Abstract
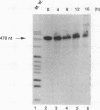
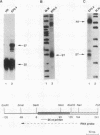
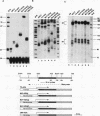
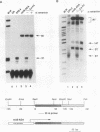
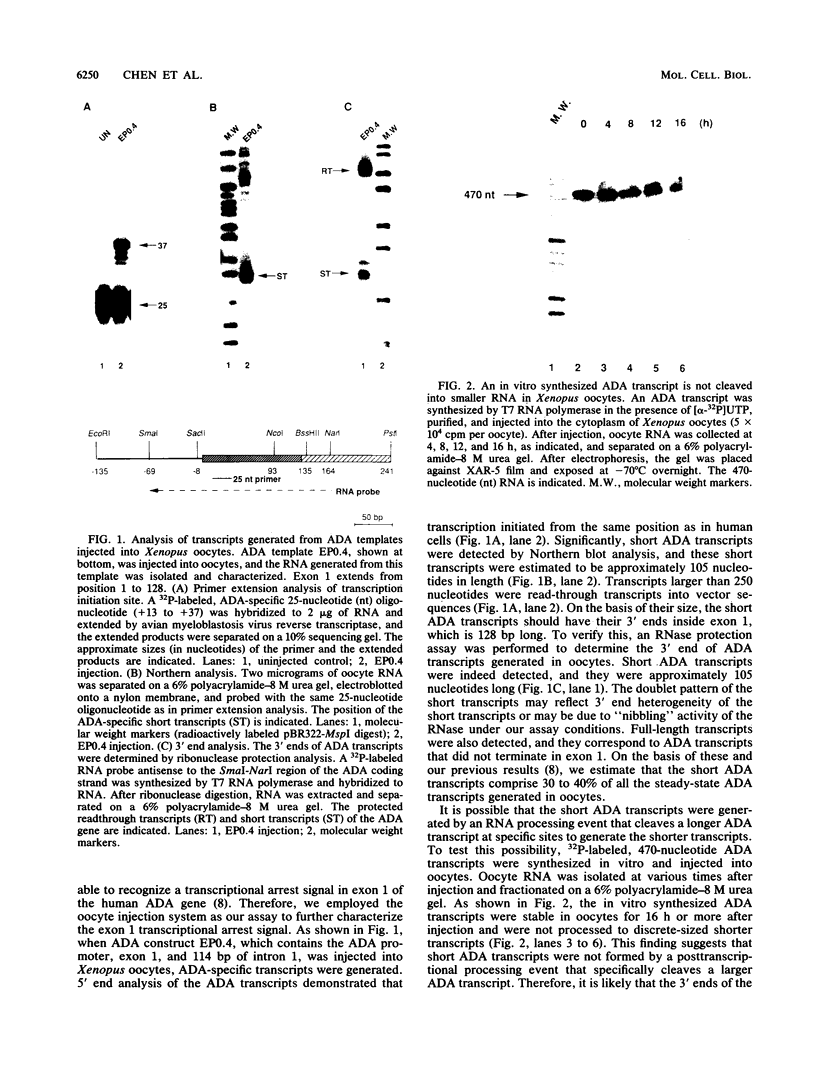
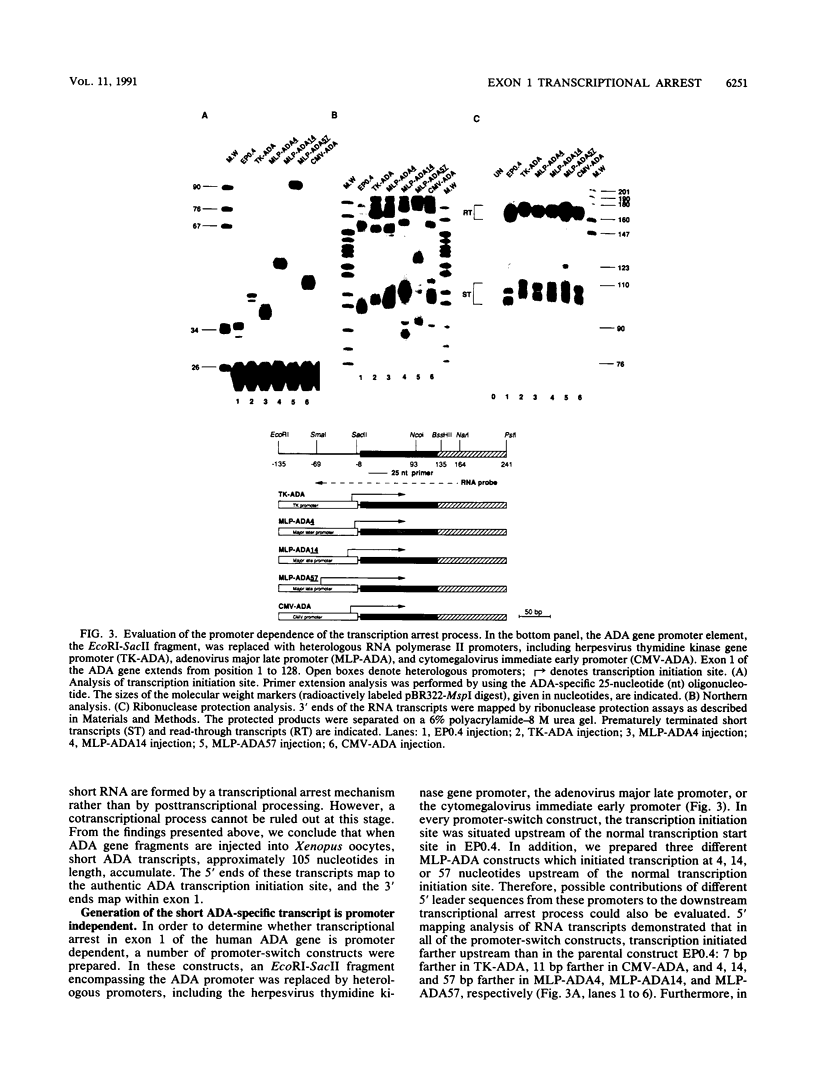
We have previously demonstrated that a transcriptional arrest site exists in exon 1 of the human adenosine deaminase (ADA) gene and that this site may play a role in ADA gene expression (Z. Chen, M. L. Harless, D. A. Wright, and R. E. Kellems, Mol. Cell. Biol. 10:4555-4564, 1990). Sequences involved in this process are not known precisely. To further define the template requirements for transcriptional arrest within exon 1 of the human ADA gene, various ADA templates were constructed and their abilities to confer transcriptional arrest were determined following injection into Xenopus oocytes. The exon 1 transcriptional arrest signal functioned downstream of several RNA polymerase II promoters and an RNA polymerase III promoter, implying that the transcriptional arrest site in exon 1 of the ADA gene is promoter independent. We identified a 43-bp DNA fragment which functions as a transcriptional arrest signal. Additional studies showed that the transcriptional arrest site functioned only in the naturally occurring orientation. Therefore, we have identified a 43-bp DNA fragment which functions as a transcriptional arrest signal in an orientation-dependent and promoter-independent manner. On the basis of our findings, we hypothesize that tissue-specific expression of the ADA gene is governed by factors that function as antiterminators to promote transcriptional readthrough of the exon 1 transcriptional arrest site.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aronow B., Lattier D., Silbiger R., Dusing M., Hutton J., Jones G., Stock J., McNeish J., Potter S., Witte D. Evidence for a complex regulatory array in the first intron of the human adenosine deaminase gene. Genes Dev. 1989 Sep;3(9):1384–1400. doi: 10.1101/gad.3.9.1384. [DOI] [PubMed] [Google Scholar]
- Barik S., Ghosh B., Whalen W., Lazinski D., Das A. An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell. 1987 Sep 11;50(6):885–899. doi: 10.1016/0092-8674(87)90515-0. [DOI] [PubMed] [Google Scholar]
- Ben-Asher E., Aloni Y. Transcription of minute virus of mice, an autonomous parvovirus, may be regulated by attenuation. J Virol. 1984 Oct;52(1):266–276. doi: 10.1128/jvi.52.1.266-276.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender T. P., Thompson C. B., Kuehl W. M. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987 Sep 18;237(4821):1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Chen Z., Harless M. L., Wright D. A., Kellems R. E. Identification and characterization of transcriptional arrest sites in exon 1 of the human adenosine deaminase gene. Mol Cell Biol. 1990 Sep;10(9):4555–4564. doi: 10.1128/mcb.10.9.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinsky J. M., Maa M. C., Ramamurthy V., Kellems R. E. Adenosine deaminase gene expression. Tissue-dependent regulation of transcriptional elongation. J Biol Chem. 1989 Aug 25;264(24):14561–14565. [PubMed] [Google Scholar]
