Abstract
During aging, intracranial volume remains unchanged and represents maximally attained brain size, while various interacting biological phenomena lead to brain volume loss. Consequently, intracranial volume and brain volume in late life reflect different genetic influences. Our genome-wide association study in 8,175 community-dwelling elderly did not reveal any genome-wide significant associations (p<5*10−8) for brain volume. In contrast, intracranial volume was significantly associated with two loci: rs4273712 (p=3.4*10−11), a known height locus on chromosome 6q22, and rs9915547, tagging the inversion on chromosome 17q21 (p=1.5*10−12). We replicated the associations of these loci with intracranial volume in a separate sample of 1,752 older persons (p=1.1*10−3 for 6q22 and p=1.2*10−3 for 17q21). Furthermore, we also found suggestive associations of the 17q21 locus with head circumference in 10,768 children (mean age 14.5 months). Our data identify two loci associated with head size, with the inversion on 17q21 also likely involved in attaining maximal brain size.
Main text
During early development and maturation, brain growth is the major force for increasing intracranial volume.1–2 This process, which begins in utero, continues throughout childhood and ends in early adulthood resulting in an intracranial volume that remains essentially unchanged throughout the remainder of life. During early life, intracranial volume is highly associated with brain volume. However, unlike intracranial volume, brain volume starts to decrease after early adulthood.3 The greatest loss occurs in advanced age and is associated with disease states under polygenic and environmental influences, such as cerebrovascular and neurodegenerative diseases that result in brain atrophy.3–4 As a consequence, the degree of association between intracranial volume and brain volume declines with advancing age. While intracranial volume and brain volume are both highly heritable,5–6 the genetic influences on these measures may differ.
To assess the genetic influence on these two measures, we performed a genome-wide association study on cross-sectional measures of intracranial volume and brain volume in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium.7 This consortium brings together six population-based cohort studies, of which five participated in the current analysis:8–9 the Aging Gene-Environment Susceptibility-Reykjavik Study (AGES-RS), the Atherosclerosis Risk in Communities (ARIC) Study, the Austrian Stroke Prevention Study (ASPS), the Framingham Heart Study (FHS), and the Rotterdam Study (RS). The current report includes 8,175 persons of European descent with genome-wide genotype data and a quantitative measure of intracranial volume based on magnetic resonance imaging (MRI). For brain volume, we further excluded persons diagnosed with dementia or who had a cortical infarct on MRI. Moreover, brain volume was expressed as a percentage of intracranial volume to exclude genetic influences on intracranial volume. The ASPS only has data available regarding brain volume and not intracranial volume, giving a total of 8,673 persons for the current GWAS on brain volume (Table 1).
Table 1.
Characteristics of the study populations
| Total | AGES | ARIC | ASPS | FHS | RS I | RS II | RS III | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Persons available for intracranial volume GWAS | 8175 | 2671 | 359 | NA | 2319 | 421 | 679 | 1726 | ||
| Mean age | 67.5 (7.7) | 76.2 (5.4) | 72.7 (4.3) | 65.1 (8.0)† | 64.0 (11.4) | 73.0 (7.7) | 67.5 (5.6) | 56.1 (5.5) | ||
| Female sex (%) | 56% | 59% | 60% | 58%† | 54% | 52% | 49% | 55% | ||
| Mean intracranial volume, mL (standard deviation) | 1301 (132) | 1499 (146) | 1463 (142) | NA | 1254 (131) | 1127 (116) | 1126 (117) | 1135 (117) | ||
| Excluded (due to pathology or technical artifacts) | 227 | 0* | 47 | NA | 77 | 20 | 26 | 57 | ||
| Persons available for brain volume GWAS | 8673 | 2671 | 312 | 725 | 2242 | 401 | 653 | 1669 | ||
| Mean brain volume, percentage of intracranial volume (standard deviation) |
78 (4) | 72 (4) | 74 (4) | 79 (4) | 79 (4) | 78 (4) | 82 (3) | 85 (3) | ||
AGES: Aging Gene-Environment Susceptibility-Reykjavik Study, ARIC: Atherosclerosis Risk in Communities Study, ASPS: Austrian Stroke Prevention Study, CHS: Cardiovascular Health Study, FHS: Framingham Heart Study, RS: Rotterdam Study, mL: milliliter
Both the intracranial volume and brain volume analyses in AGES were performed after exclusion of persons with pathology or technical artifacts
Values are reported for persons available for the brain volume GWAS
Data for replication are from a separate sample of 1,752 older subjects from AGES-RS, who had undergone MRI, but not genome-wide genotyping. Additionally, we investigated whether any of the significant loci associated with intracranial volume or brain volume were also associated with head circumference in 10,768 children with an average age of 14.5 months from the EGG-consortium.10 Rather than a formal replication this analysis adds veracity to the association of the discovered loci with human brain development. Finally, since head size is a morphological phenotype related to adult height we verified whether any significant loci for intracranial volume were already implicated for height in the GIANT-consortium;11 we performed additional height-adjusted analyses for the SNPs achieving genome-wide significance; and additionally we examined the association with intra-cranial volume of all known loci for adult height.
