Abstract
Chloroplast genome sequences are of broad significance in plant biology, due to frequent use in molecular phylogenetics, comparative genomics, population genetics, and genetic modification studies. The present study used a second-generation sequencing approach to determine and assemble the plastid genomes (plastomes) of four representatives from the agriculturally important Lolium-Festuca species complex of pasture grasses (Lolium multiflorum, Festuca pratensis, Festuca altissima, and Festuca ovina). Total cellular DNA was extracted from either roots or leaves, was sequenced, and the output was filtered for plastome-related reads. A comparison between sources revealed fewer plastome-related reads from root-derived template but an increase in incidental bacterium-derived sequences. Plastome assembly and annotation indicated high levels of sequence identity and a conserved organization and gene content between species. However, frequent deletions within the F. ovina plastome appeared to contribute to a smaller plastid genome size. Comparative analysis with complete plastome sequences from other members of the Poaceae confirmed conservation of most grass-specific features. Detailed analysis of the rbcL–psaI intergenic region, however, revealed a “hot-spot” of variation characterized by independent deletion events. The evolutionary implications of this observation are discussed. The complete plastome sequences are anticipated to provide the basis for potential organelle-specific genetic modification of pasture grasses.
Keywords: Italian ryegrass, meadow fescue, tall fescue, perennial ryegrass, chloroplast DNA, phylogenetics
Sequences of plant plastid genomes (plastomes) have been long used for a broad range of plant biology applications. Due to the properties of high sequence conservation and abundance (such that thousands of copies may be present in each cell) (Zoschke et al. 2007) plastomes, or regions thereof, have been used for phylogenetic studies (Burke et al. 2012; Wu and Ge 2012; Zhang et al. 2011b), comparative genomics (Gao et al. 2010), DNA barcoding activities (Hollingsworth et al. 2009), and various biotechnology applications (Clarke et al. 2011). The plastome is a circular molecule that, in most taxa, varies between 108 and 218 kb in size and generally displays a high level of conservation between land plant species. The typical plastome is composed of two identical inverted repeats (IR, 20–76 kb) that separate a large single-copy (LSC, 60–90 kb) and a small single-copy (SSC, 7–27 kb) region (Chumley et al. 2006). Generation of a complete plastome sequence from representative species can hence provide information related to evolutionary history, and permit the development of sequence-based tools such as molecular genetic marker assays for ecological studies and DNA barcode-based diagnostics for species identification. Traditionally, plastome sequencing and assembly has involved the extraction of high-purity chloroplast DNA, which is then cloned into vectors and sequenced using dideoxynucleotide terminator chemistry. However, as second-generation technology-based sequencing projects have become more cost-effective and feasible, a number of recent studies have exploited these methods to assemble plastome sequences from total cellular DNA templates. Target species for this approach have included rice (Oryza spp.) (Nock et al. 2011), pear (Pyrus pyrifolia [Burm.] Nak.) (Terakami et al. 2012), duckweed (Lemnoideae spp.) (Wang and Messing 2011), and date palm (Phoenix dactylifera L.) (Yang et al. 2010).
Grasses of the genera Lolium and Festuca (Poaceae family, Pooideae subfamily) include a number of agriculturally important pasture grasses that are widely cultivated in temperate regions, including perennial ryegrass (Lolium perenne L.), Italian ryegrass (L. multiflorum Lam.), meadow fescue (Festuca pratensis Huds. syn. L. pratense Huds. [Darbysh.]), and tall fescue (F. arundinacea Schreb. syn. L. arundinaceum Schreb. [Darbysh.]). In addition, the Festuca genus contains a number of grass species grown for turf or ornamental purposes such as sheep’s fescue (F. ovina L.) and red fescue (F. rubra L.). The Festuca genus is recognized as containing more than 600 species with multiple ploidy levels and near-global distribution, whereas the Lolium genus contains only 10 recognized diploid taxa (Clayton and Renvoize 1986). The two genera have been differentiated on the basis of inflorescence structure, but controversy has surrounded the taxonomic classification of some Lolium and Festuca species. Phylogenetic analysis consistently identifies the Lolium genus as being nested within the Schedonorus subgenus of the Festuca genus (Catalán et al. 2004; Charmet et al. 1997; Hand et al. 2010; Inda et al. 2008; Torrecilla and Catalán 2002; Xu and Sleper 1994), and the species complex has been subject to reclassification (Darbyshire 1993; Soreng and Terrell 1997).
Regardless of the taxonomic classification system that is used, it is clear that the Lolium and Festuca genera represent a closely allied complex of related and partially interfertile species. Further complexity is present within the allohexaploid species tall fescue, which may also be more accurately described as a species complex, as three eco-geographic races (morphotypes) are recognized (Hand et al. 2010), at least two of which (Continental and Mediterranean) differ in terms of ancestral genome origin. Of the three, the Continental morphotype has been the most widely cultivated and studied at the genetic level. Within the Lolium-Festuca species complex, complete plastome sequences have so far been assembled from perennial ryegrass and hexaploid tall fescue (the Continental cultivar KY31) (Cahoon et al. 2010; Diekmann et al. 2009).
Complete plastome sequences from members of the Lolium-Festuca species complex would be valuable for further study of these taxa and for a broader understanding of Poaceae plastome evolution. Since publication of the first plastome sequence from a grass species (Hiratsuka et al. 1989), subsequent comparisons with that of tobacco (Nicotiana tabacum L.), which is regarded as the reference for angiosperms, has revealed six main structural alterations. These changes include three inversions in the LSC region, loss of an intron in the rpoC1 gene, a sequence insertion in the rpoC2 gene (Shimada et al. 1990), rearrangement of the accD gene (Ogihara et al. 1992), absence of ORFs within the IR (Downie et al. 1994), and translocation of the rpl23 gene to a region of the LSC region, between rbcL and psaI (Ogihara et al. 1988). As more grass plastome sequences became available, these structural alterations have been studied further and the timing of their origin has been discussed (Harris et al. 2013; Katayama and Ogihara 1996; Morris and Duvall 2010). However, a definitive understanding of variation present within Poaceae plastomes is yet to be obtained.
This study describes the assembly of complete plastomes from a selection of four representative species within the Lolium−Festuca species complex, using sequence generated from total cellular DNA based on second-generation technology. The resulting plastomes were compared with those of other Poaceae species to compare higher-level organization and gene content and to support phylogenomic analysis of evolutionary divergence within the Lolium-Festuca species complex.
