Abstract
Background
The hOGG1 gene encodes a DNA glycosylase enzyme responsible for DNA repair. The Ser326Cys polymorphism in this gene may influence its repair ability and thus plays a role in carcinogenesis. Several case-control studies have been conducted on this polymorphism and its relationship with the risk of hepatocellular carcinoma (HCC) among East Asians. However, their results are inconsistent.
Methods
We performed a meta-analysis of published case-control studies assessing the association of the hOGG1 Ser326Cys polymorphism with HCC risk among East Asians. PubMed, EMBASE, SCI, BIOSIS, CNKI and WanFang databases were searched. A random-effect model was used to calculate odds ratios (ORs) and 95% confidence intervals (95% CIs). Analyses were conducted for additive, dominant and recessive genetic models.
Results
Eight studies were identified involving 2369 cases and 2442 controls assessing the association of the hOGG1 Ser326Cys polymorphism with HCC risk among East Asians. Applying a dominant genetic model, only in the Chinese population, the Cys allele was significantly associated with increased risk of HCC (OR 1.56, 95% CI 1.12–2.17). However, two studies influenced this finding according to sensitivity analysis. Furthermore, considerable heterogeneity and bias existed among Chinese studies.
Conclusion
There is limited evidence to support that the hOGG1 Ser326Cys polymorphism is associated with HCC risk among East Asians. Well-designed and large-sized studies are required to determine this relationship.
Introduction
Hepatocellular carcinoma (HCC) is the sixth most prevalent cancer and the third most frequent cause of cancer-related death worldwide [1]. The highest prevalence of HCC is in East Asia due to the high prevalence of chronic infection with hepatitis B virus (HBV) [2]. Other well-established risk factors for HCC include chronic infection with hepatitis C virus (HCV), exposure to aflatoxin B1, male gender, drinking, smoking, non-alcoholic fatty liver disease and diabetes [3], [4], [5], [6], [7]. In the past two decades, more and more GWAS (genome-wide association studies) and other gene-disease association studies have found that some variants in human genes are associated with HCC, indicating that genetic background also plays a role in hepatocellular carcinogenesis.
Human 8-hydroxyguanine glycosylase 1 (hOGG1) is a DNA glycosylase enzyme responsible for the excision of 8-oxoguanine, a mutagenic base byproduct which occurs as a result of exposure to reactive oxygen [8]. The hOGG1 gene, located on chromosome 3p26.2, is composed of eight exons and seven introns. Polymorphisms in this gene may alter glycosylase function and an individual’s ability to repair damaged DNA, possibly resulting in genetic instability that can foster carcinogenesis [8]. Among many polymorphisms identified in the hOGG1 gene, much interest has been focused on the Ser326Cys (C>G) polymorphism (rs1052133). It is in exon 7 of the hOGG1 gene, which takes the form of a single amino acid substitution, from serine to cysteine at condon 326. Although the evidence is inconclusive that this functional polymorphic variation influences the activity of hOGG1 [8], many epidemiologic studies have been conducted to examine its relationship with cancer risk.
In the past years, several studies have investigated the association of the hOGG1 Ser326Cys polymorphism with HCC risk among East Asians [9], [10], [11], [12], [13], [14], [15], [16]. Some found that the Cys allele was associated with increased risk of HCC [13], [16]. However, the others found no association [9], [10], [11], [12], [14], [15]. Such inconsistency could be due partly to insufficient power, the small effect of the Ser326Cys polymorphism on HCC risk and false-positive results. We therefore performed a meta-analysis of published studies to investigate whether the Ser326Cys polymorphism has an effect on HCC susceptibility.
Methods
Searching
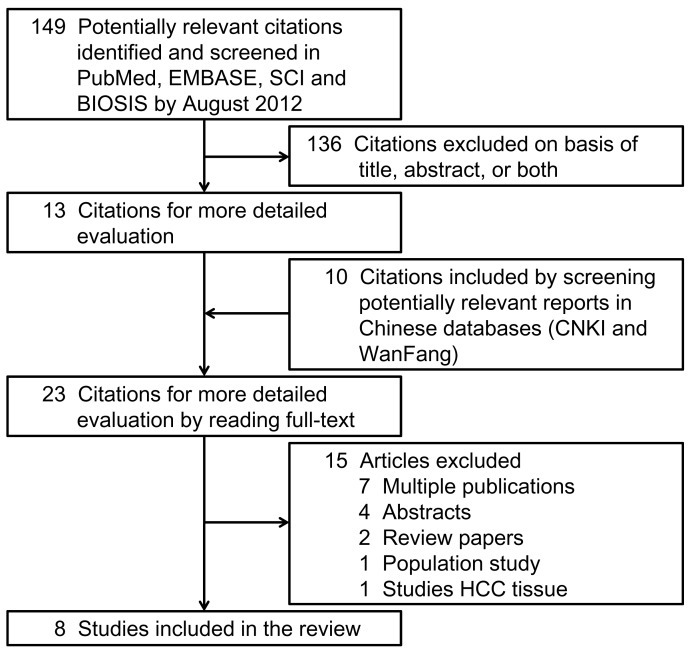
We searched PubMed, EMBASE, ISI Science Citation Index, BIOSIS, and Chinese electronic databases including CNKI and WanFang. The last search update was performed in August 2012. The search strategy was based on combinations of terms for hOGG1 and HCC (see Methods S1) without language restriction. References of retrieved reviews and articles for more detailed evaluation after reading the titles and abstracts were also screened. All case-control designed studies were considered eligible if they aimed to investigate the relation between the hOGG1 Ser326Cys polymorphism and HCC risk. Conference abstracts and review articles were excluded. Figure 1 describes the study selection process that led to the final 8 studies in this meta-analysis.
