Abstract
Background
Aberrant methylation of CpG islands acquired in tumor cells in promoter regions plays an important role in carcinogenesis. Accumulated evidence demonstrates P16INK4a gene promoter hypermethylation is involved in non-small cell lung carcinoma (NSCLC), indicating it may be a potential biomarker for this disease. The aim of this study is to evaluate the frequency of P16INK4a gene promoter methylation between cancer tissue and autologous controls by summarizing published studies.
Methods
By searching Medline, EMBSE and CNKI databases, the open published studies about P16INK4a gene promoter methylation and NSCLC were identified using a systematic search strategy. The pooled odds of P16INK4A promoter methylation in lung cancer tissue versus autologous controls were calculated by meta-analysis method.
Results
Thirty-four studies, including 2 652 NSCLC patients with 5 175 samples were included in this meta-analysis. Generally, the frequency of P16INK4A promoter methylation ranged from 17% to 80% (median 44%) in the lung cancer tissue and 0 to 80% (median 15%) in the autologous controls, which indicated the methylation frequency in cancer tissue was much higher than that in autologous samples. We also find a strong and significant correlation between tumor tissue and autologous controls of P16INK4A promoter methylation frequency across studies (Correlation coefficient 0.71, 95% CI:0.51–0.83, P<0.0001). And the pooled odds ratio of P16INK4A promoter methylation in cancer tissue was 3.45 (95% CI: 2.63–4.54) compared to controls under random-effect model.
Conclusion
Frequency of P16INK4a promoter methylation in cancer tissue was much higher than that in autologous controls, indicating promoter methylation plays an important role in carcinogenesis of the NSCLC. Strong and significant correlation between tumor tissue and autologous samples of P16INK4A promoter methylation demonstrated a promising biomarker for NSCLC.
Introduction
Lung cancer, accounting for 13% (1.6 million) of the total cases and 18% (1.4 million) of the deaths, was the most commonly diagnosed cancer as well as the leading cause of cancer death worldwide in 2008 [1]. Benefiting from the tobacco control, lung cancer death rate is decreasing in western developed countries. However, it is increasing in developing countries such as China, where smoking prevalence is still increasing [1]. Non-small cell lung cancer, accounting for 80% of primary lung carcinomas, was the most common type with a 5-year survival rate ranging from 2 to 47% for different clinical stages and histopathology [2]. About twenty percent of NSCLC patients are suitable for surgery at the time of diagnosis, and the other 80%, receiving conventional chemoradiation, can only survive a short period of time [3]. Therefore, the early diagnosis is essential to the prolonged survival of this disease.
Tumor suppressor gene promoter methylation is considered as an important mechanism for its inactivation, which occurs in the early stage of the tumorigenesis for many types of cancer [2], [4]. Thus, detection of aberrant methylation of tumor suppressor genes could be a potential method for the early diagnosis of various types of cancer, including NSCLC. The aberrant methylation status of primary tumors can be detected by methylation specific PCR(MSP), which could detect one methylated allele in the presence of 103–104 unmethylated alleles [5]. And many studies have also shown that cancer-specific methylation of tumor suppressor genes can be found in autologous clinical samples such as plasma, serum, sputum or bronchoalveolar lavage fluid(BALF) of NSCLC, indicating that it can be potential biomarkers for non-invasive diagnosis of this disease [6]–[8]. But the frequency of DNA methylation in tumor suppressor genes between cancer tissue and autologous clinical samples ranged a lot among the published studies with small sample size. Accordingly, we performed a meta-analysis on the basis of published articles of P16INK4a promoter methylation and lung cancer in order to better identify the correlation of methylation status between cancer tissue and autologous samples.
Materials and Methods
Studies Identification
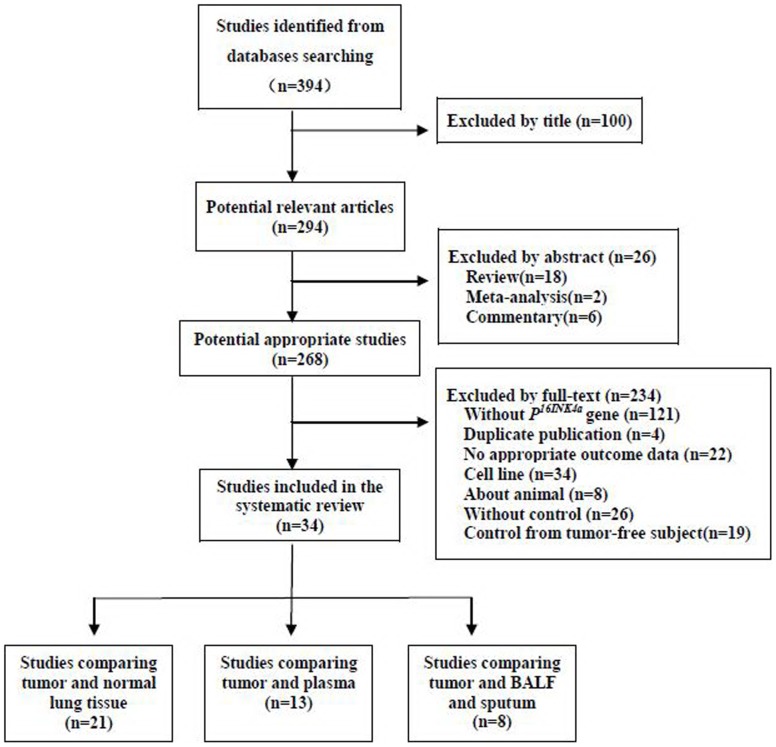
The selection procedure of studies was illustrated in the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) statement flow chart (Fig. 1). Studies about P16INK4a gene promoter methylation in NSCLC, published before January 2012, were identified through an electronic sensitive search of Medline, EMBSE and CNKI databases. The searching strategy was performed using “Non-Small-Cell Lung Carcinoma” AND “methylation” as the Medical Subject Headings (MeSH) and corresponding free text word searching term. The title and abstract of initial identified articles were evaluated for appropriateness to the inclusion criteria. Then all potentially relevant articles were assessed in full-text paper and all references of included articles were further scanned for additional analysis.
Figure 1. PRISMA flowchart of the literature search strategy for systematic review.