Materials and Methods
Plant material and DNA extraction
Sampling of diploid Lolium-Festuca species was designed to include important agricultural species, along with representatives of each major subgenus within the Festuca genus (Inda et al. 2008; Torrecilla and Catalán 2002). Seed was sourced from either Heritage Seeds Australia, Seed Force Australia, the USDA germplasm collection, or the Genetic Resources Unit of the Institute for Biological, Environmental and Rural Studies (Aberystwyth, Wales). Details of the four selected accessions are provided in Table 1. This study represents an extension of a pre-existing shotgun whole-genome sequencing activity for each of the included diploid species, which aimed to enrich the proportion of sampled nuclear genome through selection of target tissue for DNA extraction. For meadow fescue and Italian ryegrass, DNA was consequently extracted from root tissue of plants grown in sand pots, and roots were washed with distilled water and then ground in liquid nitrogen prior to extraction. As root tissue was unavailable from the F. altissima and F. ovina accessions due to quarantine restrictions, DNA was extracted from leaf tissue for these samples. All DNA extractions were performed using the DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany).
Table 1. Details of the four species selected for plastome sequencing.
| Species | Festuca Subgenus | Source | Cultivar/Accession | Plant Tissue |
|---|---|---|---|---|
| Lolium multiflorum | − | Seed Force | Accelerate | Root |
| Festuca pratensis | Schedonorus | USDA germplasm collection | Mimer (PI 310482) | Root |
| Festuca altissima | Drymanthele | IBERS | BS4384 | Leaf |
| Festuca ovina | Festuca | IBERS | BL2643 | Leaf |
USDA, United States Department of Agriculture; IBERS, Institute for Biological, Environmental and Rural Studies.
Paired-end library preparation and sequencing
For each sample, paired-end libraries with approximately 200-bp inserts were prepared using the Illumina TruSeq DNA Sample Preparation Kit (Illumina Inc., San Diego, CA). Library quantification was performed using the KAPA Library Quantification Kit (KAPA Biosystems, Boston, MA). Paired-end cluster generation and sequencing was performed on the Hi-Sequation 2000 (Illumina), with each library being allocated to one lane of a flow cell. All generated sequence reads were subjected to quality control using a custom PERL script and were trimmed if they met any of the following criteria: more than three consecutive nucleotides designated as unattributable base identities (N); more than three nucleotides had a PHRED quality score less than or equal to 20; or a median PHRED quality score less than 20. Furthermore, reads were discarded when the length was less than 25 nucleotides.
Estimation of bacterial and plastid genome contribution
For each sequenced genotype, the proportions of sequence reads of probable bacterial or plastid genome origin were estimated using a reference alignment approach. A subset of reads from each genotype was mapped to a set of generated reference sequences consisting of complete or partial genomes for 43 bacterial species and five chloroplast genomes (Supporting Information, Table S1). Alignment was performed using the Burrows-Wheeler Alignment (BWA) tool (Li and Durbin 2009) with both the maximum number of gap openings and edit distance in the seed set as five. All other parameters were left as default values. File format conversion was performed using the utilities for the Sequence Alignment/Map (i.e., SAM) format (SAMtools) (Li et al. 2009), and the alignment was ultimately viewed with Tablet 1.12.02.06 (Milne et al. 2010).
Plastome assembly
Sequence reads of plastome origin initially were filtered from the sequencing output by alignment of all reads to the published chloroplast genomes of perennial ryegrass, tall fescue, bread wheat (Triticum aestivum L.), barley (Hordeum vulgare L.), and Brachypodium distachyon L. (GenBank accession numbers NC009950, FJ466687, NC002762, EF115541, and EU325680). Alignment was performed using BWA, as described previously.
For each species, all reads that aligned to any of the chloroplast genome references were extracted and de novo assembled using SOAPdenovo v1.05 (127mer version; (http://soap.genomics.org.cn) (Li et al. 2010). A range of k-mer values were tested (ranging from 51 to 91), and the final assembly was selected when the contigs had an additive length similar to that of the perennial ryegrass chloroplast genome (135,282 bp). Contigs generated in the SOAPdenovo assembly were further assembled in Sequencher 4.7 (GeneCodes Corp., Ann Arbor, MI) with a specification of minimum match percentage of 97% and a minimum overlap of 50 bp. Contigs were ordered on the basis of the pre-existing annotation of the perennial ryegrass chloroplast genome. Gaps between contigs were presumed to derive from plastome regions that are sufficiently divergent between the reference genomes and the sequenced species, such that no reads mapped to these regions, hence preventing inclusion in the de novo assembly. The initial attempt to fill gaps therefore exploited the sampling strategy and iteratively used contigs of the next most closely related species as a reference for filtering sequence reads of plastome-origin via BWA mapping. Any gaps remaining after completion of this process were subsequently amplified by polymerase chain reaction (PCR) using primers designed to flanking regions and sequenced using dideoxynucleotide terminator chemistry. PCR conditions were as previously described (Hand et al. 2010), and cycling conditions included an enzyme activation step of 95° for 15 min, followed by 30 cycles of 95° for 1 min, 65° for 30 sec, 72° for 1 min with the annealing temperature decreasing for 1° per cycle until the final temperature was 55°, and a final extension step of 72° for 10 min. PCR products were purified and directly sequenced, as described previously (Hand et al. 2010).
As the plastome contains two identical copies of an inverted repeat (IRa and IRb), the assembly was initially performed to generate a contig that included only the LSC, IRb and SSC sections. The IRa section was subsequently added manually to the contig, and IR boundaries were confirmed using PCR.
Genome annotation, alignment, and phylogenomic analysis
Each complete plastome was annotated using the online software DOGMA with default parameters (Wyman et al. 2004). Predicted coding regions were visually inspected, compared with the published chloroplast genomes of perennial ryegrass and tall fescue, and adjusted accordingly. Complete plastome sequences of 12 species were aligned using the LAGAN program within the mVISTA online suite of computational tools (Brudno et al. 2003). Default parameters were applied, and the annotation framework of the perennial ryegrass chloroplast genome was used. Percentage identity between each plastome, all relative to that of perennial ryegrass, was subsequently visualized through a VISTA plot (Frazer et al. 2004).
Plastome-based phylogeny was reconstructed for the six Lolium-Festuca species using whole plastome alignment generated by LAGAN, as described previously. Plastomes of the Pooideae species creeping bentgrass (Agrostis stolonifera L.) and barley (GenBank accession numbers NC008591 and EF115541) were also included as outgroups. The phylogenetic tree was constructed through the method of maximum parsimony as implemented by MEGA 5.10 (Tamura et al. 2011). Sites with gaps or missing data were excluded from the analysis, and statistical support was achieved through bootstrapping using 1000 replicates.