Figure 1. Flow diagram of the study selection process.
Two of the authors (WW & YL) independently identified and reviewed each relevant study. Disagreements were reconciled through group discussion. When more than one record was identified for the same study population, we included the most recent publication or population including more information.
Data Abstraction
Following the Meta-analysis Of Observational Studies in Epidemiology (MOOSE) statement for reporting meta-analyses of observational studies [17], we used a standardized reporting form to abstract data from each included study. For each study, the following information was extracted independently by two investigators (WW & YL): the first author’s name, year of publication, study design, ethnicity, definition and numbers of cases and controls, confounding factors by matching or adjustment, genotyping method, frequency of genotypes, odds ratios (ORs) and 95% confidence intervals (95% CIs) for HCC associated with the hOGG1 Ser326Cys polymorphism, and consistency of genotype frequencies with Hardy-Weinberg equilibrium (HWE) in control subjects.
Statistical Analysis
We referred to a previous study to perform statistical analysis [18]. ORs with 95% CIs were calculated to assess the strength of the association between the hOGG1 Ser326Cys polymorphism and HCC risk. The association was examined under three genetic models: the additive model (Cys/Cys vs. Ser/Ser), the dominant model (Ser/Cys+Cys/Cys vs. Ser/Ser) and the recessive model (Cys/Cys vs. Ser/Cys+Ser/Ser). HWE was tested using the chi-squared test and it was considered statistically significant when the P value is less than 0.05. Sensitivity analyses were carried out using the one-study remove approach to assess the impact of each study on the combined effect.
Heterogeneity assumption was checked by the I2 statistic and a chi-square based Q test. A P value of more than 0.05 for the Q test indicated a lack of heterogeneity among the studies, so the summary OR estimate of each study was calculated by the fixed-effect model (the Mantel-Haenszel method) [19]. Otherwise, the random-effect model (DerSimonian and Laird method) was used [20]. Egger’s test and Begg’s graphical methods were used to provide diagnosis of the potential publication bias [21]. All statistical analyses were performed with STATA software (version 10.0, StataCorp LP, College Station, Texas, USA) and RevMan software (version 5.1, Cochrane Collaboration). This meta-analysis has a protocol (see Methods S2). The performance and report of this meta-analysis comply with PRISMA Statement (see Methods S3).
Results
Eligible Studies
There were 8 studies identified on the hOGG1 Ser326Cys polymorphism and HCC susceptibility (Figure 1) [9], [10], [11], [12], [13], [14], [15], [16]. These 8 independent studies were published from 2004 to 2012 with 5 in Chinese language [9], [11], [12], [13], [14] and 3 in English [10], [15], [16]. In total, 2369 cases and 2442 controls were included. Table 1 shows the detailed characteristics of each study. All studies were conducted in East Asia, an area with high incidence of HCC. 6 studied Chinese population [9], [11], [12], [13], [14], [16], 1 studied Japanese population [10], and 1 studied Korean population [15]. 1 study did not supply age and sex information [9]. In the other 7 studies supplying this information, all but one had matched age and sex in case and control groups [10]. For control subjects, 4 studies recruited among hospital patients with HCC-unrelated diseases [9], [10], [11], [13], 2 studies among people with a comparable HBV background [12], [16], 1 study among people with chronic liver diseases (97% were infected with HCV and/or HBV) [10], 1 study among HBV chronically infected people [15], and 1 study among healthy people [14]. Only 1 study extracted DNA from formalin-fixed or paraffin-embedded liver tissues [9]. The others extracted from blood samples. All studies but one were consistent with HWE (P<0.001) [13].