- Christianson T. W., Clayton D. A. A tridecamer DNA sequence supports human mitochondrial RNA 3'-end formation in vitro. Mol Cell Biol. 1988 Oct;8(10):4502–4509. doi: 10.1128/mcb.8.10.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J., Sussman D. J., Zeller R., Leder P. The c-myc gene encodes superimposed RNA polymerase II and III promoters. Cell. 1987 Dec 24;51(6):1001–1008. doi: 10.1016/0092-8674(87)90586-1. [DOI] [PubMed] [Google Scholar]
- Collart M. A., Tourkine N., Belin D., Vassalli P., Jeanteur P., Blanchard J. M. c-fos gene transcription in murine macrophages is modulated by a calcium-dependent block to elongation in intron 1. Mol Cell Biol. 1991 May;11(5):2826–2831. doi: 10.1128/mcb.11.5.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S., Manley J. L. RNA polymerase II transcription termination is mediated specifically by protein binding to a CCAAT box sequence. Mol Cell Biol. 1989 Nov;9(11):5254–5259. doi: 10.1128/mcb.9.11.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg M. B., Baltimore D., Frankel A. D. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc Natl Acad Sci U S A. 1991 May 1;88(9):4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort P., Rech J., Vie A., Piechaczyk M., Bonnieu A., Jeanteur P., Blanchard J. M. Regulation of c-fos gene expression in hamster fibroblasts: initiation and elongation of transcription and mRNA degradation. Nucleic Acids Res. 1987 Jul 24;15(14):5657–5667. doi: 10.1093/nar/15.14.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley J. D., Waterfield M. D. Contributory effects of de novo transcription and premature transcript termination in the regulation of human epidermal growth factor receptor proto-oncogene RNA synthesis. J Biol Chem. 1991 Jan 25;266(3):1746–1753. [PubMed] [Google Scholar]
- Hay N., Skolnik-David H., Aloni Y. Attenuation in the control of SV40 gene expression. Cell. 1982 May;29(1):183–193. doi: 10.1016/0092-8674(82)90102-7. [DOI] [PubMed] [Google Scholar]
- Hernandez N., Lucito R. Elements required for transcription initiation of the human U2 snRNA gene coincide with elements required for snRNA 3' end formation. EMBO J. 1988 Oct;7(10):3125–3134. doi: 10.1002/j.1460-2075.1988.tb03179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz R. J., Li J., Greenblatt J. An elongation control particle containing the N gene transcriptional antitermination protein of bacteriophage lambda. Cell. 1987 Nov 20;51(4):631–641. doi: 10.1016/0092-8674(87)90132-2. [DOI] [PubMed] [Google Scholar]
- Innis J. W., Kellems R. E. A heat-labile factor promotes premature 3' end formation in exon 1 of the murine adenosine deaminase gene in a cell-free transcription system. Mol Cell Biol. 1991 Nov;11(11):5398–5409. doi: 10.1128/mcb.11.11.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobovits A., Smith D. H., Jakobovits E. B., Capon D. J. A discrete element 3' of human immunodeficiency virus 1 (HIV-1) and HIV-2 mRNA initiation sites mediates transcriptional activation by an HIV trans activator. Mol Cell Biol. 1988 Jun;8(6):2555–2561. doi: 10.1128/mcb.8.6.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S. Y., Calman A. F., Luciw P. A., Peterlin B. M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987 Dec 3;330(6147):489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. Intrinsic sites of transcription termination and pausing in the c-myc gene. Mol Cell Biol. 1988 Oct;8(10):4389–4394. doi: 10.1128/mcb.8.