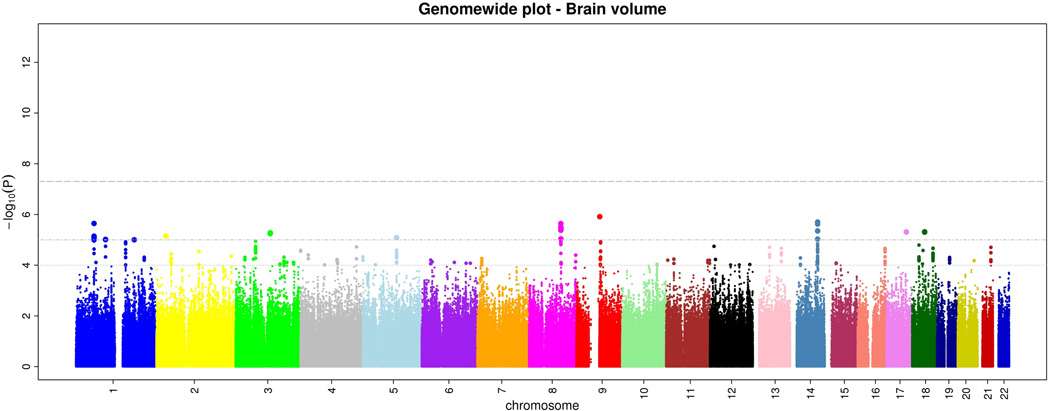
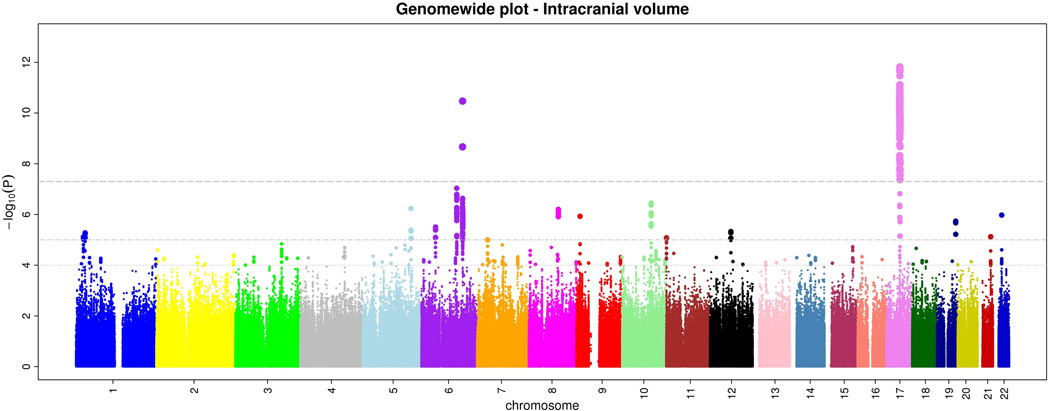
Within CHARGE, after quality control each study imputed their genome-wide genotype data to 2.2 million HapMap CEU SNPs using automated software. Subsequently, each study ran genome-wide association tests for intracranial volume and brain volume using linear regression. Each study used the same basic additive genetic model of dosages of the risk allele with a 1 degree-of-freedom trend test, adjusting for age, sex, and familial relationships (for FHS). From each study we examined quantile-quantile (Q-Q) plots for brain volume and intracranial volume to ensure that the p-value distributions conformed to a null distribution at all but the extreme tail (Supplementary Figure 1). Finally, results across studies were meta-analyzed using inverse-variance weighting after performing genomic control in each study. Examination of the Q-Q plots post meta-analysis showed an excess of extreme p-values but no evidence of systematic inflation of the genomic control inflation factor (λ=1.04 for intracranial volume and 1.02 for brain volume; Supplementary Figure 2). The genome-wide plots of p-values for the individual SNPs against their genomic position are shown in Figures 1a and b. We did not observe any genome-wide significant association for brain volume, although 46 SNPs reached p<10−5 (Supplementary Table 1).
Figure 1.
a and b. Genome-wide signal intensity (Manhattan) plots showing the individual p-values (based on the fixed-effects meta-analysis) against their genomic position for brain volume (a) and intracranial volume (b). Within each chromosome, shown on the x-axis, the results are plotted left to right from the p-terminal end. The horizontal lines indicate thresholds for p=10−4, p=10−5, and p=5*10−8 (genome-wide significant).
For intracranial volume, 720 SNPs located in two loci surpassed our preset threshold of genome-wide significance (p<5*10−8). An additional 200 SNPs showed associations with p<10−5 (Supplementary Table 2).
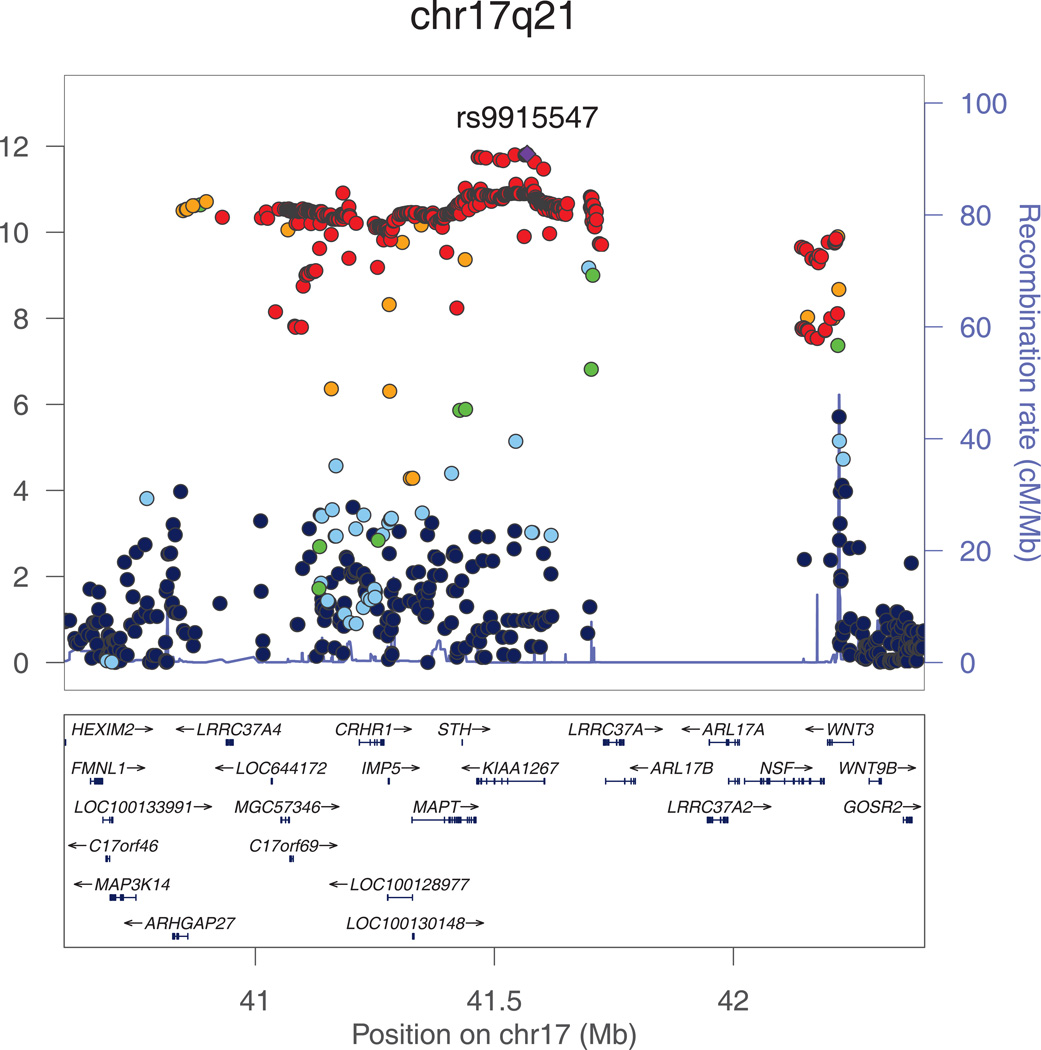
The most significant association with intracranial volume was identified for rs9915547 on chromosome 17. The minor allele of this SNP (MAF=0.22) was significantly associated with smaller intracranial volume (p=1.5*10−12). This SNP belongs to an almost 1Mb large cluster of SNPs in very strong LD with each other located in a region that has previously been identified as the chromosome 17q21 inversion (Figure 2a). The chromosome 17q21 inversion consists of two haplotypes (H1 and H2) with the minor allele of rs9915547 tagging the H2 haplotype. Adjusting for height did not change this result (padjusted for height=1.8*10−12).