Analysis of grass-specific plastome features
Structural features of the plastome that had previously been identified as grass-specific were examined for 12 Poaceae species, including each of the four species selected for this study. The structure of rpoC2 was analyzed by aligning this gene from each species using Sequencher 4.7 (GeneCodes), and the insertion event within this gene was detected using previously defined sequence boundaries (Katayama and Ogihara 1996). The rpl32′ pseudogene was deemed to be present within a plastome if the rpl32 gene predicted by DOGMA was located between rbcL and psaI. Similarly, identification of an accD pseudogene was determined based on prediction of an incomplete form by DOGMA. To further investigate the extent of inter-specific variation, the intergenic region between rbcL and psaI of the 12 Poaceae species, was aligned using Sequencher 4.7 (GeneCodes) and manually edited as required. Only deletions greater than 40 bp were recorded and represented within the diagram.
Results
Sequencing output
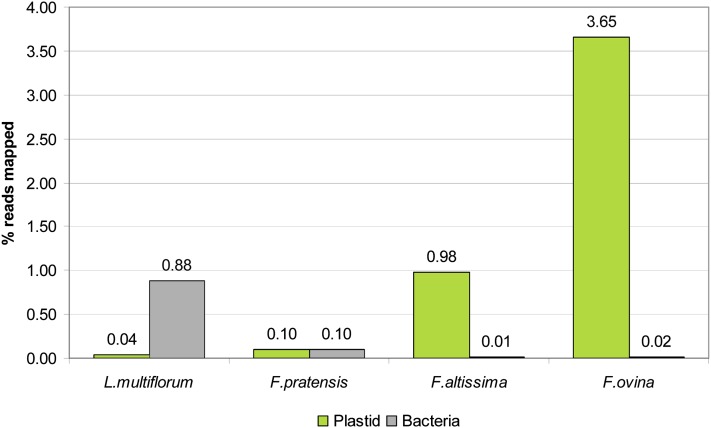
Variation was observed between samples with respect to the proportion of reads predicted to originate from either plastid or bacterial genomes (Figure 1). For instance, the library constructed from Italian ryegrass root tissue contained substantially more bacterium-derived DNA than libraries constructed for any of the other species. The bacterial reference genome which was predominantly responsible for this large discrepancy belonged to the species Flavobacterium johnsoniae. A direct comparison of the root- and leaf-extracted DNA templates revealed that the former contained fewer (average of 26-fold) plastome reads, but a greater number of reads from bacterial genomes (average of 28-fold).
Figure 1 .
Proportion of sequence reads identified as originating from either plastid or bacterial genomes, based on sequencing output from each species. The percentages of reads mapped to each of the reference genome sets are displayed above each bar.
Plastome assembly and organization
Complete plastomes for each of the included species were successfully assembled following sequencing of total DNA templates and submitted to GenBank under the accession numbers JX871939-JX871942. As expected, the read filtering process recovered a greater number of plastid genome reads from the leaf-extracted DNA template (3.6 × 106 and 7.9 × 106 reads from F. altissima and F. ovina, respectively) than the root-extracted DNA template (1.1 × 105 and 4.7 × 105 reads from Italian ryegrass and meadow fescue respectively). Throughout the assembly process, a larger number of unclosed gaps were present for F. altissima and F. ovina, as compared to plastomes of the other two species. However, all gaps present within the initial assembly were capable of closure using PCR.
The complete plastomes of the four sequenced species ranged in size from 133,165 bp to 135,291 bp, similar to those of perennial ryegrass and tall fescue (Table 2). For each plastome, the LSC and SSC regions were c. 80 kb and 12 kb in size, respectively. The LSC and SSC regions were separated by the pair of inverted repeats (IRa and IRb), which were both c. 21 kb long (Table 2).
Table 2. A comparison of plastome size and organization for six Lolium-Festuca species.
| % of Plastome Identified as: |
|||||||
|---|---|---|---|---|---|---|---|
| Plastome Size | LSC Size | SSC Size | IR Size | Gene Space | Intron Space | Intergenic Space | |
| L. perenne | 135,282 | 79,972 | 12,428 | 21,441 | 54.34 | 11.91 | 33.75 |
| L. multiflorum | 135,175 | 79,848 | 12,485 | 21,421 | 53.56 | 12.01 | 33.49 |
| F. pratensis | 135,291 | 79,934 | 12,511 | 21,423 | 53.54 | 11.91 | 33.61 |
| F. arundinacea | 136,048 | 80,560 | 11,300 | 22,600 | 54.20 | 9.31 | 36.49 |
| F. altissima | 135,272 | 79,828 | 12,598 | 21,423 | 53.64 | 11.99 | 33.44 |
| F. ovina | 133,165 | 78,329 | 12,386 | 21,225 | 54.50 | 12.19 | 32.33 |
All genome sizes are denoted in base pairs.
The annotation process identified 114 different genes within the plastomes of the sequenced species. Of these, 20 were duplicated within the IR regions, which brought the total to 134 genes. A total of 82 genes were present within the LSC and 12 within the SSC region. The gene space accounted for approximately 54% of each genome, the remaining sequence being attributed to intergenic spacers (c. 33%) and introns (c. 12%) (Table 2). These proportions are similar to those previously identified in the perennial ryegrass chloroplast genome, although the tall fescue plastome appears to contain a larger (36.49%) amount of intergenic space and a lower proportion (9.31%) of introns.
Annotation details for the four species in this study differ slightly from those of the published perennial ryegrass and tall fescue plastomes. In comparison, the perennial ryegrass chloroplast genome lacks three tRNA genes, and the hypothetical reading frame ycf68 (which is located within the intron of trnI-GAU), although present, was not annotated. Furthermore, the tall fescue plastome lacks the rps14 gene, ycf4 and two tRNA genes. Of the 77 different protein-coding genes identified within the four sequenced species, 16 display length polymorphism based on comparison between the six species (Table 3). The most notable of these are rpoC2, rps18, and ycf68, which differ in length by 38, 21, and 18 codons, respectively. For all six species that were compared, the IR region had expanded to include a portion of the ndhH gene, as observed in certain other grasses. The size of this gene portion is also variable among the six Lolium-Festuca species, varying from 60 to 67 codons in length.