Table 1. Characteristics of studies included in this meta-analysis.
| Study | Ethnicity | Screening of controls | Genotype method | Case | Control | Confounding factors adjusted or stratified | Cys(%) | HWE(P value) | ||||
| Male (%) | Age(mean, sd) | Cys-Cys/Cys-Ser/Ser-Ser | Male (%) | Age(mean, sd) | Cys-Cys/Cys-Ser/Ser-Ser | |||||||
| Zhu, 2004 | Chinese | Hospital | PCR-RFLP | NA | NA | 57/99/37 | NA | NA | 50/62/22 | Sex, age, smoking, drinking, HBV, HCV, family history of HCC | 60.4 | >0.7 |
| Sakamoto, 2006 | Japanese | Hospital | PCR-CTPP | 67% | 69 | 43/110/56 | 65% | 61 | 79/123/73 | Sex, age, smoking, drinking, HBV, HCV | 51.1 | >0.05 |
| CLD† | 54% | 61 | 100/176/105 | 49.3 | >0.1 | |||||||
| Zhang, 2006‡ | Chinese | Hospital | Sequencing | 84% | 49.9±18.0 | 18/38/30 | 82% | 49.9±17.0 | 13/35/42 | HBV | 33.9 | >0.2 |
| Wang, 2008 | Chinese | HBVcomparable§ | Taqman | 84% | 50.2±11.3 | 52/92/31 | matched | matched | 34/58/27 | Sex, age, smoking, drinking, HBV, HCV, family history of HCC | 52.9 | >0.8 |
| Ji, 2011 | Chinese | Hospital | Taqman | 79% | 47.8±11.1 | 103/40/357 | 74% | 48.9±11.2 | 29/51/427 | Drinking, HBV | 10.7 | <0.001 |
| Tang, 2011 | Chinese | Healthy | PCR-RFLP | 100% | matched | 47/82/21 | 100% | matched | 56/72/22 | None | 61.3 | >0.8 |
| Jung, 2012* | Korean | HBV | SNPstream | 82% | 53.3±8.3 | 207/343/156 | 83% | 52.6 | 116/181/89 | Sex, age | 53.3 | >0.2 |
| Yuan, 2012 | Chinese | HBVcomparable§ | PCR-RFLP | 76% | 52.1±7.6 | 66/217/67 | 75% | 51.1±9.2 | 50/206/144 | Sex, age, drinking, HBV, family history of HCC | 38.3 | >0.05 |
HWE, Hardy-Weinberg equilibrium; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; HBV, hepatitis B virus; HCV, hepatitis C virus; HCC, hepatocellular carcinoma; CLD, chronic liver diseases; CTPP, confronting two-pair primers; NA, not available.
In this control group, 97% of subjects were infected with HCV and/or HBV.
Sex and age information was from 91 case subjects and 91 control subjects.
Case group and control group had comparable background of HBV infection.
Sex and age information was from 708 case subjects and 388 control subjects.
Meta-analysis Results
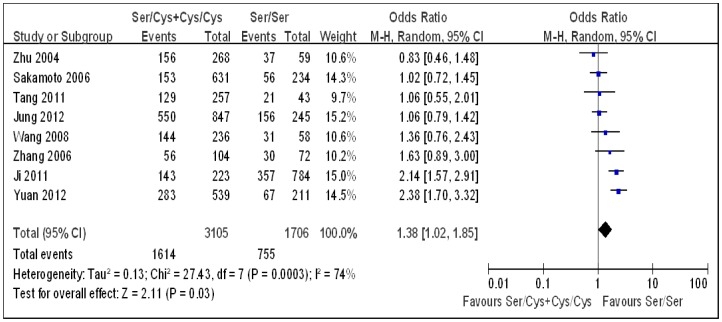
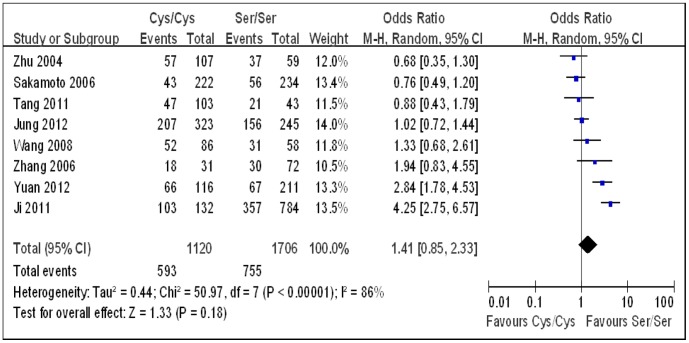
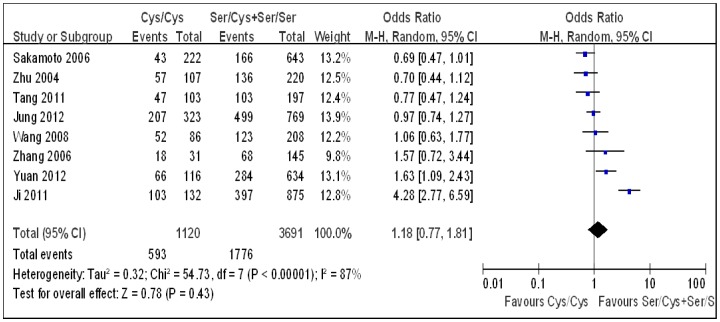
Under each genetic model, heterogeneity assessment showed significant variation across studies. Therefore, a random-effect model was used to analyze the summary ORs. As Sakamoto et al. used two control groups (hospital controls and chronic liver disease controls) [10], first we combined the two groups to carry out the overall analysis. Combining all studies, a significant positive association between the hOGG1 Ser326Cys polymorphism and HCC risk was observed under the dominant genetic model (OR 1.38, 95% CI 1.02–1.85; Table 2, Figure 2). No significant association was found