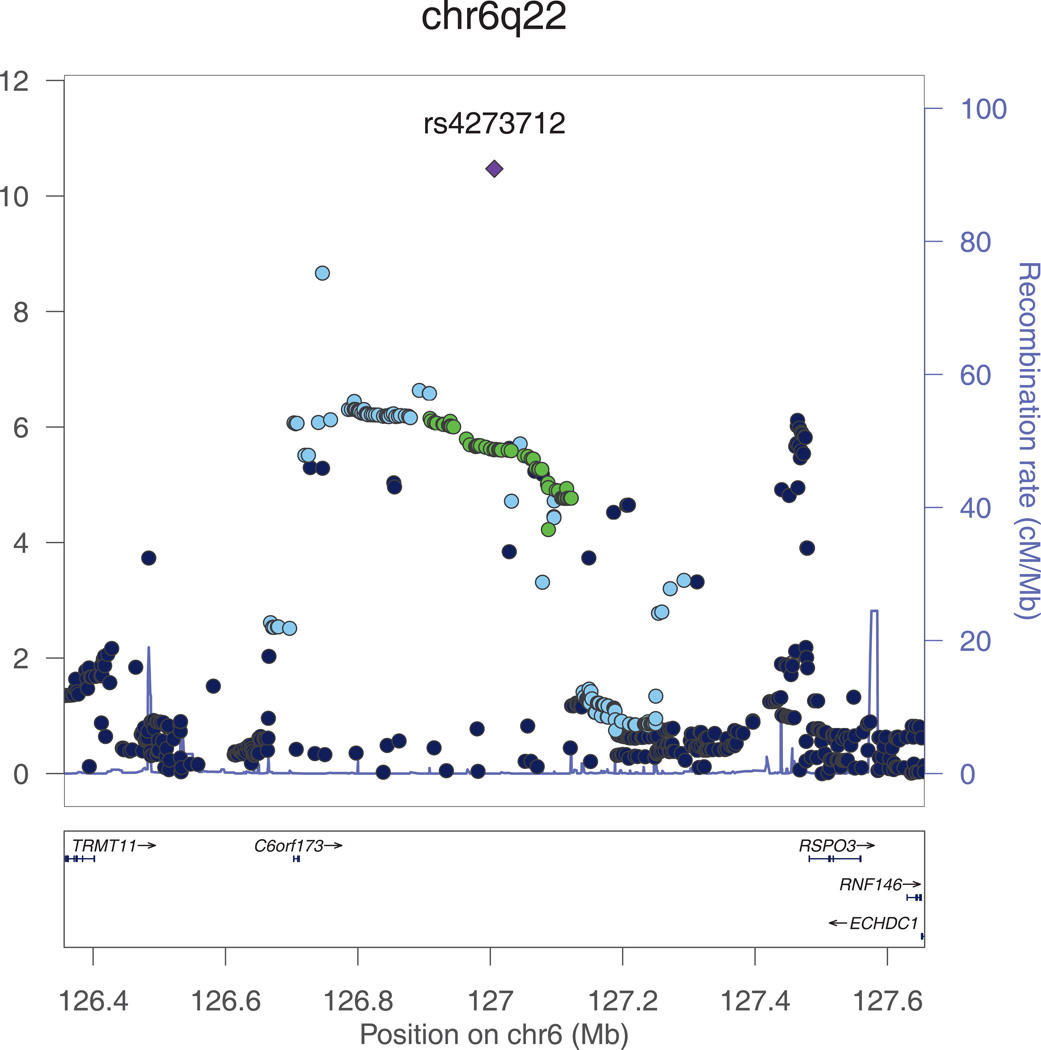
Figure 2. a and b. Regional plots for the genome-wide significant associations with intracranial volume.
All SNPs (circles) are plotted with their meta-analysis p-values against their genomic position. Circles are color-coded according to their linkage disequilibrium with the top SNP (purple rectangle) as follows: r2<0.2 darkblue; 0.2<r2<0.4 lightblue; 0.4<r2<0.6 green; 0.6<r2<0.8 orange; 0.8<r2 red. The blue line at the bottom of the graph represents the estimated recombination rates. Gene annotations are shown as the black arrows.
We note that several copy number variations have been reported at chr6q22, the closest of which are downstream of rs4273712 at position 127124970-127135701 and 127136191-127141042.
The second genome-wide significant association was found for rs4273712 on chromosome 6q22, with RSPO3 and C6Orf173 being the closest genes (Figure 2b). The minor allele of this SNP (MAF=0.27) was associated with a larger intracranial volume (p=3.4*10−11). This locus has previously been associated with adult height.11 Interestingly, copy number variations (CNV) have been reported at this locus, the closest of which are at position 127124970-127135701 and 127136191-127141042.
Both rs9915547 (p=0.23) and rs4273712 (p=0.67) were not associated with brain volume in our data.
We sought to replicate the genome-wide significant associations in a sample of 1752 older persons from AGES-RS, who were not included in the discovery sample. We genotyped rs4273712 and rs9303525 (pdiscovery=1.6*10−12), which tags the inversion in perfect LD with rs9915547, and replicated their association with intracranial volume: P=1.1*10−3 for rs4273712 and 1.2*10−3 for rs9303525); the meta-meta-analysis of the replication sample with the discovery samples yielded p-values of 1.8*10−13 for rs4273712 and 7.6*10−15 for rs9303525 (Table 2).
Table 2.