(Data from some studies was used more than once, as they reported data in multiple controls.).
Data Extraction and Quality Assessment
The inclusion criteria of the meta-analysis was as follows: the patients were limited to non-small cell lung carcinoma without restriction of stages. The methods used for methylation detection were confined to methylation-specific polymerase chain reaction(MSP), real-time MSP(RT-MSP) and quantitative MSP(q-MSP). The results were the P16INK4A gene promoter methylation status in tumor tissue and corresponding autologous controls, including non-tumor lung tissue(NLT), plasma, sputum and bronchoalveolar lavage fluid(BALF) of NSCLC patients. Information on the name of the first author, year of publication, region of the included subjects and methylation status of P16INK4A gene in cancer tissue and controls were recorded from each study. Detailed information about each article was extracted by two reviewers (JG and YW) and then checked by the third reviewer (SZ) as described in the Cochrane Handbook for systematic reviews [9].
Statistical Analysis
STATA/SE 11.0 (StataCorp LP, http://www.stata.com) and MetaAanlyst 3.13 (http://www.biomedcentral.com) were used for statistical analysis. Methylation status in tumor tissue and controls was calculated as methylation rate. The odds of P16INK4A promoter methylation in lung cancer tissue versus autologous controls was expressed as the odds ratio (OR) and its 95% confidence intervals (CI). Statistical heterogeneity across studies was assessed by chi-square (χ2) test [10], and the inconsistency was calculated by I2 [11]. If heterogeneity was significant (χ2, p<0.1 or I2>50%), meta-regression analysis was employed for further evaluation of the source of heterogeneity. And subgroup analysis according to the source of heterogeneity was performed for further evaluation. Finally, if significant heterogeneity across studies was detected and no appropriate clinical explanation of the heterogeneity was found, the random-effect method (Dersimonian-Laird method) was used to pool the data. Inversely, without significant heterogeneity between studies, fixed-effect method was purchased. And sensitivity analysis was also performed to assess the contribution of each study to the final results of the meta-analysis. The Begger’s funnel plot and Egger’s test were used to evaluate the possible publication bias [12]. The correlation of P16INK4A gene promoter methylation between tumor tissue and autologous clinical control samples were compared by Spearman’s rank correlation test.
Results
Study Characteristics
A total of three hundred and ninety-four studies were initially identified by searching the electronic databases. And 268 potential applicable articles, published from 2000 and 2012, were retrieved in full-text. Of those, 234 studies were excluded for the reasons: about other genes methylation status without P16INK4A, duplicated publication, no appropriate outcome data, about cell lines, about animals, without proper controls. Finally, thirty-four studies [6]–[8], [13]–[43] that reported data of methylation frequency in non-small-cell lung carcinoma tissue, and autologous controls were finally pooled in the meta-analysis (Fig. 1). Of the 34 included articles, 25 were conducted in Asia-Pacific(18 in Chinese mainland, 3 in Taiwan, 1 in Hong Kong, 2 in Korea, 1 in Japan), 4 in USA and 5 in Europe (3 in Italy, 1 in Greece, 1in England). Some of the included studies reported methylation status separately according to gender, histopathology types, smoking status and tumor stages. The general characteristics of included studies were summarized in table 1.