Table 3. List of plastome genes that vary in size between the six Lolium-Festuca species included in the comparative study.
| Gene Size in Codons |
|||||||
|---|---|---|---|---|---|---|---|
| Gene Name | L. perenne | L. multiflorum | F. pratensis | F. arundinacea | F. altissima | F. ovina | Size Difference |
| atpA | 508 | 508 | 508 | 508 | 508 | 505 | 3 |
| atpI | 248 | 248 | 248 | 248 | 248 | 247 | 1 |
| cemA | 231 | 231 | 231 | 231 | 231 | 226 | 5 |
| clpI | 217 | 217 | 217 | 215 | 217 | 215 | 2 |
| infA | 108 | 108 | 108 | 114 | 114 | 114 | 6 |
| ndhD | 503 | 503 | 503 | 501 | 501 | 501 | 2 |
| ndhF | 742 | 742 | 742 | 739 | 740 | 740 | 3 |
| ndhHa | 60 | 60 | 60 | 60 | 63 | 67 | 7 |
| psbF | 40 | 40 | 40 | 40 | 43 | 40 | 3 |
| rbcL | 478 | 478 | 478 | 488 | 478 | 480 | 10 |
| rpl32 | 60 | 60 | 60 | 68 | 60 | 64 | 8 |
| rpoC2 | 1467 | 1467 | 1474 | 1505 | 1474 | 1474 | 38 |
| rps15 | 91 | 91 | 91 | 91 | 93 | 91 | 2 |
| rps16 | 90 | 93 | 90 | 89 | 86 | 86 | 7 |
| rps18 | 157 | 157 | 157 | 150 | 157 | 171 | 21 |
| ycf68 | − | 127 | 127 | 127 | 145 | 145 | 18 |
The length of each gene is given in codons, along with the size difference between the shortest and longest variant for each gene.
This ndhH gene fragment is present within the IRb region.
Whole plastome comparison
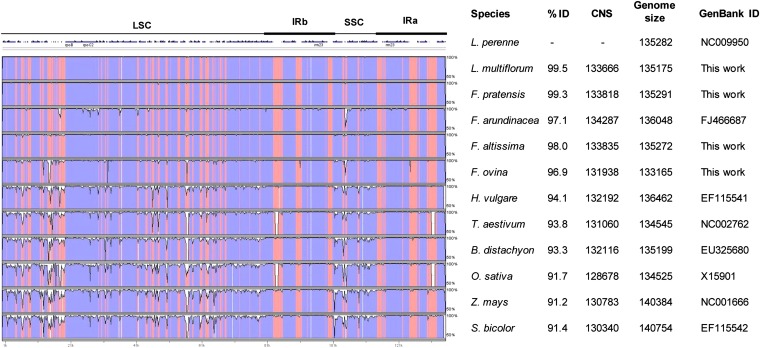
Genome-wide comparison of the Lolium-Festuca plastome sequences reveals a high level of conservation (96.9%–99.5%) as visualized by the VISTA plot (Figure 2). Interestingly, the plastid genome of F. altissima showed greater sequence similarity to that of perennial ryegrass (98.0%) than the previously published plastome of tall fescue (97.1%). In general, the IRs displayed lower levels of sequence divergence than the single-copy regions. Divergence was most apparent within intergenic regions, closer analysis of which revealed frequent deletions within the F. ovina plastome, ranging between 16 and 503 bp in length. These small, intergenic deletions presumably account for the smaller genome size observed for this species.
Figure 2 .
Alignment of complete plastome sequences from 12 Poaceae species. Alignment and comparison was performed using mVISTA, and percentage identity between the plastomes was visualized in the form of a VISTA plot. Each measure of similarity (% ID) is relative to the plastome of perennial ryegrass, which was used as a reference. Blue-shaded regions indicate coding regions, as defined by annotation of the perennial ryegrass. Pink regions represent conserved noncoding sequence. Regions of the plastome are illustrated above the VISTA plot. CNS, conserved nucleotide sequence.
Obvious “hot-spots” of variation occur between the psbC and rpoB genes, the intergenic regions between rps4 and clpP, and a small area within the SSC region, between the ndhF and ccsA genes. The similarity of plastome organization that the six species share with other members of the Poaceae family is also apparent from the VISTA plot. Within the broader Poaceae, the genes also appear to be well conserved, and the major sites of divergence are located within the aforementioned regions. Compared with other Poaceae species, the wheat and rice plastomes possess an IR-located deletion between the rpl23 and ndhB genes of c. 1 kb, which is likely to partially contribute to smaller observed plastome sizes.
Grass-specific plastome features
Each plastome generated in this study was examined for the presence of previously identified structural features that are unique to grasses and subsequently was compared with other Poaceae plastomes. As with most other Poaceae species sequenced to date, each plastome contained three previously defined inversions within the LSC and failed to contain an intron within the rpoC1 gene. Lolium and Festuca plastomes also displayed an insertion within the rpoC2 gene compared with the reference dicotyledonous plant plastome sequence (of N. tabacum). The size of this insertion varies within the Poaceae and ranges from 341 to 438 bp in length (Table 4).
Table 4. Details of the variable grass-specific plastome features for 12 Poaceae species.
| rpoC2 Insertion Size, bp | rpl32′ Presence | accD Presence | rbcL-psaI Region, bp | |
|---|---|---|---|---|
| L. perenne | 341 | Absent | Pseudo | 1183 |
| L. multiflorum | 341 | Absent | Pseudo | 1182 |
| F. pratensis | 362 | Absent | Pseudo | 1205 |
| F. arundinacea | 419 | Absent | Pseudo | 1206 |
| F. altissima | 362 | Absent | Pseudo | 1179 |
| F. ovina | 362 | Absent | Absent | 885 |
| H. vulgare | 407 | Present | Pseudo | 1603 |
| T. aestivum | 413 | Present | Absent | 879 |
| B. distachyon | 383 | Absent | Absent | 461 |
| O. sativa | 386 | Present | Pseudo | 1693 |
| Z. mays | 438 | Presenta | Absent | 888 |
| S. bicolor | 417 | Present | Absent | 861 |
Detected in this study although not annotated in GenBank.
Variation also was detected in the presence of an rpl23 translocation product (rpl23′) and an accD pseudogene in the region between rbcL and psaI (Table 4). The rpl23′ element was absent from the plastomes of all Lolium-Festuca species, as well as B. distachyon. The presence of this pseudogene, however, was confirmed in all other analyzed species. Remnants of the accD gene were detected in almost all Lolium-Festuca species, with the exception of F. ovina. This pseudogene was also identified in barley and rice but was not predicted in the other species.