Genome-wide significant loci for intra-cranial volume and results at various stages of the analysis.
| SNP | Discovery | Replication | Meta-meta-analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | Locus | Strand | Minor allele |
Major allele |
MAF | Beta* | P | MAF | Beta* | P | Beta* | P |
| rs4273712 | 6q22 | + | G | A | 0.27 | 12.2 | 3.4*10−11 | 0.27 | 14.7 | 1.1*10−3 | 12.5 | 1.8*10−13 |
| rs9303525 | 17q21 | + | G | A | 0.22 | −14.7 | 1.6*10−12 | 0.18 | −16.5 | 1.2*10−3 | −14.9 | 7.6*10−15 |
Beta represents the difference in intracranial volume (in milliliters) per copy increase of the minor allele.
Finally, given the hypothesis that intracranial volume is reflective of the maximum brain size during life, we reasoned that these loci may play a role in brain growth. Therefore, we investigated associations of these loci with head circumference in 10,768 children from the EGG-consortium,10 where head circumference is much more likely to be a direct marker for brain growth.1 We found that rs9915547 was significantly associated with head circumference (p=5.3*10−3). In this birth cohort, the most significant SNP located within the 17q21 inversion was rs11655470 with p=1.4*10−6, though this SNP does not accurately tag the inversion itself (r2 with the inversion is 0.22 according to HapMap CEU).10 Indeed, within CHARGE rs11655470 was only weakly associated with intracranial volume at p=0.04. To investigate whether these signals were independent, we performed conditional analyses for rs9915547 and rs11655470 and found p=6.3*10−15 and p=0.38, respectively. The attenuation for rs11655470 suggests that the association is primarily driven by rs9915547, which tags the inversion.
No association was found for rs4273712 with head circumference in the EGG-consortium (p=0.16). Because rs4273712 is located in a known height locus, we also looked up the association with intracranial volume of all 180 known loci for adult height.11 These results are presented in Supplementary Table 3. Apart from the 6q22 locus only rs1351394 on chromosome 12 was nominally significantly associated with intracranial volume (p=4.7*10−6; threshold for Bonferroni-adjusted significance 0.05/180=2.8*10−4). Interestingly, another SNP at this locus (rs1042725) was genome-wide significantly associated with head circumference in the EGG-consortium (p=2.8*10−10).10 The r2 between these rs1042725 and rs1351394 is 0.91 based on CEU HapMap2.
Several genes are located in the 17q21 inversion, which is tagged by two haplotypes (H1 and H2). The two haplotypes show different expression patterns of these genes, some of which have been implicated in neuro-degenerative disorders in late life, particularly among individuals with the H1 haplotype.12 This region includes MAPT which encodes the tau protein, a major hallmark of various dementias, including Alzheimer’s disease and fronto-temporal dementia.12–14 Mutations in MAPT have been consistently associated with fronto-temporal dementia, progressive supra-nuclear palsy and Parkinson’s disease.15 This region also includes GRN which encodes progranulin, mutations in which can also cause fronto-temporal degeneration.16–17 Additional genes in the 17q21 inversion region include corticotrophin releasing hormone receptor 1 (CRHR1), a gene that has been associated with early brain development and may mediate response to environmental stress;18 and saitohin (STH) which has been associated with an increased risk of late-onset Alzheimer’s disease and with progressive supranuclear palsy.14–15 In addition, smaller intracranial volume has been consistently associated with an increased risk for late-life dementia19 suggesting that at least some of the findings related to intracranial volume and dementia risk may reflect the impact of this inversion on early development. Finally, several microdeletions within the inversion have been shown to cause microencephaly and various childhood mental retardation syndromes.20–21
The population genetics and the evolutionary history of this inversion are the object of ongoing research with some studies suggesting that this inversion is about 3 million years old and almost exclusively present in the Caucasian population,22 although more recent work suggests contests this notion.23 Moreover, findings that the H2 haplotype is undergoing positive selection in some Caucasians populations,22 have also recently been contested with the alternative explanation that the higher H2 frequency is a founder effect.23 The influence of the inversion on brain development as shown in our study might be hypothesized as one mechanism to explain the putative positive selection. However, this is less likely since the H2 haplotype is associated with a smaller intracranial volume, whereas intracranial volume has increased throughout evolution of the primates to homo sapiens.24 Still, our finding provides further insights into the role of the 17q21 inversion in human biology. More research is needed to determine its exact role.
Thus far, in order to explain our findings regarding 17q21 we have focused primarily on neurological mechanisms, assuming that brain growth drives intracranial volume. An alternative explanation is that variations in bone growth determine both intracranial volume and thus maximum brain volume. However, given that this inversion does not seem to influence height11 and that height is particularly influenced by bone growth, this alternative explanation appears unlikely.
Two loci implicated in height were also associated with intracranial volume. This suggests potential overlap in the function for the chromosomes 6 and 12 loci and brain development. Rs4273712 on chromosome 6 is close to the gene RSPO3 encoding a protein of the thrombospondin type 1 repeat supergene family, which appear to play a role in the development of the nervous system.25 Another potentially interesting gene in this region is RNF146, which lies further downstream from RSPO3 and has shown to be upregulated in brains of Alzheimer’s disease patients.26 We also note that reported CNVs could explain the association pattern seen at chr6q22, with the region showing high linkage disequilibrium (LD) without any SNP in clear LD with rs4273712.
The chromosome 12 locus tags a gene which encodes a protein belonging to the non-histone chromosomal high mobility group (HMG) protein family. This gene has been implicated in height, adipogenesis and mesenchymal differentiation. Though this locus did not reach genome-wide significance with intracranial volume, it was associated at genome-wide significance with head circumference in children.10 This further demonstrates overlap in genetic variants that influence head size in childhood, maximum head size in adulthood, and possibly brain growth.
In conclusion, this genome-wide association study identified the chromosome 17 inversion as strongly associated with intracranial volume. We further found that this inversion may play a role in early life brain growth. A second genome-wide significant association was found for rs4273712 on chromosome 6 near RSPO3 and RNF146, both potentially interesting genes in neuronal development and degeneration.
Online Methods
Participating Cohorts
Analyses were performed among participants of studies in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium.7 Details on the 6 participating discovery samples, the replication and extension samples can be found in the Supplementary Note. Each study has an Institutional Review Board that approved the consent procedures, examination components, data security processes, genotyping protocols and current study design. All participants gave written informed consent for study participation and for use of DNA for genetic research.