Table 1. General characteristics of included studies.
| Sample size (n) | Histology | Control type | |||||||||
| Author | Year publication | Location | Age(y) | Gender(M/F) | T | C | Method | Sq | Ad | Ots | |
| Seike [13] | 2000 | Japan | 63.7(40–80) | 15/6 | 21 | 21 | MSP | 9 | 12 | 0 | NLT |
| Su [16] | 2000 | China | 58.9 | Na | 72 | 10 | MSP | 39 | 31 | 2 | NLT |
| He [31] | 2001 | China | Na | Na | 30 | 30 | MSP | 17 | 11 | 2 | NLT |
| Zochbauer [6] | 2001 | USA | Na | 76/31 | 107 | 104 | MSP | 43 | 45 | 19 | NLT |
| Bearzatto [30] | 2002 | Italy | 64 | 28/7 | 35 | 35 | RT-MSP | 10 | 18 | 7 | Plasm |
| Chen [25] | 2002 | Taiwan | Na | Na | 67 | 21 | MSP | Na | Na | Na | Sputum |
| He [32] | 2002 | China | Na | Na | 21 | 21 | MSP | 12 | 9 | 0 | BALF |
| Ng [17] | 2002 | Hong Kong | 60.2 | 25/8 | 33 | 33 | MSP | 13 | 15 | 5 | Plasm,BALF |
| Cai [33] | 2003 | China | 59.5 | 46/23 | 69 | 69 | MSP | 25 | 36 | 8 | Plasm |
| Harden [26] | 2003 | USA | 67(40–87) | 50/40 | 90 | 90 | q-MSP | 33 | 36 | 21 | NLT |
| Liu [18] | 2003 | China | Na | Na | 98 | 110 | MSP | 58 | 40 | 0 | Plasm sputum |
| Guo [27] | 2004 | USA | 66.1(42–83) | Na | 20 | 20 | MSP | 1 | 18 | 1 | NLT BLAF |
| Liu [14] | 2004 | China | Na | Na | 40 | 40 | MSP | 23 | 17 | 0 | Plasm |
| Zhang [34] | 2004 | China | Na | Na | 40 | 40 | MSP | 23 | 14 | 3 | NLT |
| Russo [19] | 2005 | USA | Na | Na | 48 | 48 | MSP | Na | Na | Na | Plasm |
| Georgiou [23] | 2007 | Greece | 63(38–76) | 32/3 | 35 | 35 | MSP | 15 | 17 | 3 | NLT BALF |
| Li [35] | 2006 | China | Na | 38/11 | 49 | 49 | MSP | 22 | 24 | 3 | Plasm |
| Rosalia [29] | 2006 | Italy | 60.2(51–74) | 20/9 | 29 | 18 | MSP | 5 | 23 | 1 | Sputum |
| Ulivi [15] | 2006 | Italy | Na | 49/12 | 61 | 61 | RT-MSP | 16 | 36 | 9 | Plasm |
| Wang [36] | 2006 | China | 32–73 | 42/5 | 47 | 47 | MSP | 31 | 7 | 9 | NLT |
| Belinsky [20] | 2007 | England | 62(37–80) | 49/23 | 72 | 72 | MSP | 22 | 29 | 21 | Plasm Sputum |
| Hong [22] | 2007 | Korea | Na | 63/18 | 81 | 81 | RT-MSP | 40 | 34 | 7 | NLT |
| Hsu(1) [21] | 2007 | Taiwan | 69 | 45/18 | 63 | 63 | q-MSP | 41 | 13 | 9 | NLT Plasm |
| Hsu(2) [24] | 2007 | Taiwan | Na | Na | 82 | 82 | MSP | 37 | 23 | 22 | NLTSputum |
| Kim [28] | 2007 | Korea | 63±8.4 | 80/19 | 99 | 99 | MSP | 61 | 38 | 0 | NLT |
| Yang [37] | 2007 | China | 56(31–77) | 34/15 | 49 | 49 | MSP | 26 | 23 | 0 | NLT |
| Zhang [38] | 2007 | China | Na | Na | 29 | 29 | MSP | 7 | 16 | 6 | NLT |
| Guo [39] | 2008 | China | 59±13 | 72/34 | 106 | 106 | MSP | 41 | 27 | 39 | Plasm |
| Wang [8] | 2008 | China | Na | 17/11 | 28 | 18 | MSP | 7 | 15 | 6 | NLT |
| Chen [40] | 2010 | China | 59.7(32–79) | 102/18 | 120 | 120 | MSP | 66 | 26 | 28 | NLT Plasm |
| Guo [41] | 2010 | China | 59.2 | 23/5 | 28 | 28 | MSP | Na | Na | Na | NLT |
| Zhang [42] | 2006 | China | 52.3(37–73) | 33/15 | 48 | 48 | MSP | 25 | 20 | 3 | NLT |
| Zhang [7] | 2011 | China | 61(32–79) | 162/38 | 200 | 200 | MSP | 104 | 59 | 37 | NLT |
| Sun [43] | 2012 | China | 65 | 96/24 | 120 | 120 | MSP | 32 | 72 | 16 | Sputum |
M = male; F = female; T = tumor; C = control; Sq = squamous cell carcinoma; Ad = adenocarcinoma; Ots = others; BALF = bronchoalveolar lavage; NLT:non-tumor lung issue; Na = not available.
Pooled Results from the Meta-analysis
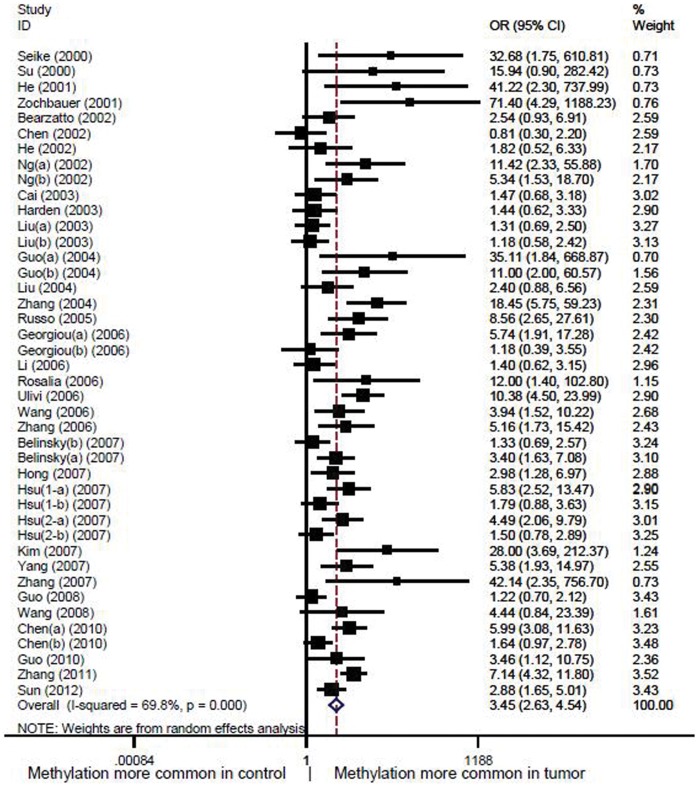
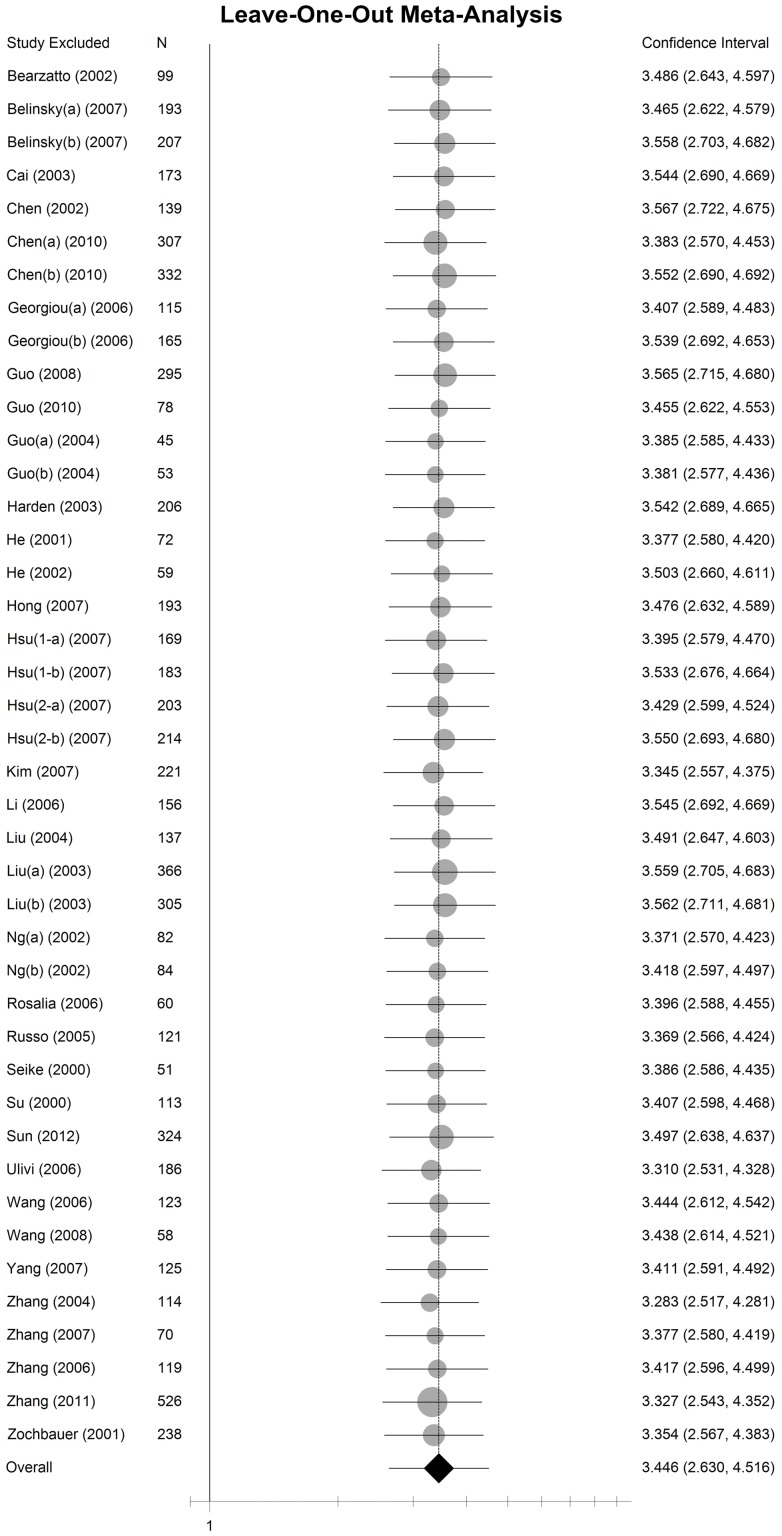