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M., Aloni Y. The block to transcription elongation at the SV40 attenuation site is decreased in vitro by oligomers complementary to segments of the attenuator RNA. Gene. 1989 Dec 7;84(1):65–72. doi: 10.1016/0378-1119(89)90140-6. [DOI] [PubMed] [Google Scholar]
- Krystal G., Birrer M., Way J., Nau M., Sausville E., Thompson C., Minna J., Battey J. Multiple mechanisms for transcriptional regulation of the myc gene family in small-cell lung cancer. Mol Cell Biol. 1988 Aug;8(8):3373–3381. doi: 10.1128/mcb.8.8.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Bartsch I., Grummt I. Specific interaction of the murine transcription termination factor TTF I with class-I RNA polymerases. Nature. 1990 Apr 5;344(6266):559–562. doi: 10.1038/344559a0. [DOI] [PubMed] [Google Scholar]
- Lamb N. J., Fernandez A., Tourkine N., Jeanteur P., Blanchard J. M. Demonstration in living cells of an intragenic negative regulatory element within the rodent c-fos gene. Cell. 1990 May 4;61(3):485–496. doi: 10.1016/0092-8674(90)90530-r. [DOI] [PubMed] [Google Scholar]
- Lattier D. L., States J. C., Hutton J. J., Wiginton D. A. Cell type-specific transcriptional regulation of the human adenosine deaminase gene. Nucleic Acids Res. 1989 Feb 11;17(3):1061–1076. doi: 10.1093/nar/17.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London L., Keene R. G., Landick R. Analysis of premature termination in c-myc during transcription by RNA polymerase II in a HeLa nuclear extract. Mol Cell Biol. 1991 Sep;11(9):4599–4615. doi: 10.1128/mcb.11.9.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maa M. C., Chinsky J. M., Ramamurthy V., Martin B. D., Kellems R. E. Identification of transcription stop sites at the 5' and 3' ends of the murine adenosine deaminase gene. J Biol Chem. 1990 Jul 25;265(21):12513–12519. [PubMed] [Google Scholar]
- Maderious A., Chen-Kiang S. Pausing and premature termination of human RNA polymerase II during transcription of adenovirus in vivo and in vitro. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5931–5935. doi: 10.1073/pnas.81.19.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A DNA-binding protein is required for termination of transcription by RNA polymerase I in Xenopus laevis. Mol Cell Biol. 1990 Jun;10(6):2793–2800. doi: 10.1128/mcb.10.6.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechti N., Piechaczyk M., Blanchard J. M., Jeanteur P., Lebleu B. Sequence requirements for premature transcription arrest within the first intron of the mouse c-fos gene. Mol Cell Biol. 1991 May;11(5):2832–2841. doi: 10.1128/mcb.11.5.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H., Asselin C., Dufort D., Yang J. Q., Gupta K., Marcu K. B., Nepveu A. A cis-acting element in the promoter region of the murine c-myc gene is necessary for transcriptional block. Mol Cell Biol. 1989 Dec;9(12):5340–5349. doi: 10.1128/mcb.9.12.