Measurement of intracranial volume and brain volume on magnetic resonance imaging
Each study performed brain magnetic resonance imaging (MRI) on scanners ranging from 0.5 to1.5 T in field strength. Each study used its own custom-made or freely downloadable fully automatic or semi-quantitative MRI post-processing software to measure intra-cranial volume and brain volume. Brain volume was expressed as percentage of intracranial volume in order to correct for individual head-size differences in all studies. Due to the software used, the Austrian Stroke Prevention Study only had measurements available for brain volume. All software have been extensively validated against manual tracings, the gold standard for automated MRI post-processing algorithms. We note that any remaining heterogeneity across studies due the differences in post-processing algorithms would lead to lower power; this in turn could lead to false-negatives, but would not invalidate the associations that were found.
Specific details for each study’s MRI protocol are provided in the Supplementary Note. The correlation between intracranial volume (in milliliters) and brain volume (in milliliters) was 0.83 for AGES, 0.88 for ARIC, 0.89 for FHS, 0.91 for RS I, 0.92 for RS II, 0.95 for RS III. In contrast when using brain volume expressed as percentage of intracranial volume, as was done in our GWAS, the correlations strongly attenuated and became −0.35 for AGES, −0.29 for ARIC, −0.01 for FHS, 0.04 for RS I, −0.19 for RS II, and −0.17 for RS III.
Genotyping, quality controls, and imputation
The consortium was formed after the individual studies had finalized their GWAS platforms, and the 6 studies included used different platforms: the Illumina Human 370-Duo BeadChip® for AGES-Reykjavik Study; the Affymetrix GeneChip SNP Array 6.0® for ARIC; the Illumina Human610-Quad BeadChip® for ASPS; the Affymetrix GeneChip Human Mapping 500K Array Set® and 50K Human Gene Focused Panel® for FHS; and the Illumina HumanHap550 Duo BeadChip® for the Rotterdam Study.
As detailed previously,8–9 participant-specific quality controls included filters for call rate, heterozygosity, and number of Mendelian errors per individual. SNP-specific quality controls included filters for call rate, minor allele frequency, Hardy-Weinberg equilibrium, and differential missingness by outcome or genotype (mishap test in PLINK, http://pngu.mgh.harvard.edu/purcell/plink/).
The set of genotyped input SNPs used for imputation in each study was selected based on their highest quality GWA data. We used a call rate >95% in ARIC, >97% in FHS, and >98% in AGES-Reykjavik, ASPS, and Rotterdam; a minor allele frequency >0.01 in each study; a Hardy-Weinberg p>1x10−6 in AGES-Reykjavik, ARIC, ASPS, FHS, and Rotterdam; and a test of differential missingness by the “mishap” test in PLINK p>1x10−9 in each study. We used the Markov Chain Haplotyping (MaCH) package (http://www.sph.umich.edu/csg/abecasis/MACH, version 1.0.15 or 1.0.16 software) for AGES-Reykjavik, ARIC, ASPS, FHS and Rotterdam imputed to plus strand of NCBI build 36, HapMap release #22. Imputation of genotypes provides a dosage value for every SNP between 0 and 2 indicating the expected value of a SNP being homozygous for the reference allele. For each imputed SNP an assessment of the informativeness of the imputation was estimated (as the ratio of the empirically observed dosage variance to the expected binomial dosage variance, O/E ratio).
For the primary meta-analysis using inverse-variance weighting less weight is given to imputed SNPs with low O/E ratio resulting in higher variance of the estimate.
Studies were screened for latent population substructure,8–9 including cryptic relatedness, using suitable programs: EIGENSTRAT in ARIC, FHS and AGES-Reykjavik, and IBD matrix in ASPS and Rotterdam. When appropriate, components related to ICV or brain volume were included as covariates in the linear regression.
We studied the quantile-quantile (QQ) plots to ensure that the p-value distributions in each of the cohorts conformed to a null distribution at all but the extreme tail.
We also calculated the genomic inflation factor lambda, which measures over-dispersion of test-statistics from association tests indicating population stratification and can be used to apply genomic control.
Analysis within studies
Each study ran a 1-degree of freedom additive association models of the GWAS with intracranial volume and brain volume. Alleles were used as dosages of 0, 1, or 2 copies of the risk allele. Intracranial volume was expressed in milliliters, whereas brain volume was expressed as percentage of intracranial volume to correct for individual head size differences. We note that the SIENAX software used in ASPS only provides this estimate of atrophy and no absolute volumes in milliliters. All models were adjusted for age and sex and – in the Framingham Heart Study – for familial relationships. In secondary analyses, we additionally adjusted for height.
Discovery meta-analysis
After quality control and filtering within each study, AGES-Reykjavik had either genotyped or imputed data for 2,408,992 SNPs, ARIC for 2,298,221 SNPS, ASPS for 2,317,924 SNPs, FHS for 2,543,888 SNPs, the Rotterdam study I for 2,543,888 SNPs, the Rotterdam study II for 2,543,888 SNPs, and the Rotterdam study III for 2,543,888 SNPs. We restricted the present meta-analysis to the 2,229,753 autosomal SNPs common to all studies. For our discovery meta-analysis we used the technique of inverse-variance weighting assuming fixed-effects, after applying genomic control within each individual study. Beta estimates were weighted by their inverse variance and a combined estimate was obtained by summing the weighted betas and dividing by the summed weights. Hence results for SNPs imputed with low certainty were down-weighted because the low reliability-of-imputation ensures a large variance. In contrast, studies with large sample sizes and with directly genotyped or well-imputed SNPs had a greater effect on the meta-analyses p-value because of small variances. We undertook the meta-analysis at Rotterdam using the R-package MetABEL.
We estimated the genomic inflation factor lambda after meta-analysis. The estimate of lambda was 1.039 for ICV and 1.019 for brain volume indicating no significant inflation of p-values. We also examined Q-Q plots of the results from the meta-analysis: for intracranial volume, the original QQ-plot was driven for a large part by the chromosome 17 inversion and the numerous SNPs in close LD in that region. Therefore, we recalculated the QQ-plot after excluding the chromosome 17 inversion.