In the meta-analysis, data from 2 652 non-small cell lung cancer patients including 5 175 samples were pooled with an odds ratio of 3.45 (95% CI: 2.63–4.54) in tumor tissue versus autologous controls under random-effect method (Fig. 2). The sensitivity analysis indicated that the odds ratio range from 3.28(95% CI: 2.52–4.28) to 3.57(95% CI: 2.72–4.68) by omitting a single study under the random-effect model (Fig. 3). Only very slight change of odds ratio was seen in the sensitivity analysis, which demonstrated that the pooled odds ratio was not sensitive to a single study.
Figure 2. Forest plot of P16INK4A promoter methylation in cancer tissue versus autologous controls.
The squares and horizontal lines represent the study-specific OR and 95% CI. The area of the squares reflects the weight (inverse of the variance). The diamond represents the pooled OR and 95% CI.
Figure 3. The sensitivity analysis by omitting a single study under the random-effect method.
The circles and horizontal lines represents the pooled OR and 95% CI by omitting a certain study. The area of the circles reflects the weight (by sample size). The diamond represents the pooled OR and 95% CI by including all of studies.
Meta-regression and Subgroup Analysis
As the significant heterogeneity was found across the studies (I2 = 69.8%, χ2 = 135.7, P<0.0001), the meta-regression was performed for further evaluation of the source of heterogeneity with the Knapp-Hartung modification method. We assumed the heterogeneity may arise from the control types, age of the subjects, ethnicity of the patients, histology types, smoking status, tumor stages, sample size and the methods of methylation detection. However, complete subtype data can be only obtained in the control types, ethnicity, sample size and methylation detection methods. So, the regression was carried out by including each of complete subtypes data in the covariates. In the results of the meta-regression, no source of significant heterogeneity was found in all of them except for the control type (coefficient = −0.36, P = 0.018, Table 2). The τ2 decreased from 0.48 to 0.37, which indicates 23% [(0.48–0.37)/0.48] of heterogeneity can be explained by different control types. However, the adjustment for all the other factors with complete data mentioned above reduced the residual variance across studies only by 6%, which indicates that different ethnicity, sample size and methylation detection methods can explain only a slight proportion of the heterogeneity among studies. But for conservative, we still performed subgroup analysis according to the potential heterogeneity sources. In the subgroup analysis, the significant odds of the P16INK4A promoter methylation in tumor tissue was only changed in non-smokers (OR = 4.53, 95% CI: 0.68–30.26, P = 0.120) and sputum autologous control (OR = 1.49, 95% CI: 0.86–2.57, P = 0.151, Table 3). However, the changed of results should be interpreted with caution as only a small subject was included in non-smokers and sputum control subgroup analysis (Table 3).
Table 2. Meta-regression analysis.
| Heterogeneity sources | Coef.(95%CI) | t | p | τ2 | I2 Res(%) | R2(%) Adjusted |
| Control type | −0.36(−0.65,0.063) | −2.4 | 0.018 | 0.37 | 63.77 | 17.67 |
| Ethnicity | 0.35(−0.31,1.02) | 1.07 | 0.29 | 0.45 | 67.72 | 1.06 |
| Sample size | −0.0036(−0.011,0.004) | −0.96 | 0.34 | 0.48 | 68.83 | −5.23 |
| Method | −0.12(−0.61,0.38) | −0.47 | 0.64 | 0.48 | 68.84 | −6.17 |
Table 3. Subgroup analysis.