under the additive (OR 1.41, 95% CI 0.85–2.33; Table 2, Figure 3) and the recessive models (OR 1.18, 95% CI 0.77–1.81; Table 2, Figure 4). Then, we used each control group in the study by Sakamoto et al. to carry out the overall analysis. The results hardly changed (data not shown). As the hOGG1 Ser326Cys polymorphism in control subjects did not fulfill HWE in the study by Ji et al. [13], we excluded this study and repeated the above analyses. No significant association was found under any genetic models (Table 2). Sensitivity analyses confirmed that several other studies also contributed to the observed significant association between the hOGG1 Ser326Cys polymorphism and HCC risk under the dominant genetic model (Table 3) [11], [12], [16].
Table 2. Meta-analysis of published association between the hOGG1 Ser326Cys polymorphism and HCC risk.
| Genetic model (No. of studies) | Case | Control | Analysis model | Summary OR (95% CI) | P † | P ‡ |
| Overall§ (8) | ||||||
| Additive model | 1348 | 1478 | Random | 1.41 (0.85–2.33) | <0.00001 | 0.86 |
| Dominant model | 2369 | 2442 | Random | 1.38 (1.02–1.85) | <0.001 | 0.52 |
| Recessive model | 2369 | 2442 | Random | 1.18 (0.77–1.81) | <0.00001 | 0.69 |
| Chinese population (6) | ||||||
| Additive model | 886 | 916 | Random | 1.67 (0.91–3.08) | <0.0001 | 0.08 |
| Dominant model | 1454 | 1400 | Random | 1.56 (1.12–2.17) | 0.01 | 0.03 |
| Recessive model | 1454 | 1400 | Random | 1.36 (0.76–2.42) | <0.00001 | 0.72 |
| Consistent with HWE§ (7) | ||||||
| Additive model | 888 | 1022 | Random | 1.18 (0.79–1.77) | <0.001 | 0.87 |
| Dominant model | 1869 | 1935 | Random | 1.27 (0.94–1.73) | 0.003 | 0.80 |
| Recessive model | 1869 | 1935 | Random | 0.96 (0.75–1.24) | 0.03 | 0.74 |
| HBV/HCV comparable control* (4) | ||||||
| Additive model | 678 | 665 | Random | 1.32 (0.76–2.30) | 0.001 | 0.77 |
| Dominant model | 1440 | 1286 | Random | 1.37 (0.90–2.11) | 0.002 | 0.97 |
| Recessive model | 1440 | 1286 | Random | 1.04 (0.76–1.43) | 0.04 | 0.84 |
| Hospital control# (4) | ||||||
| Additive model | 701 | 735 | Random | 1.42 (0.52–3.89) | <0.00001 | 0.60 |
| Dominant model | 988 | 1006 | Random | 1.33 (0.82–2.14) | 0.004 | 0.38 |
| Recessive model | 988 | 1006 | Random | 1.32 (0.49–3.51) | <0.00001 | 0.97 |
OR, odds ratio; CI, confidence interval.
P value for the Q test.
P value for Egger’s test.
In the study by Sakamoto, subjects from the hospital control and the CLD (chronic liver diseases) control were pooled together.
Including studies by Sakamoto, Wang, Jung and Yuan. In the study by Sakamoto, data from the CLD (chronic liver diseases) group was used.
In the study by Sakamoto, data from the hospital group was used.
Figure 2. Forest plots for the hOGG1 Ser326Cys polymorphism and risk of hepatocellular carcinoma using the dominant genetic model (Ser/Cys+Cys/Cys vs. Ser/Ser).