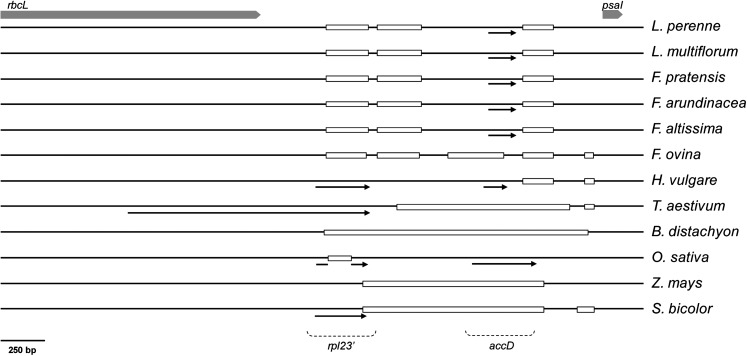
Alignment of the rbcL–psaI region from each Poaceae taxon revealed that differences in annotation result from deletions of varying size (Figure 3). All Lolium-Festuca species included in the alignment possess two deletions (of c. 250 bp each) in the position occupied by the annotated rpl23′ element in other species. The only remaining evidence for presence of rpl23′ within these species is a c. 55 bp region that precedes the two deletions. The accD pseudogene is absent from F. ovina due to a 317-bp deletion which is not present in the other Lolium-Festuca species. Of the 12 candidate species, barley and rice possess the smallest number of deletions, and contain evidence for both the rpl23′ and accD pseudogenes. Plastomes of the remaining species contain large (1–1.5 kb) deletions which have removed either one (wheat, maize and sorghum) or both of the pseudogenes (B. distachyon) in this region. This alignment also revealed frequent single base indels within the tall fescue plastome sequence, as compared to the other grass species.
Figure 3 .
Major deletions within the rbcL–psaI intergenic region across 12 Poaceae species. Annotated genes from each species are depicted by a black arrow, and gene identity is illustrated at the bottom of the diagram. Deletions greater than 40 bp in length are represented by a rectangle. The boundaries of the intergenic region are defined at the top of the figure by shaded block arrows representing rbcL and psaI genes.
Phylogenomic analysis
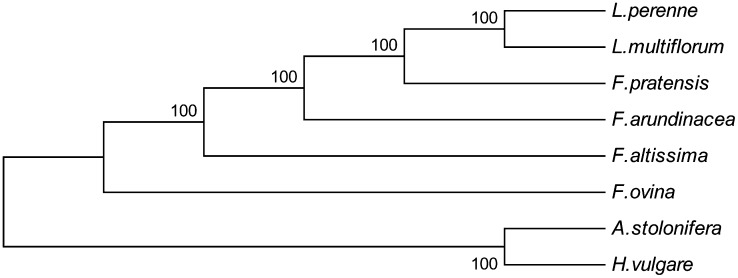
Phylogenomic analysis of representatives from the Lolium-Festuca species complex produced a single, well-supported tree using maximum parsimony (Figure 4). The tree is fully resolved with the two Lolium species sister to each other and paraphyletic with Festuca. The two outgroup species (A. stolonifera and H. vulgare) are basal to the remaining species in a separate, resolved clade.
Figure 4 .
Phylogeny of Lolium-Festuca species based on whole plastome sequence. Plastomes of Agrostis stolonifera and Hordeum vulgare were included as outgroup species. The phylogenetic tree was drawn using maximum parsimony and bootstrap support was achieved using 1000 replicates.
Discussion
Optimization of plastome sequencing methodology
Through extraction and sequencing of total DNA from a given sample, the genomes from each organelle (nucleus, plastid and mitochondria) are captured, along with any microbial genomes that may have been inadvertently coextracted. In the present study, generation of total DNA template from both root and leaf tissue permitted comparison of proportions of sequence reads generated from the various genomes for each source. Substantial variation was observed between species with respect to the proportions of bacterial and plastome reads. Nonetheless, a greater proportion of plastome-derived reads, as expected, was obtained from the leaf tissue-derived template, presumably due to greater abundance of chloroplasts within leaf cells. The number of plastome copies per leaf cell depends upon age and physiological state, but a ratio of more than 10,000 has been reported (Boffey and Leech 1982).
In addition to generating a smaller proportion of plastome-related reads, sequencing of the root-extracted template also suffered the disadvantage of producing a greater number of reads of bacterial origin. Possible sources for the contamination include rhizobacteria growing in close association with the root system, or bacterial pathogens present within the plant tissue (Egli et al. 1975; Franche et al. 2009; Okon and Labandera-Gonzalez 1994; Vauterin et al. 1995). In addition, a large proportion of the bacterial contamination possibly occurred through the inability to remove all soil particles attached to the root tissue prior to DNA extraction. Flavobacterium johnsoniae, which was predominantly responsible for the higher levels of contamination associated with the Italian ryegrass library, is commonly found in soil and freshwater (McBride et al. 2009; Stanier 1947), consistent with the assumption of soil-borne contamination. This result has broader implications for the design of whole-genome sequencing projects. The use of root tissue template for DNA extraction would be beneficial for such enterprises to maximize yield of sequence reads generated from the nuclear genome. The success of this approach, however, is obviously dependent upon extensive cleaning of source tissue to prevent inadvertent sequencing of microbial genomes.
Although presenting its own unique challenges, plastome assembly from reads representing total DNA is now undoubtedly a more rapid and efficient strategy than the previous method based on organelle fractionation.Filtering of plastome-derived reads from the remaining pool is an essential first step, which has been achieved for most studies to date through alignment of all reads to a reference plastid genome (Wang and Messing 2011; Yang et al. 2010; Zhang et al. 2011a). The success of this filtration step hence depends upon the evolutionary distance between the reference and target species, such that a more closely related reference genome would be able to recover a greater proportion of reads. This prediction was accurate for the present study, as a larger number of gaps were apparent within the plastome assemblies for those species (F. altissima and F. ovina) that are more distantly related to the perennial ryegrass reference than more closely related species (Italian ryegrass and meadow fescue). Once plastome reads have been filtered, the de novo assembly process is complicated by the presence of the IR regions, which are unable to be independently assembled due to sequence identity. PCR amplification was consequently required to verify the presence and precise boundaries of the IRs. Hence, although shallow sequencing of total DNA template provides a rapid and cost-effective approach for plastome assembly, PCR and first-generation sequencing are still necessary to close gaps within the assembly and, at the very least, provide a level of quality control.
Comparison of Lolium-Festuca plastomes
Comparative analysis revealed that the plastomes generated in this study are highly similar in terms of organization and sequence identity, with major differences being attributable to divergence within intergenic regions. In particular, the F. ovina plastome contains a larger number of deletions within the intergenic regions, in comparison with those of the other Lolium-Festuca species. This degree of differentiation correlates with its taxonomic position because F. ovina is the only fine-leaved Festuca species to be selected in this study. Another variable region between the plastomes of the targeted species is within the IR boundary. The IR region has expanded to include a portion of the ndhH gene, in common with other species of the Pooideae subfamily (Ogihara et al. 2000; Saski et al. 2007). The extent of this expansion, however, differs between species, such that the duplicated ndhH coding region ranges in size from a region specifying 32 amino acids in wheat, to 69 in barley, with the Lolium-Festuca species possessing 60−67 amino acids.