| NSCLC | Control | ||||||
| Subgroup | M+ | Total | M+ | Total | OR | 95% CI | p |
| Sex | |||||||
| Male | 151 | 331 | 58 | 331 | 5.72 | 2.50–13.10 | 0 |
| Female | 34 | 88 | 11 | 88 | 5.74 | 2.41–13.70 | 0 |
| Race | |||||||
| Asia-pacific | 972 | 2028 | 488 | 1903 | 3.23 | 2.37–4.40 | 0 |
| Caucasus | 293 | 624 | 151 | 620 | 4.32 | 2.37–7.87 | 0 |
| Histology | |||||||
| Sq | 228 | 348 | 151 | 332 | 2.81 | 1.96–4.05 | 0 |
| Ad | 224 | 421 | 140 | 421 | 2.53 | 1.85–3.44 | 0 |
| Other NSCLC | 19 | 44 | 7 | 43 | 4.97 | 1.57–15.76 | 0.006 |
| Smoking status | |||||||
| Nonsmoker | 6 | 32 | 2 | 32 | 4.53 | 0.68–30.26 | 0.12 |
| Smoker | 84 | 220 | 24 | 209 | 7.28 | 3.89–13.62 | 0 |
| Stage | |||||||
| Early (I–II) | 137 | 405 | 45 | 394 | 4.62 | 2.29–9.30 | 0 |
| Late (III–IV) | 118 | 222 | 48 | 228 | 5.19 | 3.28–8.23 | 0 |
| Method | |||||||
| MSP | 1114 | 2294 | 569 | 2166 | 3.49 | 2.58–4.70 | 0 |
| RT-MSP | 70 | 142 | 25 | 142 | 5.58 | 1.64–18.94 | 0.006 |
| q-MSP | 81 | 216 | 45 | 216 | 2.44 | 1.07–5.54 | 0.033 |
| Control type | |||||||
| Normal lung tissue | 555 | 1363 | 155 | 1287 | 5.49 | 3.77–8.00 | 0 |
| Blood | 441 | 823 | 300 | 819 | 2.56 | 1.71–3.84 | 0 |
| Sputum | 205 | 357 | 126 | 287 | 1.49 | 0.86–2.57 | 0.151 |
| BALF | 64 | 109 | 58 | 130 | 2.97 | 1.16–7.65 | 0.024 |
Correlation of P16INK4A Gene Promoter Methylation between Tumor Tissue and Autologous Clinical Samples
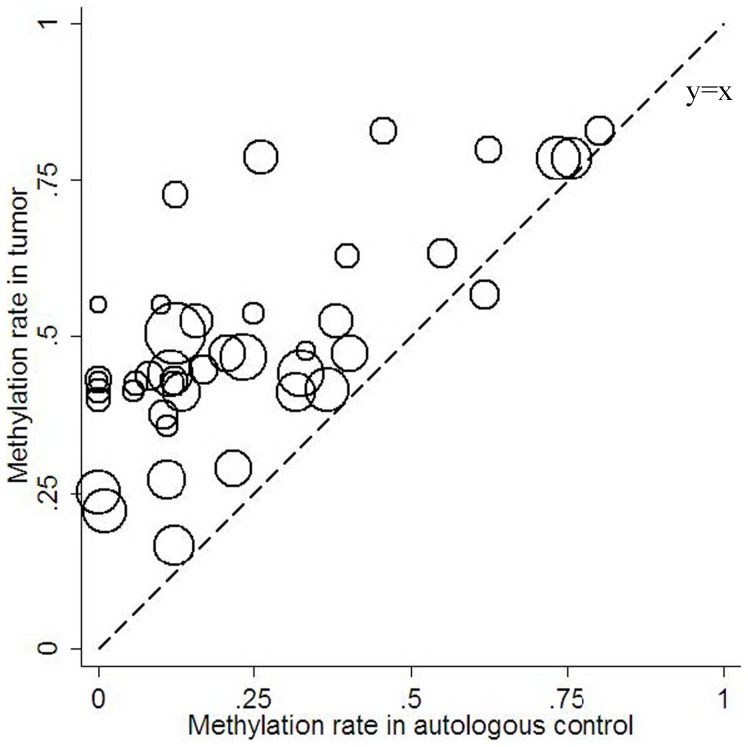
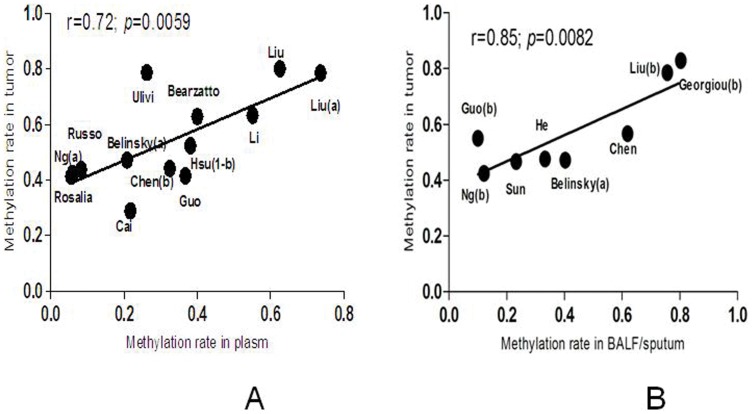
Generally, the frequency of P16INK4A promoter methylation ranged from 17% to 80% (median 44%) in the lung cancer tissue and 0 to 80% (median 15%) in the autologous controls according to the included studies. The methylation frequency in cancer tissue was much higher than that in clinical controls. We also find a strong and significant correlation between tumor tissue and autologous samples of P16INK4A promoter methylation across studies (Correlation coefficient 0.71, 95% CI:0.51–0.83, P<0.0001,). Fig. 4 demonstrates that most studies lie above the equal line between tumor tissue and controls, which illustrates the tumor tissue excess. In plasma samples, the methylation frequency ranged from 6% to 74% (median 33%), which showed a significant correlation of P16INK4A promoter methylation with cancer tissue (Correlation coefficient 0.72, 95% CI: 0.27–0.91, P = 0.0059, Fig 5A). The similar correlation was also found between the cancer tissue and sputum/BALF (Correlation coefficient 0.85, 95% CI: 0.35–0.97, P = 0.0082, Fig. 5B). The strong and significant correlation between tumor tissue and clinical autologous controls indicated that detection of methylation status in the clinical samples such as plasma, sputum or BALF can be a potential method for diagnosis of NSCLC without invasion.
Figure 4. Methylation frequency in tumor tissue versus autologous controls.
Figure 5. Correlation of P16INK4A promoter methylation between tumor tissue and autolougs clinical samples (A:plasm; B:BALF/sputum).