The squares and horizontal lines correspond to the study specific odds ratios and 95% confidence intervals. The diamond represents the summary odds ratio and 95% confidence interval.
Figure 3. Forest plots for the hOGG1 Ser326Cys polymorphism and risk of hepatocellular carcinoma using the additive genetic model (Cys/Cys vs. Ser/Ser).
The squares and horizontal lines correspond to the study specific odds ratios and 95% confidence intervals. The diamond represents the summary odds ratio and 95% confidence interval.
Figure 4. Forest plots for the hOGG1 Ser326Cys polymorphism and risk of hepatocellular carcinoma using the recessive genetic model (Cys/Cys vs. Ser/Cys+Ser/Ser).
The squares and horizontal lines correspond to the study specific odds ratios and 95% confidence intervals. The diamond represents the summary odds ratio and 95% confidence interval.
Table 3. Sensitivity analysis using the one-study remove approach.
| Study omitted | Additive model | Dominant model | Recessive model | |||
| OR | 95% CI | OR | 95% CI | OR | 95% CI | |
| Zhu, 2004 | 1.55 | 0.91–2.66 | 1.46 | 1.08–1.98 | 1.28 | 0.80–2.04 |
| Sakamoto, 2006 | 1.55 | 0.90–2.66 | 1.44 | 1.05–2.00 | 1.29 | 0.81–2.05 |
| Zhang, 2006 | 1.35 | 0.78–2.35 | 1.35 | 0.97–1.86 | 1.15 | 0.73–1.82 |
| Wang, 2008 | 1.42 | 0.81–2.49 | 1.37 | 0.99–1.91 | 1.20 | 0.75–1.95 |
| Ji, 2011 | 1.18 | 0.79–1.77 | 1.27 | 0.94–1.73 | 0.96 | 0.75–1.24 |
| Tang, 2011 | 1.50 | 0.86–2.59 | 1.41 | 1.03–1.95 | 1.26 | 0.79–2.02 |
| Jung, 2012 | 1.48 | 0.82–2.68 | 1.44 | 1.04–2.00 | 1.23 | 0.72–2.07 |
| Yuan, 2012 | 1.26 | 0.74–2.17 | 1.26 | 0.95–1.67 | 1.13 | 0.70–1.83 |
| Combined | 1.41 | 0.85–2.33 | 1.38 | 1.02–1.85 | 1.18 | 0.77–1.81 |
OR, odds ratio; CI, confidence interval.
Subgroup analyses were performed by dividing studies into groups according to ethnicity and source of controls. Only in the Chinese population, significant association between the hOGG1 Ser326Cys polymorphism and HCC risk was observed under the dominant genetic model (OR 1.56, 95% CI 1.12–2.17; Table 2). However, this association was lost if the study by Ji et al. or by Yuan et al. was removed (OR 1.41, 95% CI 0.93–2.14; OR 1.39, 95% CI 0.96–2.02; respectively) [13], [16].
Begg’s funnel plot and Egger’s test were performed to assess the publication bias of the literatures. The results indicated that bias may exist among Chinese studies (P Egger’s test = 0.03; Table 2).
Discussion
The association between the hOGG1 Ser326Cys polymorphism and HCC risk was not clear due to inconsistent data generated by a range of independent studies. Therefore we performed a meta-analysis of published studies to clarify the inconsistency and to establish a comprehensive picture of this gene-disease association. All studies included in this analysis were conducted in East Asia, an area with high prevalence of HCC. By pooling 8 studies with 2369 cases and 2442 controls, our meta-analysis showed a statistically significant but very weak association between the hOGG1 Ser326Cys polymorphism and HCC risk when applying a dominant genetic model. Further subgroup analyses revealed that this association only existed in the Chinese population. Other subgroup analyses excluding study not fulfilling HWE or regarding the source of controls did not produce any significant findings. In addition, considerable heterogeneity was detected across studies. And the heterogeneity cannot be fully explained by ethnicity, source of controls and whether fulfilling HWE or not. The study by Yuan et al. is the main source of heterogeneity [16]. This study, with a relatively large sample size (350 cases and 400 controls), studied Chinese population and after adjusting confounding factors exhibited an odds ratio of 2.38 (95% CI 1.80–3.14) [16], indicating a moderate association.
The significant positive findings from meta-analyses are not robust because they are sensitive to four studies in the overall analysis [11], [12], [13], [16] and to two studies in the Chinese subgroup analysis [13], [16] according to the leave-one-out sensitivity analysis. The Egger test suggested the existence of bias among Chinese studies. This may be due to reporting bias, other biases or genuine heterogeneity, and it is difficult to determine which is the case [22]. Taken together, there is limited evidence to support the association between the hOGG1 Ser326Cys polymorphism and HCC risk.