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok M., Maderious A., Chen-Kiang S. Premature termination by human RNA polymerase II occurs temporally in the adenovirus major late transcriptional unit. Mol Cell Biol. 1984 Oct;4(10):2031–2040. doi: 10.1128/mcb.4.10.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy V., Maa M. C., Harless M. L., Wright D. A., Kellems R. E. Sequence requirements for transcriptional arrest in exon 1 of the murine adenosine deaminase gene. Mol Cell Biol. 1990 Apr;10(4):1484–1491. doi: 10.1128/mcb.10.4.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnasabapathy R., Sheldon M., Johal L., Hernandez N. The HIV-1 long terminal repeat contains an unusual element that induces the synthesis of short RNAs from various mRNA and snRNA promoters. Genes Dev. 1990 Dec;4(12A):2061–2074. doi: 10.1101/gad.4.12a.2061. [DOI] [PubMed] [Google Scholar]
- Reddy R., Henning D., Das G., Harless M., Wright D. The capped U6 small nuclear RNA is transcribed by RNA polymerase III. J Biol Chem. 1987 Jan 5;262(1):75–81. [PubMed] [Google Scholar]
- Reines D., Wells D., Chamberlin M. J., Kane C. M. Identification of intrinsic termination sites in vitro for RNA polymerase II within eukaryotic gene sequences. J Mol Biol. 1987 Jul 20;196(2):299–312. doi: 10.1016/0022-2836(87)90691-7. [DOI] [PubMed] [Google Scholar]
- Resnekov O., Aloni Y. RNA polymerase II is capable of pausing and prematurely terminating transcription at a precise location in vivo and in vitro. Proc Natl Acad Sci U S A. 1989 Jan;86(1):12–16. doi: 10.1073/pnas.86.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnekov O., Kessler M., Aloni Y. RNA secondary structure is an integral part of the in vitro mechanism of attenuation in simian virus 40. J Biol Chem. 1989 Jun 15;264(17):9953–9959. [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. Postinitiation transcriptional control in Drosophila melanogaster. Mol Cell Biol. 1990 Nov;10(11):6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988 Sep 9;54(6):795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Selby M. J., Bain E. S., Luciw P. A., Peterlin B. M. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989 Apr;3(4):547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- Skarnes W. C., Tessier D. C., Acheson N. H. RNA polymerases stall and/or prematurely terminate nearby both early and late promoters on polyomavirus DNA. J Mol Biol. 1988 Sep 5;203(1):153–171. doi: 10.1016/0022-2836(88)90099-x. [DOI] [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990 Jun;5(6):777–785. [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson R. J. Expression of the c-myb and c-myc genes is regulated independently in differentiating mouse erythroleukemia cells by common processes of premature transcription arrest and increased mRNA turnover. Mol Cell Biol. 1988 Sep;8(9):3938–3942. doi: 10.1128/mcb.8.9.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., Bishop J. M. DNA sequences that mediate attenuation of transcription from the mouse protooncogene myc. Proc Natl Acad Sci U S A. 1989 Jan;86(2):505–509. doi: 10.1073/pnas.86.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vegvar H. E., Lund E., Dahlberg J. E. 3' end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986 Oct 24;47(2):259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]