Publication Bias
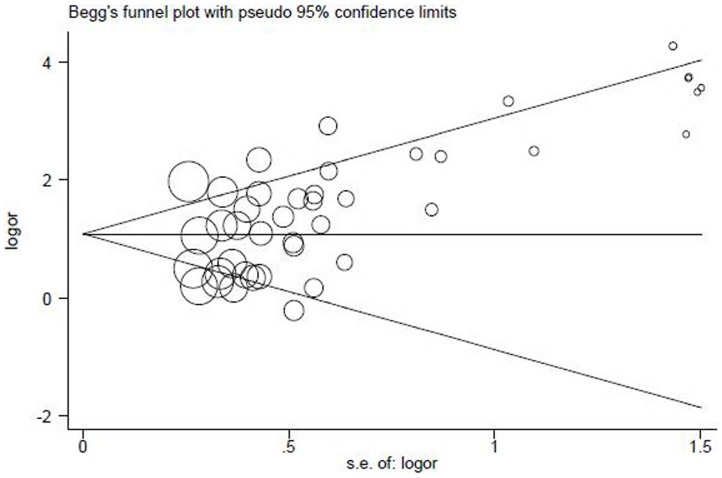
A Begg’s funnel plot and Egger’s test were used to evaluate possible publication bias [13]. As demonstrated in Fig. 6, the shape of the funnel plot showed a slight asymmetry at the bottom, with a trend towards reporting bigger odds ratio. However, Egger’s test did not illustrate any evidence of statistical publication bias (t = 0.78, P = 0.44).
Figure 6. Begg’s funnel plot for assessment of publication bias.
Each hollow circle represents a separate study for the indicated association. The area of the hollow circle reflects the weight (inverse of the variance). Horizontal line stands for the mean magnitude of the effect.
Discussion
Hypermethylation of CpG inlsnds in promoter regions is one of the important mechanisms for inactivation of tumor-suppressor genes, involving apoptosis, cell cycle, DNA repair and etc. Deregulation of the cell cycle control system was considered important in the procedure of tumorigenesis. P16INK4 is known as one the most important tumor suppressor genes, which plays an important role in regulating the cell cycle. This gene generates several transcript variants that regulate the G1-S transition of the cell cycle [44]. In NSCLC, this gene product has been shown to be absence in about 32–70% of the cancer cells [45], [46]. However, mutations of the P16INK4 gene are only found to be 0–10% [25], which indicating at least 22%–60% loss expression of P16INK4 is associated with other mechanisms, including promoter hypermethylation.
In NSCLC, promoter hypermethylation of P16INK4a gene which encodes a cyclin-dependent kinase inhibitor, has been found in variety of studies with a frequency of 17% [26] to 83% [23] in the tumor tissue and 6% [29] to 80% [23] in autologous clinical samples. The frequency of aberrant methylation of this gene ranged from 6% [17] to 74% [18] in serum or plasma and 10% [27] to 80% [23] in sputum or BALF. Although many studies have reported the prevalence of P16INK4a gene methylation in NSCLC, the association between cancer tissue and autologous clinical samples was not definitive with the reasons of small sample size. Thus, a meta-analysis was performed to quantify the methylation-disease association, by pooling data from published studies, which can increase the statistical power.
In the present study, we included a total of thirty-four articles that reported data of methylation frequency in non-small cell lung carcinoma tissue and autologous samples. The frequency of P16INK4A promoter methylation ranged from 17% to 80% (median 44%) in the lung cancer tissue and 0 to 80% (median 15%) in the autologous controls, which shows a great variety of methylation rate between studies. In general, the pooled odds ratio of methylation was 3.45 (95% CI: 2.63–4.54) in tumor tissue versus autologous samples under random-effect method, indicating the P16INK4A promoter methylation plays an important role in the tumorigenesis of NSCLC.
In subgroup analysis, the methylation odds in tumor tissue ranged from 1.49(0.86–2.57) to 5.49(3.77–8.00) when comparing to different autologous sample sets (non-tumor lung tissue, plasma, sputum and BALF). The methylation odds in tumor tissue was not significant when comparing to sputum (P = 0.151) indicating no statistical different frequency of P16INK4A promoter methylation was observed between sputum and cancer tissue in non-small cell lung cancer patients. However, the results should be interpreted with caution as only a small subject was included in sputum control subgroup analysis. In other subgroups, the methylation odds in tumor tissue ranged from 2.53 (1.85–3.44) to 7.28(3.89–13.62) according to clinical characteristics such as sex, ethnicity, histology, smoking status and stages. And the highest odds 7.28(3.89–13.62) in tumor tissue was found in smokers, demonstrating smoking may play an important role in the methylation of P16INK4A promoter regions, which was in accordance with previous studies [47]. The lowest odds 2.53(1.85–3.44) in tumor tissue was shown in the adenocarcinoma, suggesting the influence of P16INK4A promoter methylation was reduced in this kind of histology type.
Generally, a strong and significant correlation between tumor tissue and autologous samples in P16INK4A promoter methylation was found across studies(Correlation coefficient 0.71, 95% CI: 0.51–0.83, P<0.0001), which suggested the higher frequency of methylation in autologous sample was found, the higher prevalence of methylation can be observed in cancer tissue in patients with NSCLC. And this indicated that detection of methylation status in autologous samples such as plasma, sputum or BALF can be a potential method for diagnosis of NSCLC without invasion. And according to Esteller [48], the detection of promoter hypermethylaiton in tumor suppressor genes had important clinical use, such as diagnostic tool, biomarker for prognosis, predictor for treatment responses and etc.
However, several limitations required consideration of this study. The first limitation is heterogeneity. In this meta-analysis a significant heterogeneity was existed between studies (I2 = 69.8%, χ2 = 135.7, P<0.0001). Although, the meta-regression was performed for further evaluation of the source of heterogeneity with the Knapp-Hartung modification method, complete data can only be obtained in the subtypes of control types, ethnicity, sample size and methylation detection methods. In the results of the meta-regression, only a small part of heterogeneity can be explained by different ethnicity, sample size and methylation detection methods, indicating that some other source of heterogeneity must be exist among studies. Second, although no evident of publication bias was found in this study by Egger’s test, the small number of studies and possible existence of unpublished articles are inevitable and completely ruling out this possibility in all aspects is difficult [49]. The third limitation is the co-variate analysis of methylation. Demonstrating by the previous studies, promoter hypermethylation was associated with many clinical, demographic and molecular features, such as gender, age, smoking status and ethnicity [21], [23], [28]. And methylation events themselves may also be linked and interact with each other, suggesting methylation analysis of a single gene may be far from enough [50]. Fourth, as known that the promoter methylation is correlated with the reduction of gene expression. However, only three articles included in this meta-analysis provided the P16 gene expression status by using immunohistochemical analysis. The individual patient data (IPD) for the relationship between methylation status and expression of this gene was not given in the original articles. For the P16INK4A mutation, with carefully examination of the included studies, we found only two studies [13], [25] reported the P16INK4A mutation in exon 1, 2a and 2b regions. And none of the included 34 articles reported the mutation status of P16INK4A in promoter region.