8-oxoguanine is one of the most common DNA lesions resulting from reactive oxygen species [23]. It has the ability to pair with adenine instead of cytosine during DNA replication, and therefore plays a role in carcinogenesis [24]. In human, hOGG1 is responsible for the repair of 8-oxoguanine. The conduction of studies to examine the hOGG1 Ser326Cys polymorphism and cancer risk, is based on the notion that this polymorphism may influence the enzyme activity of hOGG1 and thus influence the process of carcinogenesis through 8-oxoguanine. Some studies suggested that the 326Cys allele confers decreased ability to repair 8-oxoguanine [25], [26], [27]. Other studies, however, found no difference in activity by the hOGG1 Ser326Cys polymorphism [28], [29], [30], [31], [32], [33], [34], [35]. So, whether the hOGG1 Ser326Cys polymorphism has an impact on the repair of 8-oxoguanine is inconclusive.
High levels of 8-oxogudanine are found in HCC patients (liver tissue: adjacent nontumor tissue>tumor tissue>chronic viral hepatitis>control) and are closely associated with inflammatory infiltration [36], [37]. Peng et al. found that levels of 8-oxogudanine were high and levels of hOGG1 were low in peripheral leukocytes from adolescents in a high risk region for HCC in China. Individuals with the 326Ser allele rather than the 326Cys allele had a significantly higher concentration of leukocyte 8-oxogudanine level [38]. Tang et al. studied the urea 8-oxogudanine level in HCC patients, and did not find a relationship with the hOGG1 Ser326Cys polymorphism [14]. However, these studies had a relatively small sample size, and were unable to control for other factors that may affect 8-oxogudanine levels. Together with our meta-analysis, there is lacked evidence to support a link between the hOGG1 Ser326Cys polymorphism and HCC development.
Our meta-analysis has several limitations. Firstly, only 8 published studies were included, thus the meta-analysis was restricted to a relatively small population. All the 8 studies studied East Asian population, a population with high-HCC risk. So, our findings are not suitable for other populations, especially in Caucasian population which has a low-HCC risk. Recently, one study involving Caucasian population failed to find any association between the hOGG1 Ser326Cys polymorphism and HCC risk [39]. As it was reported as meeting abstract and further information was not available by contacting the authors, it was not included in our meta-analysis. Secondly, polymorphisms that affect disease susceptibility may do so only in the presence of a relevant exposure; in the case of hOGG1, these include hepatitis virus, smoking, alcohol consumption, meat intake, and other factors that are thought to induce DNA damage [8]. However, only three included studies reported the hOGG1 Ser326Cys polymorphism in populations exposed to some of the above factors [10], [11], [16]. And the numbers of involved subjects are too small to draw a conclusion. Thirdly, the heterogeneity of control groups should be noticed. In most meta-analysis, controls are roughly divided into hospital controls and population controls. Considering the overwhelming impact of HBV and HCV on HCC development, we divided controls into hospital controls, healthy controls and HBV/HCV background comparable controls. However, hospital controls are from patients with different diseases, patients with HBV or HCV have various statuses such as inactive carrier for long years and liver cirrhosis. These conditions are different in the included studies, and thus may exaggerate or underestimate the real effect of the hOGG1 Ser326Cys polymorphism on HCC risk. Fourthly, like most meta-analysis, this study is based on unadjusted estimates, while a more precise analysis might be conducted if individual data were available, which could allow for an adjusted estimate by confounding factors. At last, quality of reporting is low in most included studies, although lack of reporting should not be assumed to imply poor quality of a study [22].
We reviewed full-text and supplementary data of GWAS studies of HCC identified in the Catalogue of Published Genome-Wide Association Studies (http://www.genome.gov/gwastudies/). The hOGG1 Ser326Cys polymorphism has not been highlighted in these studies.
In conclusion, there is limited evidence to support that the hOGG1 Ser326Cys polymorphism is associated with HCC risk among East Asian populations. Well-designed and large-sized studies are required to determine this relationship.
Supporting Information
Search strategies.
(DOCX)
Protocol for this meta-analysis.
(DOCX)
Checklist to confirm compliance with PRISMA guidelines for systematic reviews and meta-analyses.
(DOCX)
Funding Statement
The authors have no support or funding to report.