The phylogenomic dendrogram generated using whole plastome sequence is congruent with other published studies that are based on sequences of the trnL-trnF spacer region, and the matK gene (Catalán et al. 2004; Hand et al. 2010; Inda et al. 2008). However, the level of divergence between the tall fescue plastome and those of other Lolium-Festuca species as observed through the VISTA plot was an unexpected result. Studies of phylogeny based on DNA sequence, molecular genetic marker polymorphism, and morphological variation have consistently positioned tall fescue within the Schedonorus subgenus of Festuca, and hence more closely related to meadow fescue and the Lolium species, than to F. altissima and F. ovina (Charmet et al. 1997; Clayton and Renvoize 1986; Torrecilla and Catalán 2002; Xu and Sleper 1994). It is possible that this incongruence is related to the quality of the tall fescue plastome sequence, a proposition further supported by observation of many single nucleotide indels within the rbcL–psaI region, compared with other Poaceae species. At the time of publication of the tall fescue plastome sequence, the magnitude of divergence was not obvious, but the comparisons made in the present study reveal clear inconsistencies. To determine whether the unexpected sequence dissimilarity is a byproduct of the sequencing strategy or a genuine evolutionary anomaly, additional plastome sequences should be generated from multiple tall fescue individuals. Given the relative ease with which plastomes can now be sequenced and assembled, as demonstrated here, such resequencing studies are a highly feasible future objective.
Comparative plastome analysis within the Poaceae family
Plastomes of the Lolium-Festuca species possess known grass-specific structural alterations in that each contains the sequence insertion within rpoC2 but has lost the ORF of ycf2 within the IR, and an intron within rpoC1. Each of these features appears to be common throughout the Poaceae, although the plastome of the grass species Anomochloa marantoidea Brogn. still contains the rpoC1 intron (Morris and Duvall 2010), suggesting that loss of this intron is a more recent evolutionary event. The region of the plastome between rbcL and psaI has been identified as a “mutational hot-spot” and displays a greater level of interspecies variation than the other grass-specific alterations. One particular feature within this region is the presence of a pseudogene resembling rpl23, which has presumably originated after a translocation event, involving insertion of part of the IR into this region of the LSC (Shimada and Sugiura 1989). Studies focused on the presence of this pseudogene have previously indicated variability within the Lolium and Festuca genera. The rpl23′ pseudogene has been reported to be present in tall fescue and F. ovina but absent in the plastomes of perennial ryegrass and F. rubra (Katayama and Ogihara 1996). This result was obtained using Southern hybridization and was also subsequently achieved through BLAST analysis of the sugarcane rpl23′ sequence to the perennial ryegrass and tall fescue plastome sequences (Morris and Duvall 2010). The generation of complete plastome sequences in the present study, however, has revealed two deletions common to all included Lolium-Festuca species compared with other Poaceae species, which have effectively removed the majority of the rpl23′ pseudogene. Based on the available data, it is possible to propose that these deletions occurred at a point predating the origin of the Festuca genus, and hence no Lolium-Festuca species are likely to contain rpl23′. Conflicting previous results with respect to presence of the rpl23′ pseudogene in Festuca and Lolium species are therefore possibly due to an inadequate level of resolution associated with the Southern hybridization analyses, due to positive signals arising from residual homology with partial remnants of rpl23′ for both tall fescue and F. ovina.
Similarly, a more ancient deletion event that is possibly common to the entire Pooideae subfamily has occurred within the region in which accD pseudogenes have been predicted. The timing of this 173-bp deletion event is more difficult to determine, as it is nested within much larger deleted regions of both the wheat and B. distachyon plastomes. A more recent deletion has occurred within F. ovina, or an ancestor of this taxon, which has removed an even larger component of the accD pseudogene.
Alignment of the rbcL–psaI region for selected Poaceae family taxa has revealed a number of other deletions that appear to be common to particular subfamilies (such as the c. 1-kb deletion within Panicoideae species maize and sorghum) and others that are diagnostic for either whole tribes, genera or individual species. To further understand the evolutionary timing of these mutational events, comparison of a larger number of species is required. An apparent prediction of this analysis, however, is that the variable presence of the rpl23′ and accD pseudogenes is the product of a number of independent mutation events. Consequently, the sole use of the presence of these genes as a phylogenetic feature will not necessarily generate genuine species relationships, and should therefore be avoided.
Future applications
Apart from the obvious significance for genome evolutionary and phylogenetic studies, the complete plastome sequences generated in this study are anticipated to be valuable in the field of plant genetic modification. Generation of transformed plants through chloroplast-directed genetic engineering may provide benefits that include high levels of transgene expression, an apparent lack of associated gene silencing, and transgene containment due to restriction to the maternal line (and hence absence of pollen-mediated dispersal) (Han et al. 2008). Consequently, plastome-mediated transformation has been successfully used to improve resistance or tolerance to herbicides, insect predation, disease, drought and salt stress (Degray et al. 2001; Kota et al. 1999; Lee et al. 2003; Morris and Duvall 2010; Ruhlman et al. 2010; Tu et al. 2010). Within the Lolium-Festuca complex, transgenic approaches have been implemented in the agriculturally important species perennial ryegrass, Italian ryegrass, tall fescue and meadow fescue, with the aim of improving traits such as abiotic stress tolerance (Cao et al. 2009; Han et al. 2008), digestibility (Chen et al. 2004; Tu et al. 2010), resistance to fungal diseases (Dong et al. 2007; Takahashi et al. 2005) and allergenicity (Bhalla et al. 2001; Petrovska et al. 2005). Although the transformation approaches used for Lolium-Festuca species have not yet involved plastid-directed modification engineering, the availability of plastome sequences from each of these species, as described in this study, now allows for this to be a future possibility.
Ideally, development of transformation techniques using plastome-directed vectors will use species-specific plastome sequence data, rather than relying upon heterologous information. A number of studies have correlated reduced transformation efficiency with decreased sequence identity between flanking sequence within the transformation vector, and the target plastome (Degray et al. 2001; Nguyen et al. 2005; Ruhlman et al. 2010). However, variation between target plastome sites makes it difficult to identify a universal integration site. This effect has been demonstrated for the grass family, such that identical intergenic regions failed to be identified between rice, wheat, barley, and creeping bentgrass despite their close evolutionary affinities (Saski et al. 2007). Consequently, efficient plastid transformation is dependent on parallel efforts in both transformation techniques and plastome sequencing (Clarke et al. 2011). To date, the most widely used site within the plastome for transgene integration has been the transcriptionally active trnI-A intergenic region within the IR, resulting in the highest reported levels of expression (De Cosa et al. 2001; Verma and Daniell 2007). This intergenic region is identical among the six Lolium-Festuca species analyzed in this study and so may prove highly effective for chloroplast-mediated transformation of Lolium-Festuca grasses.