In conclusion, the results of this study showed a higher prevalence of methylation in tumor tissue versus autologous samples in NSCLC patients, which demonstrate promoter methylation plays an important role in carcinogenesis. And the significant correlation between tumor tissue and clinical controls of P16INK4A gene promoter methylation indicated a promising biomarker for NSCLC diagnosis. However, significant methodological and validation issues remain to be addressed to provide the data that will enable this information to be considered for further clinical use [51].
Funding Statement
The authors have no support or funding to report.
References
- 1. Jemal A, Bray F (2011) Center MM, Ferlay J, Ward E, et al (2011) Global cancer statistics. CA Cancer J Clin 61(2): 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Risch A, Plass C (2008) Lung cancer epigenetics and genetics. Int J Cancer 123(1): 1–7. [DOI] [PubMed] [Google Scholar]
- 3. Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, et al. (2002) Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 346(2): 92–98. [DOI] [PubMed] [Google Scholar]
- 4. Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, et al. (1998) Aberrant methylation of p16(INK4a) is an early event in lung cancer and apotential biomarker for early diagnosis. Proc Natl Acad Sci U S A 95(20): 11891–11896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 93(18): 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, et al. (2001) Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res 61(1): 249–255. [PubMed] [Google Scholar]
- 7. Zhang CY, Jin YT, Xu HY, Zhang H, Zhang WM, et al. (2011) Relationship between promoter methylation of p16, DAPK and RAR beta genes andthe clinical data of non-small cell lung cancer. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 28(1): 23–28. [DOI] [PubMed] [Google Scholar]
- 8. Wang Y, Zhang D, Zheng W, Luo J, Bai Y, et al. (2008) Multiple gene methylation of nonsmall cell lung cancers evaluated with 3-dimensional microarray. Cancer 112(6): 1325–1336. [DOI] [PubMed] [Google Scholar]
- 9.Higgins JP, Green S (2008) Cochrane Handbook for Systematic Reviews of Interventions updated March 2011 John Wiley & Sons, Available: http://www.cochrane-handbook.org/.Accessed: 2011 DEC 12.
- 10. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7(3): 177–188. [DOI] [PubMed] [Google Scholar]
- 11. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327(7414): 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Egger M, Davey SG, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315(7109): 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seike M, Gemma A, Hosoya Y, Hemmi S, Taniguchi Y, et al. (2000) Increase in the frequency of p16INK4 gene inactivation by hypermethylation in lung cancer during the process of metastasis and its relation to the status of p53. Clin Cancer Res 6(11): 4307–4313. [PubMed] [Google Scholar]
- 14. Liu JY, An Q, Xu GD, Lei WD, Li L, et al. (2004) Hypermethylation of p16 gene in clinical specimens of patients with lung cancer. Zhonghua Zhong Liu Za Zhi 26(2): 75–77. [PubMed] [Google Scholar]
- 15. Ulivi P, Zoli W, Calistri D, Fabbri F, Tesei A, et al. (2006) P16INK4A and CDH13 hypermethylation in tumor and serum of non-small cell lung cancer patients. J Cell Physiol 206(3): 611–6115. [DOI] [PubMed] [Google Scholar]
- 16. Su C, Ye Y, Wang D (2000) Correlation between methylation on SmaI locus of the CDKN2/p16 gene CpG island and lung cancer. Zhonghua Zhong Liu Za Zhi 22(6): 471–473. [PubMed] [Google Scholar]
- 17. Ng CS, Zhang J, Wan S, Lee TW, Arifi AA, et al. (2002) Tumor p16M is a possible marker of advanced stage in non-small cell lung cancer. J Surg Oncol 79(2): 101–106. [DOI] [PubMed] [Google Scholar]
- 18. Liu Y, An Q, Li L, Zhang D, Huang J, et al. (2003) Hypermethylation of p16INK4a in Chinese lung cancer patients: biological andclinical implications. Carcinogenesis 24(12): 1897–1901. [DOI] [PubMed] [Google Scholar]
- 19. Russo AL, Thiagalingam A, Pan H, Califano J, Cheng KH, et al. (2005) Differential DNA hypermethylation of critical genes mediates the stage-specific tobacco smoke-induced neoplastic progression of lung cancer. Clin Cancer Res 11(7): 2466–2470. [DOI] [PubMed] [Google Scholar]
- 20. Belinsky SA, Grimes MJ, Casas E, Stidley CA, Franklin WA, et al. (2007) Predicting gene promoter methylation in non-small-cell lung cancer by evaluating sputum and serum. Br J Cancer 96(8): 1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsu HS, Chen TP, Hung CH, Wen CK, Lin RK, et al. (2007) Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer 110(9): 2019–2026. [DOI] [PubMed] [Google Scholar]
- 22. Hong YS, Roh MS, Kim NY, Lee HJ, Kim HK, et al. (2007) Hypermethylation of p16INK4a in Korean non-small cell lung cancer patients. J Korean Med Sci 22 Suppl: S32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Georgiou E, Valeri R, Tzimagiorgis G, Anzel J, Krikelis D, et al. (2007) Aberrant p16 promoter methylation among Greek lung cancer patients and smokers:correlation with smoking. Eur J Cancer Prev 16(5): 396–402. [DOI] [PubMed] [Google Scholar]
- 24. Hsu HS, Chen TP, Wen CK, Hung CH, Chen CY, et al. (2007) Multiple genetic and epigenetic biomarkers for lung cancer detection incytologically negative sputum and a nested case-control study for risk assessment. J Pathol 213(4): 412–419. [DOI] [PubMed] [Google Scholar]