Replication analysis and extension analysis
The replication sample consisted of a random subset of 1607 white participants from the AGES-Reykjavik study, who had not undergone genome-wide genotyping at the time of the study, but had MRI scans with measurement of intracranial volume available. Methods for measurements of intracranial volume were the same as for the rest of the AGES-Reykjavik study. Using TaqMan assays we genotyped rs4273712 and rs9303535 (pdiscovery=1.6*10−12) which tags the chromosome 17 inversion in perfect linkage disequilibrium with rs9915547 (r2=1 and D’=1 between these two SNPs). We ran 1-degree of freedom additive association relating these SNPs to intracranial volume Thresholds for statistical significance were set at p<0.025 (Bonferoni correction for two SNPs). Subsequently, results from the replication sample were meta-analyzed with the discovery sample using fixed-effect inverse variance meta-analysis.
For extension of our samples we examined genome-wide significant loci from the discovery analysis in an existing meta-analysis on head circumference during infancy in 10,768 children (mean age 14.5 months).10 Here too, thresholds for statistical significance were set at p<0.025. Head circumference was measured manually.
Association of reported height loci with intracranial volume
Because head size is a morphological phenotype related to adult height, we also examined all loci that were recently reported to be associated with adult height for their association with intracranial volume.11 A total of 180 SNPs were tested, resulting in a Bonferoni-corrected p-value of 0.05/180=2.8*10−4 as threshold for significance.
Supplementary Material
Acknowledgments
Aging Gene-Environment Susceptibility-Reykjavik Study: The research has been funded by NIA contract N01-AG-12100 with contributions from NEI, NIDCD and NHLBI, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament).
Atherosclerosis Risk in Communities Study: The research is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, and grants R01-HL087641 and R01-HL093029; National Human Genome Research Institute contract U01-HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
Austrian Stroke Prevention Study: The research reported in this article was funded by the Austrian Science Fond (FWF) grant number P20545-P05 and P13180. The Medical University of Graz supports the databank of the ASPS. The authors thank the staff and the participants of the ASPS for their valuable contributions. We thank Birgit Reinhart for her long-term administrative commitment and Ing Johann Semmler for the technical assistance at creating the DNA-bank.
Framingham Heart Study: Framingham Heart Study: From the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. This work was supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. This study was also supported by grants from the National Institute of Neurological Disorders and Stroke (NS17950), the National Heart, Lung and Blood Institute (HL093029), and the National Institute of Aging (AG08122, AG16495, AG033193, AG033040, AG031287, P30AG013846).
Rotterdam Study: The generation and management of GWAS genotype data for the Rotterdam Study is supported by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project nr. 050-060-810. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters for their help in creating the GWAS database, and Karol Estrada and Maksim V. Struchalin for their support in creation and analysis of imputed data. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The Rotterdam Scan Study is supported by the Netherlands Organization of Scientific Research (NWO) project nrs. 918-46-615; 904-61-096; 904-61-133; 948-00-010; and Nederlandse Hartstichting 2009B102; and Internationaal Parkinson Fonds. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists.
EGG consortium: Financial support was received from the Academy of Finland (project grants 104781, 120315, 1114194 and Center of Excellence in Complex Disease Genetics), University Hospital Oulu, Biocenter, University of Oulu, Finland, NHLBI grant 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01), ENGAGE project and grant agreement HEALTH-F4-2007-201413, the Medical Research Council (studentship grant G0500539, PrevMetSyn/Salve/MRC), the Wellcome Trust (project grant GR069224), UK. The DNA extractions, sample quality controls, biobank up-keeping and aliquotting was performed in the National Public Health Institute, Biomedicum Helsinki, Finland and supported financially by the Academy of Finland and Biocentrum Helsinki. The Generation R Study is conducted by the Erasmus Medical Center in close collaboration with the School of Law and Faculty of Social Sciences of the Erasmus University Rotterdam, the Municipal Health Service Rotterdam area, Rotterdam, the Rotterdam Homecare Foundation, Rotterdam and the Stichting Trombosedienst & Artsenlaboratorium Rijnmond (STAR), Rotterdam. We gratefully acknowledge the contribution of general practitioners, hospitals, midwives and pharmacies in Rotterdam. The Generation R Study is made possible by financial support from the Erasmus Medical Center, Rotterdam, the Erasmus University Rotterdam and the Netherlands Organization for Health Research and Development (ZonMw 21000074). Vincent Jaddoe received an additional grant from the Netherlands Organization for Health Research and Development (ZonMw 90700303, 916.10159). Additional support was provided by a grant from the Dutch Kidney Foundation (C08.2251).
We would like to thank all participating subjects and families from the Children’s Hospital in Philadelphia. The research was financially supported by an Institute Development Award from the Children’s Hospital of Philadelphia, a Research Development Award from the Cotswold Foundation and NIH grant 1R01HD056465-01A1. We are grateful to the Raine Foundation, to the Raine Study Families, and to the Raine Study research staff. We gratefully acknowledge the assistance of the Western Australian Genetic Epidemiology Resource and the Western Australian DNA Bank (both National Health and Medical Research Council of Australia National Enabling Facilities). The authors also acknowledge the support of the Healthway Western Australia, the National Health and Medical Research Council of Australia (Grant 572613) and the Canadian Institutes of Health Research (Grant MOP 82893). We gratefully acknowledge the assistance of the Wind Over Water Foundation, the Telethon Institute for Child Health Research, and the Raine Medical Research Foundation of the University of Western Australia. We wish to acknowledge the following: Helmholtz Zentrum Muenchen - German Research Center for Environment and Health, Institute of Epidemiology, Neuherberg; Department of Pediatrics, University of Leipzig; Department of Pediatrics, Marien-Hospital, Wesel; Bad Honnef; Department of Human Exposure Research and Epidemiology, UFZ-Centre for Environmental Research Leipzig-Halle; Department of Environmental Immunology, UFZ- Centre for Environmental Research Leipzig-Halle; IUF-Institut für Umweltmedizinische Forschung, Düsseldorf; Department of Pediatrics, Technical University, Munich. The UK Medical Research Council (Grant ref:74882), the Wellcome Trust (Grant ref:076467), and the University of Bristol provide core support for the Avon Longitudinal Study of Parents and Children (ALSPAC). We are extremely grateful to all the families who took part in the ALSPAC study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionist and nurses.
