptional activation of the allantoate permease gene in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):602–608. doi: 10.1128/mcb.9.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Genbauffe F., Lea H. Z., Cooper T. G. Transcriptional regulation of the DAL5 gene in Saccharomyces cerevisiae. J Bacteriol. 1987 Aug;169(8):3521–3524. doi: 10.1128/jb.169.8.3521-3524.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roon R. J., Even H. L., Larimore F. Glutamate synthase: properties of the reduced nicotinamide adenine dinucleotide-dependent enzyme from Saccharomyces cerevisiae. J Bacteriol. 1974 Apr;118(1):89–95. doi: 10.1128/jb.118.1.89-95.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzocchio C. Of moulds and men, or two fingers are not better than one. Trends Genet. 1990 Oct;6(10):311–313. doi: 10.1016/0168-9525(90)90238-2. [DOI] [PubMed] [Google Scholar]
- Schultz J., Carlson M. Molecular analysis of SSN6, a gene functionally related to the SNF1 protein kinase of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Oct;7(10):3637–3645. doi: 10.1128/mcb.7.10.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Berk A. J., Berget S. M. Transcription maps of adenovirus. Methods Enzymol. 1980;65(1):750–768. doi: 10.1016/s0076-6879(80)65071-x. [DOI] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Ubiquitous upstream repression sequences control activation of the inducible arginase gene in yeast. Proc Natl Acad Sci U S A. 1987 Jun;84(12):3997–4001. doi: 10.1073/pnas.84.12.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. F., Martin D. I., Zon L. I., D'Andrea A. D., Wong G. G., Orkin S. H. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989 Jun 8;339(6224):446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- Wiame J. M., Grenson M., Arst H. N., Jr Nitrogen catabolite repression in yeasts and filamentous fungi. Adv Microb Physiol. 1985;26:1–88. doi: 10.1016/s0065-2911(08)60394-x. [DOI] [PubMed] [Google Scholar]
- Wright R. M., Poyton R. O. Release of two Saccharomyces cerevisiae cytochrome genes, COX6 and CYC1, from glucose repression requires the SNF1 and SSN6 gene products. Mol Cell Biol. 1990 Mar;10(3):1297–1300. doi: 10.1128/mcb.10.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. S., Cooper T. G. The DAL7 promoter consists of multiple elements that cooperatively mediate regulation of the gene's expression. Mol Cell Biol. 1989 Aug;9(8):3231–3243. doi: 10.1128/mcb.9.8.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. S., Genbauffe F. S., Cooper T. G. Identification of the ureidoglycolate hydrolase gene in the DAL gene cluster of Saccharomyces cerevisiae. Mol Cell Biol. 1985 Sep;5(9):2279–2288. doi: 10.1128/mcb.5.9.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]