- 25. Chen JT, Chen YC, Wang YC, Tseng RC, Chen CY, et al. (2002) Alterations of the p16(ink4a) gene in resected nonsmall cell lung tumors and exfoliated cells within sputum. Int J Cancer 98(5): 724–731. [DOI] [PubMed] [Google Scholar]
- 26. Harden SV, Tokumaru Y, Westra WH, Goodman S, Ahrendt SA, et al. (2003) Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancerpatients. Clin Cancer Res 9(4): 1370–1375. [PubMed] [Google Scholar]
- 27. Guo M, House MG, Hooker C, Han Y, Heath E, et al. (2004) Promoter hypermethylation of resected bronchial margins: a field defect ofchanges. Clin Cancer Res 10(15): 5131–5136. [DOI] [PubMed] [Google Scholar]
- 28. Kim DS, Cha SI, Lee JH, Lee YM, Choi JE, et al. (2007) Aberrant DNA methylation profiles of non-small cell lung cancers in a Koreanpopulation. Lung Cancer 58(1): 1–6. [DOI] [PubMed] [Google Scholar]
- 29. Cirincione R, Lintas C, Conte D, Mariani L, Roz L, et al. (2006) Methylation profile in tumor and sputum samples of lung cancer patients detected by spiral computed tomography: a nested case-control study. Int J Cancer 118(5): 1248–1253. [DOI] [PubMed] [Google Scholar]
- 30. Bearzatto A, Conte D, Frattini M, Zaffaroni N, Andriani F, et al. (2002) P16(INK4A) Hypermethylation detected by fluorescent methylation-specific PCR inplasmas from non-small cell lung cancer. Clin Cancer Res 8(12): 3782–3787. [PubMed] [Google Scholar]
- 31. He JY, WJ, Zhou Y, Liang B, Wang ZG (2011) Aberrant methylation of 5'CpG island in the p16 tumor suppressor gene in non-small cell lung cancer. Journal of Guangdong Medical College 35(3): 321–322. [Google Scholar]
- 32. He Y, Zhang SL, Yang LP, Li SJ, Tang BX (2002) Aberrent methylation of p16(INK4A) in non-small cell lung cancer. Chinese Journal of Health Laboratory Technology 12(3): 294–295. [Google Scholar]
- 33. Cai ZX, Liang QZ, Liao SX, Zhang XX, Wang X (2003) Detection of Methylation in p16 Gene Promoter in Serum of Non-small Cell Lung Cancer. China Cancer 12(9): 536–538. [Google Scholar]
- 34. Zhang W, Sun YE, Cai Q, Wang GH, Lu GM (2004) The study of p16 methylation and p16 protein expression in non-small cell lung cancers(NSCLC). Medical Journal of Air Force 20(1): 25–30. [Google Scholar]
- 35. Li WF, Xu H, Zhang L (2006) Aberrant methylation of p16 gene in plasma of patients with non-small cell lung cancer. Journal of Guangdong Medical College 27(6): 855–856. [Google Scholar]
- 36. Wang YS, Jin YT, Xue SL, Yu ZC, Xu YC, et al. (2006) Research on methylation of p16 gene in non-small cell lung cancer. Acta Universitatis Medicinalis Anhui 41(6): 619–621. [Google Scholar]
- 37. Yang ZH, Cai YY, Sun LH, Qiao Y (2007) Aberrant promoter methylation of p16(INK4A) and RASSF1A genes in non-small cell lung cancers. Journal of Practical Oncology 22(2): 153–155. [Google Scholar]
- 38. Zhang L (2007) A preliminary study of radiotherapeutic target region design for NSCLC by detection of p16 gene methylation. Med J Qilu 22(4): 347–349. [Google Scholar]
- 39. Guo X J, Shen L, Zhang H, Wang Y (2008) The detection and clinical diagnostic value of promoter hypermethylation of p16 gene in serum and tissue of lung cancer patients. Hebei Medical Journal 30(6): 757–759. [Google Scholar]
- 40. Chen SH, Xue SL, Jin YT, Yu ZC, Hu HL, et al. (2010) Methylation of p16 gene in plasma and tissues from non-small cell lung cancer patients. Chin J Lab Diagn 14(7): 1035–1038. [Google Scholar]
- 41. Guo MF, Liu H, Feng L, Wang YC (2010) Methylation of p16 gene in lung cancer patients. MMJC12(5): 82–83. [Google Scholar]
- 42. Zhang ZX, Zhang L (2006) Methylation of ASC and p16 genes in promoter region associated with genesis and progress of non-small cell lung cancer. China Journal of Modern Medicine 16(9): 1338–44. [Google Scholar]
- 43. Sun N, Zhang L, Liu YY, Zheng SY, Zhao X (2012) Methylation of P16 and RASSF1A Genes in Sputum Samples Associated with Peripheral Non-Small Cell Lung Cancer. Progress in Modern Biomedicine 12(13): 2503–2510. [Google Scholar]
- 44. Liggett WH, Sidransky D (1998) Role of the p16 tumor suppressor gene in cancer. J Clin Oncol 16(3): 1197–1206. [DOI] [PubMed] [Google Scholar]
- 45. Brambilla E, Moro D, Gazzeri S, Brambilla C (1999) Alterations of expression of Rb, p16(INK4A) and cyclin D1 in non-small cell lung carcinoma and their clinical significance. J Pathol 188(4): 351–360. [DOI] [PubMed] [Google Scholar]
- 46. Gazzeri S, Gouyer V, Vour’ch C, Brambilla C, Brambilla E (1998) Mechanisms of p16INK4A inactivation in non small-cell lung cancers. Oncogene 16(4): 497–504. [DOI] [PubMed] [Google Scholar]
- 47. Zhang B, Zhu W, Yang P, Liu T, Jiang M, et al. (2011) Cigarette smoking and p16INK4alpha gene promoter hypermethylation in non-smallcell lung carcinoma patients: a meta-analysis. PLOS ONE 6(12): e28882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Esteller M (2008) Epigenetics in cancer. N Engl J Med 358(11): 1148–1159. [DOI] [PubMed] [Google Scholar]
- 49. Trinquart L, Abbe A, Ravaud P (2012) Impact of reporting bias in network meta-analysis of antidepressant placebo-controlled trials. PLOS ONE 7(4): e35219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Taby R, Issa JP (2010) Cancer epigenetics. CA Cancer J Clin 60(6): 376–392. [DOI] [PubMed] [Google Scholar]
- 51. Sapari NS, Loh M, Vaithilingam A, Soong R (2012) Clinical potential of DNA methylation in gastric cancer: a meta-analysis. PLOS ONE 7(4): e36275. [DOI] [PMC free article] [PubMed] [Google Scholar]