References
- 1. Ferlay J, Shin H, Bray F, Forman D, Mathers C, et al. (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127: 2893–2917. [DOI] [PubMed] [Google Scholar]
- 2. Venook AP, Papandreou C, Furuse J, de Guevara LL (2010) The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist 15 Suppl 45–13. [DOI] [PubMed] [Google Scholar]
- 3. Donato F, Tagger A, Gelatti U, Parrinello G, Boffetta P, et al. (2002) Alcohol and hepatocellular carcinoma: the effect of lifetime intake and hepatitis virus infections in men and women. Am J Epidemiol 155: 323–331. [DOI] [PubMed] [Google Scholar]
- 4. El-Serag H (2011) Hepatocellular carcinoma. N Engl J Med 365: 1118–1127. [DOI] [PubMed] [Google Scholar]
- 5. El-Serag H, Tran T, Everhart J (2004) Diabetes increase the risk of chronic liver disease and hepatocellular carcinoma. Gastroenterology 126: 460–468. [DOI] [PubMed] [Google Scholar]
- 6. Forner A, Llovet J, Bruix J (2012) Hepatocellular carcinoma. Lancet 379: 1245–1255. [DOI] [PubMed] [Google Scholar]
- 7. Marrero J, Fontana R, Fu S, Conjeevaram H, Su G, et al. (2005) Alcohol, tobacco and obesity are synergistic risk factors for hepatocellular carcinoma. J Hepatol 42: 218–224. [DOI] [PubMed] [Google Scholar]
- 8. Weiss J, Goode E, Ladiges W, Ulrich C (2005) Polymorphic variation in hOGG1 and risk of cancer: a review of the functional and epidemiologic literature. Mol Carcinog 42: 127–141. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Z (2004) Studies on the relationship between single nucleotide polymorphisms and susceptibility to hepatocellular carcinoma in Chinese population. Thesis (PhD), Second Military Medical University. [Google Scholar]
- 10. Sakamoto T, Higaki Y, Hara M, Ichiba M, Horita M, et al. (2006) hOGG1 Ser326Cys polymorphism and risk of hepatocellular carcinoma among Japanese. J Epidemiol 16: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang H, Hao B, He F (2006) Impact of DNA repair gene hOGG1 Ser326Cys polymorphism on the risk of hepatocellular carcinoma. World Chinese Journal of Digestology 14: 2311–2314. [Google Scholar]
- 12. Wang A, Cong W, He X, Jia H, Jin X, et al. (2008) A hOGG1 gene polymorhism and genetic susceptibility to colorectal cancer and hepatocellular carcinoma. Chin J Gastroenterol Hepatol 17: 854–857. [Google Scholar]
- 13. Ji L, Zeng X, Li L, Qiu X, Chen S, et al. (2011) Interaction between the single-nucleotide polymorphism of DNA repair gene hOGG1 and HBV infection and its susceptibility to hepatocellular carcinoma. Wei Sheng Yan Jiu 40: 705–708. [PubMed] [Google Scholar]
- 14. Tang Y, Li X, Liu T, Yang J, Luo J, et al. (2011) Genetic polymorphisms of DNA repair genes in patients with hepatocellular carcinoma. Shandong Yi Yao 51: 19–20. [Google Scholar]
- 15. Jung S, Park N, Shin J, Park B, Kim C, et al. (2012) Polymorphisms of DNA repair genes in Korean hepatocellular carcinoma patients with chronic hepatitis B: possible implications on survival. J Hepatol 57: 621–627. [DOI] [PubMed] [Google Scholar]
- 16. Yuan T, Wei J, Luo J, Liu M, Deng S, et al. (2012) Polymorphisms of base-excision repair genes hOGG1 326cys and XRCC1 280His increase hepatocellular carcinoma risk. Dig Dis Sci 57: 2451–2457. [DOI] [PubMed] [Google Scholar]
- 17. Stroup D, Berlin J, Morton S, Olkin I, Williamson G, et al. (2000) Meta-analysis of observational studies in epidemiology: a proposal for reporting. JAMA 283: 2008–2012. [DOI] [PubMed] [Google Scholar]
- 18. Yu Y, Wang W, Zhai S, Dang S, Sun M (2012) IL6 gene polymorphisms and susceptibility to colorectal cancer: a meta-analysis and review. Mol Biol Rep 39: 8457–8463. [DOI] [PubMed] [Google Scholar]
- 19. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 20. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 21. Egger M, Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Little J, Higgins J (2006) The HuGENet™ HuGE Review Handbook, version 1.0. Avalaible: http://wwwcdcgov/genomics/hugenet/participatehtm Accessed 1 September 2012.