Full list of contributors from the Early Growth Genetics (EGG) consortium
Linda S. Adair47, Wei Ang24, Mustafa Atalay48, Toos van Beijsterveldt56, Nienke Bergen1,14, Kelly Benke24, Diane Berry50, Lachlan Coin30, Oliver S.P. Davis51, Paul Elliott30, Claudia Flexeder25, Tim Frayling52, Romy Gaillard1,14, Maria Groen-Blokhuis49, Liang-Kee Goh53,54, Claire M.A. Haworth51, Dexter Hadley27, Johannes Hedebrand55, Anke Hinney55, Joel N. Hirschhorn56,57,58, John W. Holloway59,60, Claus Holst61, Jouke Jan Hottenga49, Momoko Horikoshi34,35, Ville Huikari31,32, Elina Hypponen50,62, Tuomas O. Kilpeläinen63, Mirna Kirin64, Mattew Kowgier24, Hanna-Maaria Lakka65, Leslie A. Lange66, Debbie A. Lawlor36, Terho Lehtimäki67,68, Alex Lewin30, Cecilia Lindgren35, Virpi Lindi48, Reedik Maggi35,69, Julie Marsh24, Christel Middeldorp49, Iona Millwood30,70, Jeffrey C. Murray71, Michel Nivard49, Ellen Aagaard Nohr61, Ioanna Ntalla72, Emily Oken56,57,58, Kalliope Panoutsopoulou73, Jennifer Pararajasingham51, Alina Rodriguez30,51,74, Rany M. Salem56,57,58, Sylvain Sebert30, Niina Siitonen75, David P. Strachan76, Yik-Ying Teo54, Beatriz Valcárcel30, Gonneke Willemsen49, Eleftheria Zeggini73, Dorret I. Boomsma49, Cyrus Cooper77, Matthew Gillman78, Berthold Hocher79,80, Timo A. Lakka48, Karen L. Mohlke66, George V. Dedoussis72, Ken K. Ong81, Ewan R. Pearson82, Thomas S. Price51, Chris Power50, Olli T. Raitakari75,83, Seang-Mei Saw53,54,84, Andre Scherag85, Olli Simell75,86, Thorkild I.A. Sørensen61,87, James F. Wilson64,88
47. Department of Nutrition, University of North Carolina, Chapel Hill, NC, USA.
48. Department of Physiology, Institute of Biomedicine, University of Eastern Finland, Kuopio, Finland.
49. Department of Biological Psychology, VU University, Amsterdam, The Netherlands.
50. Centre for Paediatric Epidemiology and Biostatistics/MRC Centre of Epidemiology for Child Health, University College of London Institute of Child Health, London, UK.
51. MRC Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, King's College London, UK.
52. Genetics of Complex Traits, Peninsula College of Medicine and Dentistry, University of Exeter, Exeter, UK.
53. Duke-NUS Graduate Medical School, Singapore.
54. Saw Swee Hock School of Public Health, National University of Singapore.
55. Department of Child and Adolescent Psychiatry, University of Duisburg-Essen, Essen, Germany.
56. Divisions of Genetics and Endocrinology and Program in Genomics, Children's Hospital, Boston, Massachusetts, USA.
57. Department of Genetics, Harvard Medical School, Boston, Massachusetts, USA.
58. Metabolism Initiative and Program in Medical and Population Genetics, Broad Institute, Cambridge, Massachusetts, USA.
59. Human Genetics and Medical Genomics, Human Development & Health, Faculty of Medicine, University of Southampton, Southampton, UK.
60. Clinical & Experimental Sciences, Faculty of Medicine, University of Southampton, Southamptom, UK.
61. Institute of Preventive Medicine, Copenhagen University Hospital, Copenhagen, Denmark.
62. Department of Genomics of Common Disease, School of Public Health, Imperial College London, UK.
63. Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health Sciences, University of Copenhagen, Copenhagen, Denmark.
64. Centre for Population Health Sciences, University of Edinburgh, Edinburgh, Scotland.
65. Department of Public Health, Institute of Public Health and Clinical Nutrition, University of Eastern Finland, Kuopio Campus, Finland.
66. Department of Genetics, University of North Carolina, Chapel Hill, NC, USA.
67. Department of Clinical Chemistry, Tampere University Hospital, Tampere, Finland.
68. Department of Clinical Chemistry, University of Tampere School of Medicine, Tampere, Finland.
69. Estonian Genome Center, University of Tartu, Tartu, Estonia.
70. Clinical Trial Service Unit and Epidemiological Studies Unit (CTSU), University of Oxford, UK.
71. Department of Pediatrics, University of Iowa, Iowa City, Iowa, USA.
72. Department of Dietetics - Nutrition, Harokopio University of Athens, Athens, Greece.
73. Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK.
74. Department of Psychology, Mid Sweden University, Sweden.
75. Research Centre of Applied and Preventive Cardiovascular Medicine, University of Turku, Turku, Finland.
76. Division of Population Health Sciences and Education, St George's, University of London, UK.
77. MRC Lifecourse Epidemiology Unit, University of Southampton, Southampton, UK.
78. Obesity Prevention Program, Department of Population Medicine, Harvard Medical School/Harvard Pilgrim Health Care Institute, Obesity Prevention Program, Department of Population Medicine, Harvard Medical School/Harvard Pilgrim Health Care Institute, Boston, Massachusetts, USA.
79. Institute of Nutritional Science, University of Potsdam, Nuthetal Potsdam, Germany.
80. Center for Cardiovascular Research/Institute of Pharmacology, Charité, Berlin, Germany.
81. MRC Epidemiology Unit, Institute of Metabolic Science, Cambridge, UK.
82. Biomedical Research Institute, University of Dundee, UK.
83. Department of Clinical Physiology, University of Turku and Turku University Hospital, Turku, Finland.
84. Singapore Eye Research Institute, Singapore.
85. Institute for Medical Informatics, Biometry and Epidemiology, University of Duisburg-Essen, Essen, Germany.
86. Department of Pediatrics, University of Turku and Turku University Hospital, Turku, Finland.
87. The Novo Nordisk Foundation Center for Basic Metabolic Research, Section of Metabolic Genetics, University of Copenhagen, Copenhagen, Denmark.
88. MRC Institute of Genetics and Molecular Medicine at the University of Edinburgh, Western General Hospital, Edinburgh, Scotland
Ed. Summary: M. Arfan Ikram and colleagues report a genome-wide association study for intracranial volume and brain volume. They report two loci associated with intracranial volume.