Supplementary Material
Acknowledgments
We thank Steve Petrovski and Sami Hakim for processing the samples on the HiSeq2000 instrument and Matthew Hayden for developing and sharing the PERL script. This research was funded by the Victorian Department of Primary Industries and the Dairy Futures Cooperative Research Centre. Melanie Hand was the recipient of an Australian Postgraduate Award.
Footnotes
Communicating editor: D. Zamir
Literature Cited
- Bhalla P. L., Swoboda I., Singh M. B., 2001. Reduction in allergenicity of grass pollen by genetic engineering. Int. Arch. Allergy Immunol. 124: 51–54 [DOI] [PubMed] [Google Scholar]
- Boffey S. A., Leech R. M., 1982. Chloroplast DNA levels and the control of chloroplast division in light-grown wheat leaves. Plant Physiol. 69: 1387–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M., Do C. B., Cooper G. M., Kim M. F., Davydov E., et al. , 2003. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13: 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke S. V., Grennan C. P., Duvall M. R., 2012. Plastome sequences of two New World bamboos-Arundinaria gigantea and Cryptochloa strictiflora (Poaceae)-extend phylogenomic understanding of Bambusoideae. Am. J. Bot. 99: 1951–1961 [DOI] [PubMed] [Google Scholar]
- Cahoon A. B., Sharpe R. M., Mysayphonh C., Thompson E. J., Ward A. D., et al. , 2010. The complete chloroplast genome of tall fescue (Lolium arundinaceum; Poaceae) and comparison of whole plastomes from the family Poaceae. Am. J. Bot. 97: 49–58 [DOI] [PubMed] [Google Scholar]
- Cao Y. J., Wei Q., Liao Y., Song H. L., Li X., et al. , 2009. Ectopic overexpression of AtHDG11 in tall fescue resulted in enhanced tolerance to drought and salt stress. Plant Cell Rep. 28: 579–588 [DOI] [PubMed] [Google Scholar]
- Catalán P., Torrecilla P., Rodríguez J. Á. L., Olmstead R. G., 2004. Phylogeny of the festucoid grasses of subtribe Loliinae and allies (Poeae, Pooideae) inferred from ITS and trnL-F sequences. Mol. Phylogenet. Evol. 31: 517–541 [DOI] [PubMed] [Google Scholar]
- Charmet G., Ravel C., Balfourier F., 1997. Phylogenetic analysis in the Festuca-Lolium complex using molecular markers and ITS rDNA. Theor. Appl. Genet. 94: 1038–1046 [Google Scholar]
- Chen L., Auh C.-K., Dowling P., Bell J., Lehmann D., et al. , 2004. Transgenic down-regulation of caffeic acid O-methyltransferase (COMT) led to improved digestibility in tall fescue (Festuca arundinacea). Funct. Plant Biol. 31: 235–245 [DOI] [PubMed] [Google Scholar]
- Chumley T. W., Palmer J. D., Mower J. P., Fourcade H. M., Calie P. J., et al. , 2006. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 23: 2175–2190 [DOI] [PubMed] [Google Scholar]
- Clarke J., Daniell H., Nugent J., 2011. Chloroplast biotechnology, genomics and evolution: current status, challenges and future directions. Plant Mol. Biol. 76: 207–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton W., Renvoize S., 1986. Genera Graminum, Kew Publishing, London [Google Scholar]
- Darbyshire S., 1993. Realignment of Festuca subgenus Schedonorus with the genus Lolium (Poaceae). Novon 3: 239–243 [Google Scholar]
- De Cosa B., Moar W., Lee S. B., Miller M., Daniell H., 2001. Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat. Biotechnol. 19: 71–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degray G., Rajasekaran K., Smith F., Sanford J., Daniell H., 2001. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol. 127: 852–862 [PMC free article] [PubMed] [Google Scholar]
- Diekmann K., Hodkinson T. R., Wolfe K. H., Van Den Bekerom R., Dix P. J., et al. , 2009. Complete chloroplast genome sequence of a major allogamous forage species, perennial ryegrass (Lolium perenne L.). DNA Res. 16: 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Tredway L. P., Shew H. D., Wang G.-L., Sivamani E., et al. , 2007. Resistance of transgenic tall fescue to two major fungal diseases. Plant Sci. 173: 501–509 [Google Scholar]
- Downie S. R., Katz-Downie D. S., Wolfe K. H., Calie P. J., Palmer J. D., 1994. Structure and evolution of the largest chloroplast gene (ORF2280): internal plasticity and multiple gene loss during angiosperm evolution. Curr. Genet. 25: 367–378 [DOI] [PubMed] [Google Scholar]
- Egli T., Goto M., Schmidt D., 1975. Bacterial wilt, a new forage grass disease. J. Phytopathol. 82: 111–121 [Google Scholar]
- Franche C., Lindström K., Elmerich C., 2009. Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants. Plant Soil 321: 35–59 [Google Scholar]
- Frazer K. A., Pachter L., Poliakov A., Rubin E. M., Dubchak I., 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32: W273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Su Y.-J., Wang T., 2010. Plastid genome sequencing, comparative genomics, and phylogenomics: current status and prospects. J. Syst. Evol. 48: 77–93 [Google Scholar]
- Han L., Li X., Liu J., Zeng H., 2008. Drought-tolerant transgenic perennial ryegrass (Lolium perenne L.) obtained via particle bombardment gene transformation of CBF3/DREB1A gene. Acta Hortic. 783: 273–282 [Google Scholar]
- Hand M. L., Cogan N. O. I., Stewart A. V., Forster J. W., 2010. Evolutionary history of tall fescue morphotypes inferred from molecular phylogenetics of the Lolium-Festuca species complex. BMC Evol. Biol. 10: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M., Meyer G., Vandergon T., Vandergon V., 2013. Loss of the acetyl-coA carboxylase (accD) gene in Poales. Plant Mol. Biol. Rep. 31: 21–31 [Google Scholar]
- Hiratsuka J., Shimada H., Whittier R., Ishibashi T., Sakamoto M., et al. , 1989. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 217: 185–194 [DOI] [PubMed] [Google Scholar]
- Hollingsworth P. M., Forrest L. L., Spouge J. L., Hajibabaei M., Ratnasingham S., et al. , 2009. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 106: 12794–12797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inda L. A., Segarra-Moragues J. G., Müller J., Peterson P. M., Catalán P., 2008. Dated historical biogeography of the temperate Loliinae (Poaceae, Pooideae) grasses in the northern and southern hemispheres. Mol. Phylogenet. Evol. 46: 932–957 [DOI] [PubMed] [Google Scholar]
- Katayama H., Ogihara Y., 1996. Phylogenetic affinities of the grasses to other monocots as revealed by molecular analysis of chloroplast DNA. Curr. Genet. 29: 572–581 [DOI] [PubMed] [Google Scholar]