- 23. Kanvah S, Joseph J, Schuster GB, Barnett RN, Cleveland CL, et al. (2009) Oxidation of DNA: damage to nucleobases. Acc Chem Res 43: 280–287. [DOI] [PubMed] [Google Scholar]
- 24. Cheng K, Cahill D, Kasai H, Nishimura S, Loeb L (1992) 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G-T and A-C substitutions. J Biol Chem 267: 166–172. [PubMed] [Google Scholar]
- 25. Chen SK, Hsieh WA, Tsai MH, Chen CC, Hong AI, et al. (2003) Age-associated decrease of oxidative repair enzymes, human 8-oxoguanine DNA glycosylases (hOgg1), in human aging. J Radiat Res 44: 31–35. [DOI] [PubMed] [Google Scholar]
- 26. Tarng D, Tsai T, Chen W, Liu T, Wei Y (2001) Effect of human OGG1 1245C>G gene polymorphism on 8-hydroxy-2′-deoxyguanosine levels of leukocyte DNA among patients undergoing chronic hemodialysis. J Am Soc Nephrol 12: 2338–2347. [DOI] [PubMed] [Google Scholar]
- 27. Yamane A, Kohno T, Ito K, Sunaga N, Aoki K, et al. (2004) Differential ability of polymorphic OGG1 proteins to suppress mutagenesis induced by 8-hydroxyguanine in human cell in vivo. Carcinogenesis 25: 1689–1694. [DOI] [PubMed] [Google Scholar]
- 28. Audebert M, Radicella JP, Dizdaroglu M (2000) Effect of single mutations in the OGG1 gene found in human tumors on the substrate specificity of the Ogg1 protein. Nucleic Acids Res 28: 2672–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blons H, Radicella JP, Laccourreye O, Brasnu D, Beaune P, et al. (1999) Frequent allelic loss at chromosome 3p distinct from genetic alterations of the 8-oxoguanine DNA glycosylase 1 gene in head and neck cancer. Mol Carcinog 26: 254–260. [PubMed] [Google Scholar]
- 30. Dherin C, Radicella JP, Dizdaroglu M, Boiteux S (1999) Excision of oxidatively damaged DNA bases by the human alpha-hOgg1 protein and the polymorphic alpha-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res 27: 4001–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hardie LJ, Briggs JA, Davidson LA, Allan JM, King RF, et al. (2000) The effect of hOGG1 and glutathione peroxidase I genotypes and 3p chromosomal loss on 8-hydroxydeoxyguanosine levels in lung cancer. Carcinogenesis 21: 167–172. [DOI] [PubMed] [Google Scholar]
- 32. Janssen K, Schlink K, Gotte W, Hippler B, Kaina B, et al. (2001) DNA repair activity of 8-oxoguanine DNA glycosylase 1 (OGG1) in human lymphocytes is not dependent on genetic polymorphism Ser326/Cys326. Mutat Res 486: 207–216. [DOI] [PubMed] [Google Scholar]
- 33. Kondo S, Toyokuni S, Tanaka T, Hiai H, Onodera H, et al. (2000) Overexpression of the hOGG1 gene and high 8-hydroxy-2′-deoxyguanosine (8-OHdG) lyase activity in human colorectal carcinoma: regulation mechanism of the 8-OHdG level in DNA. Clin Cancer Res 6: 1394–1400. [PubMed] [Google Scholar]
- 34. Li D, Firozi PF, Zhang W, Shen J, DiGiovanni J, et al. (2002) DNA adducts, genetic polymorphisms, and K-ras mutation in human pancreatic cancer. Mutat Res 513: 37–48. [DOI] [PubMed] [Google Scholar]
- 35. Park YJ, Choi EY, Choi JY, Park JG, You HJ, et al. (2001) Genetic changes of hOGG1 and the activity of oh8Gua glycosylase in colon cancer. Eur J Cancer 37: 340–346. [DOI] [PubMed] [Google Scholar]
- 36. Cardin R, Piciocchi M, Sinigaglia A, Lavezzo E, Bortolami M, et al. (2012) Oxidative dna damage correlates with cell immortalization and mir-92 expression in hepatocelllular carcinoma. BMC Cancer 12: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jungst C, Cheng B, Gehrke R, Schmitz V, Nischalke HD, et al. (2004) Oxidative damage is increased in human liver tissue adjacent to hepatocellular carcinoma. Hepatology 39: 1663–1672. [DOI] [PubMed] [Google Scholar]
- 38. Peng T, Shen HM, Liu ZM, Yan LN, Peng MH, et al. (2003) Oxidative DNA damage in peripheral leukocytes and its association with expression and polymorphisms of hOGG1: a study of adolescents in a high risk region for hepatocellular carcinoma in China. World J Gastroenterol 9: 2186–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miyanishi K, Kobune M, Tanaka S, Nagashima H, Sato T, et al. (2011) The comparative analyses of single nucleotide polymorphism of oxidative DNA Repair genes in patients with chronic hepatitis C. Gastroenterology. 140: S971. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Search strategies.
(DOCX)
Protocol for this meta-analysis.
(DOCX)
Checklist to confirm compliance with PRISMA guidelines for systematic reviews and meta-analyses.
(DOCX)