Footnotes
Author contributions
Study concept and design: MAI, MF, SuSe, RS, WTL, AGU, ALH, EAPS, JH, LJP, SFAG, NJT, GDS, AH, AvdL, MAvB, VWVJ, VG, PAW, CMvD, THM, HS, LJL, MMBB, CDC
Acquisition of data: MAI, MF, AVS, SD, HAV, SiSi, SR, HRT, DOMK, LHC, WJN, AB, AZ, MS, CRJ, FR, DSK, CEP, EAPS, HH, LJP, MRJ, SFAG, BSP, MAN, RA, HG, TBH, WMM, MWV, MAvB, DC, BGW, THM
Statistical analysis and interpretation of the findings: MAI, MF, AVS, SuSe, RS, SD, HRT, DOMK, ALDS, FR, ET, SFAG, MAN, RA, VWVJ, CMvD, HS, LJL, MMBB, CDC
Drafting of the manuscript: MAI, MF, SuSe, RS, HRT, DOMK, LJL, CDC
Critical revision of the manuscript: AVS, SD, HAV, SiSi, SR, LHC, WTL, WJN, ALDS, AB, AZ, MS, CRJ, FR, AGU, DSK, ALH, CEP, ET, EAPS, HH, JH, LJP, MRJ, MIM, BSP, NJT, GDS, US, MAN, RA, AH, HG, AvdL, TBH, WMM, MWV, MAvB, VG, BGW, PAW, CMvD, THM, HS, MMBB
Obtained funding and supervision: AVS, SuSe, RS, WTL, WJN, AGU, MRJ, MIM, SFAG, NJT, US, AH, AvdL, MAvB, DC, VWVJ, VG, PAW, CMvD, THM, HS, LJL, MMBB, CDC
Competing interests statement
The authors declare no competing financial interests related to this manuscript.
References
- 1.Gale CR, O'Callaghan FJ, Bredow M, Martyn CN. The influence of head growth in fetal life, infancy, and childhood on intelligence at the ages of 4 and 8 years. Pediatrics. 2006;118:1486–1492. doi: 10.1542/peds.2005-2629. [DOI] [PubMed] [Google Scholar]
- 2.Gale CR, O'Callaghan FJ, Godfrey KM, Law CM, Martyn CN. Critical periods of brain growth and cognitive function in children. Brain. 2004;127:321–329. doi: 10.1093/brain/awh034. [DOI] [PubMed] [Google Scholar]
- 3.DeCarli C, et al. Measures of brain morphology and infarction in the framingham heart study: establishing what is normal. Neurobiol Aging. 2005;26:491–510. doi: 10.1016/j.neurobiolaging.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Ikram MA, et al. Brain tissue volumes in the general elderly population. The Rotterdam Scan Study. Neurobiol Aging. 2008;29:882–890. doi: 10.1016/j.neurobiolaging.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Carmelli D, et al. Evidence for genetic variance in white matter hyperintensity volume in normal elderly male twins. Stroke. 1998;29:1177–1181. doi: 10.1161/01.str.29.6.1177. [DOI] [PubMed] [Google Scholar]
- 6.Atwood LD, et al. Genetic variation in white matter hyperintensity volume in the Framingham Study. Stroke. 2004;35:1609–1613. doi: 10.1161/01.STR.0000129643.77045.10. [DOI] [PubMed] [Google Scholar]
- 7.Psaty BM, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seshadri S, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. Jama. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikram MA, et al. Genomewide association studies of stroke. N Engl J Med. 2009;360:1718–1728. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taal HR, et al. Common variants at 12q15 and 12q24 are associated with infant head circumference. Nat Genet. 2012 doi: 10.1038/ng.2238. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Maria E, et al. The H1 Haplotype of the Tau Gene (MAPT) is Associated with Mild Cognitive Impairment. J Alzheimers Dis. 2009 doi: 10.3233/JAD-2010-1285. [DOI] [PubMed] [Google Scholar]
- 13.Zody MC, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet. 2008;40:1076–1083. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conrad C, et al. Molecular evolution and genetics of the Saitohin gene and tau haplotype in Alzheimer's disease and argyrophilic grain disease. J Neurochem. 2004;89:179–188. doi: 10.1046/j.1471-4159.2004.02320.x. [DOI] [PubMed] [Google Scholar]
- 15.Levecque C, et al. Association of polymorphisms in the Tau and Saitohin genes with Parkinson's disease. J Neurol Neurosurg Psychiatry. 2004;75:478–480. doi: 10.1136/jnnp.2003.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gijselinck I, Van Broeckhoven C, Cruts M. Granulin mutations associated with frontotemporal lobar degeneration and related disorders: an update. Hum Mutat. 2008;29:1373–1386. doi: 10.1002/humu.20785. [DOI] [PubMed] [Google Scholar]
- 17.Gijselinck I, et al. Progranulin locus deletion in frontotemporal dementia. Hum Mutat. 2008;29:53–58. doi: 10.1002/humu.20651. [DOI] [PubMed] [Google Scholar]
- 18.Hsuchou H, Kastin AJ, Wu X, Tu H, Pan W. Corticotropin-releasing hormone receptor-1 in cerebral microvessels changes during development and influences urocortin transport across the blood-brain barrier. Endocrinology. 2009;151:1221–1227. doi: 10.1210/en.2009-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortimer JA, Snowdon DA, Markesbery WR. Head circumference, education and risk of dementia: findings from the Nun Study. J Clin Exp Neuropsychol. 2003;25:671–679. doi: 10.1076/jcen.25.5.671.14584. [DOI] [PubMed] [Google Scholar]
- 20.Koolen DA, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 21.Koolen DA, et al. Clinical and molecular delineation of the 17q21.31 microdeletion syndrome. J Med Genet. 2008;45:710–720. doi: 10.1136/jmg.2008.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stefansson H, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 23.Donnelly MP, et al. The distribution and most recent common ancestor of the 17q21 inversion in humans. Am J Hum Genet. 2010;86:161–171. doi: 10.1016/j.ajhg.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roth G, Dicke U. Evolution of the brain and intelligence. Trends Cogn Sci. 2005;9:250–257. doi: 10.1016/j.tics.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Adams JC, Tucker RP. The thrombospondin type 1 repeat (TSR) superfamily: diverse proteins with related roles in neuronal development. Dev Dyn. 2000;218:280–299. doi: 10.1002/(SICI)1097-0177(200006)218:2<280::AID-DVDY4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 26.von Rotz RC, Kins S, Hipfel R, von der Kammer H, Nitsch RM. The novel cytosolic RING finger protein dactylidin is up-regulated in brains of patients with Alzheimer's disease. Eur J Neurosci. 2005;21:1289–1298. doi: 10.1111/j.1460-9568.2005.03977.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.