- Kota M., Daniell H., Varma S., Garczynski S. F., Gould F., et al. , 1999. Overexpression of the Bacillus thuringiensis (Bt) Cry2Aa2 protein in chloroplasts confers resistance to plants against susceptible and Bt-resistant insects. Proc. Natl. Acad. Sci. USA 96: 1840–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. B., Kwon H. B., Kwon S. J., Park S. C., Jeong M. J., et al. , 2003. Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol. Breed. 11: 1–13 [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Zhu H., Ruan J., Qian W., Fang X., et al. , 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20: 265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride M. J., Xie G., Martens E. C., Lapidus A., Henrissat B., et al. , 2009. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl. Environ. Microbiol. 75: 6864–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I., Bayer M., Cardle L., Shaw P., Stephen G., et al. , 2010. Tablet—next generation sequence assembly visualization. Bioinformatics 26: 401–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris L. M., Duvall M. R., 2010. The chloroplast genome of Anomochloa marantoidea (Anomochlooideae; Poaceae) comprises a mixture of grass-like and unique features. Am. J. Bot. 97: 620–627 [DOI] [PubMed] [Google Scholar]
- Nguyen T. T., Nugent G., Cardi T., Dix P. J., 2005. Generation of homoplasmic plastid transformants of a commercial cultivar of potato (Solanum tuberosum L.). Plant Sci. 168: 1495–1500 [Google Scholar]
- Nock C. J., Waters D. L. E., Edwards M. A., Bowen S. G., Rice N., et al. , 2011. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol. J. 9: 328–333 [DOI] [PubMed] [Google Scholar]
- Ogihara Y., Terachi T., Sasakuma T., 1988. Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc. Natl. Acad. Sci. USA 85: 8573–8577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara Y., Terachi T., Sasakuma T., 1992. Structural analysis of length mutations in a hot-spot region of wheat chloroplast DNAs. Curr. Genet. 22: 251–258 [DOI] [PubMed] [Google Scholar]
- Ogihara Y., Isono K., Kojima T., Endo A., Hanaoka M., et al. , 2000. Chinese spring wheat (Triticum aestivum L.) chloroplast genome: complete sequence and contig clones. Plant Mol. Biol. Rep. 18: 243–253 [Google Scholar]
- Okon Y., Labandera-Gonzalez C. A., 1994. Agronomic applications of azospirillum: an evaluation of 20 years worldwide field inoculation. Soil Biol. Biochem. 26: 1591–1601 [Google Scholar]
- Petrovska N., Wu X., Donato R., Wang Z., Ong E.-K., et al. , 2005. Transgenic ryegrasses (Lolium spp.) with down-regulation of main pollen allergens. Mol. Breed. 14: 489–501 [Google Scholar]
- Ruhlman T., Verma D., Samson N., Daniell H., 2010. The role of heterologous chloroplast sequence elements in transgene integration and expression. Plant Physiol. 152: 2088–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saski C., Lee S.-B., Fjellheim S., Guda C., Jansen R., et al. , 2007. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera and comparative analyses with other grass genomes. Theor. Appl. Genet. 115: 571–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H., Sugiura M., 1989. Pseudogenes and short repeated sequences in the rice chloroplast genome. Curr. Genet. 16: 293–301 [DOI] [PubMed] [Google Scholar]
- Shimada H., Fukuta M., Ishikawa M., Sugiura M., 1990. Rice chloroplast RNA polymerase genes: the absence of an intron in rpoC1 and the presence of an extra sequence in rpoC2. Mol. Gen. Genet. 221: 395–402 [DOI] [PubMed] [Google Scholar]
- Soreng R. J., Terrell E. E., 1997. Taxonomic notes on Schedonorus, a segregate genus from Festuca or Lolium, with a new nothogenus, x Schedololium, and new combinations. Phytologia 83: 85–88 [Google Scholar]
- Stanier R. Y., 1947. Studies on nonfruiting myxobacteria. J. Bacteriol. 53: 297–315 [DOI] [PubMed] [Google Scholar]
- Takahashi W., Fujimori M., Miura Y., Komatsu T., Nishizawa Y., et al. , 2005. Increased resistance to crown rust disease in transgenic Italian ryegrass (Lolium multiflorum Lam.) expressing the rice chitinase gene. Plant Cell Rep. 23: 811–818 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., et al. , 2011. 20011 MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakami S., Matsumura Y., Kurita K., Kanamori H., Katayose Y., et al. , 2012. Complete sequence of the chloroplast genome from pear (Pyrus pyrifolia): genome structure and comparative analysis. Tree Genet. Genomes 8: 841–854 [Google Scholar]
- Torrecilla P., Catalán P., 2002. Phylogeny of broad-leaved and fine-leaved Festuca lineages (Poaceae) based on nuclear ITS sequences. Syst. Bot. 27: 241–251 [Google Scholar]
- Tu Y., Rochfort S., Liu Z., Ran Y., Griffith M., et al. , 2010. Functional analyses of caffeic acid O-methyltransferase and Cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne). Plant Cell 22: 3357–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauterin L., Hoste B., Kersters K., Swings J., 1995. Reclassification of Xanthomonas. Int. J. Syst. Bacteriol. 45: 472–489 [Google Scholar]
- Verma D., Daniell H., 2007. Chloroplast vector systems for biotechnology applications. Plant Physiol. 145: 1129–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Messing J., 2011. High-throughput sequencing of three Lemnoideae (Duckweeds) chloroplast genomes from total DNA. PLoS ONE 6: e24670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.-Q., Ge S., 2012. The phylogeny of the BEP clade is grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol. Phylogenet. Evol. 62: 573–578 [DOI] [PubMed] [Google Scholar]
- Wyman S. K., Jansen R. K., Boore J. L., 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20: 3252–3255 [DOI] [PubMed] [Google Scholar]
- Xu W. W., Sleper D. A., 1994. Phylogeny of tall fescue and related species using RFLPs. Theor. Appl. Genet. 88: 685–690 [DOI] [PubMed] [Google Scholar]
- Yang M., Zhang X., Liu G., Yin Y., Chen K., et al. , 2010. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.). PLoS ONE 5: e12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Zhang X., Hu S., Yu J., 2011a An efficient procedure for plant organellar genome assembly, based on whole genome data from the 454 GS FLX sequencing platform. Plant Methods 7: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-J., Ma P.-F., Li D.-Z., 2011b High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). PLoS ONE 6: e20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Liere K., Börner T., 2007. From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J. 50: 710–722 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.