Abstract
Trans-lesion DNA polymerases (TLSPs) enable bypass of DNA lesions during replication and are also induced under stress conditions. Being only weakly dependent on their template during replication, TLSPs introduce mutations into DNA. The low processivity of these enzymes ensures that they fall off their template after a few bases are synthesized and are then replaced by the more accurate replicative polymerase. We find that the three TLSPs of budding yeast Saccharomyces cerevisiae Rev1, PolZeta (Rev3 and Rev7), and Rad30 are induced during meiosis at a time when DNA double-strand breaks (DSBs) are formed and homologous chromosomes recombine. Strains deleted for one or any combination of the three TLSPs undergo normal meiosis. However, in the triple-deletion mutant, there is a reduction in both allelic and ectopic recombination. We suggest that trans-lesion polymerases are involved in the processing of meiotic double-strand breaks that lead to mutations. In support of this notion, we report significant yeast two-hybrid (Y2H) associations in meiosis-arrested cells between the TLSPs and DSB proteins Rev1-Spo11, Rev1-Mei4, and Rev7-Rec114, as well as between Rev1 and Rad30. We suggest that the involvement of TLSPs in processing of meiotic DSBs could be responsible for the considerably higher frequency of mutations reported during meiosis compared with that found in mitotically dividing cells, and therefore may contribute to faster evolutionary divergence than previously assumed.
Keywords: meiosis, trans-lesion synthesis polymerases, DSB processing, DNA repair, recombination
Maintenance and faithful propagation of genetic information through replication, repair and recombination requires new DNA synthesis. To achieve accuracy and reliability, most DNA synthesis is performed by high-fidelity DNA polymerases, collectively called replicative DNA polymerases. However, other, low-fidelity DNA polymerases also exist, known as trans-lesion DNA polymerases (TLSPs). Unlike canonical DNA polymerases, these enzymes lack exonucleolytic proofreading activity and have the specialized ability to replicate across damaged DNA templates, a process known as trans-lesion synthesis (TLS) (reviewed by Friedberg et al. 2002). Nucleotide misincorporation opposite a damaged base during replication or repair results in mutation through base substitution, insertion, or deletion (Harfe and Jinks-Robertson 2000; Matsuda et al. 2000).
The necessity for TLSPs is apparent when the DNA is facing spontaneous or exogenous damage, which often blocks progression of replication. Under these circumstances, TLSPs are employed, allowing progression of DNA replication, often at the expense of an increased mutagenesis. TLSPs can also operate on undamaged DNA templates, manifesting higher error rates than replicative DNA polymerases (Friedberg et al. 2002; Matsuda et al. 2001).
In bacteria, trans-lesion DNA synthesis is well characterized during the cellular SOS-response to DNA damage. Upon damage, mutagenic SOS polymerases (polymerases IV, coded by DinB, and V, coded by UmuC/D) facilitate tolerance of the damage rather than physical removal of the lesion (Friedberg and Gerlach 1999). Orthologs of the bacterial TLSP genes were found in many eukaryotes (Ohmori et al. 2001), serving a broad spectrum of functions (Friedberg et al. 2001; Goodman 2002). In the budding yeast Saccharomyces cerevisiae, polymerase eta (PolEta) is encoded by RAD30, the deoxycytidyl-transferase Rev1 is encoded by the gene REV1, and polymerase zeta (PolZeta) is encoded by the genes REV3 and REV7 (catalytic and regulatory units, respectively). Recent findings have identified Pol31 and Pol32 (subunits of the eukaryotic replicative polymerase PolDelta) as two important subunits of PolZeta (Baranovskiy et al. 2012; Johnson et al. 2012; Makarova et al. 2012). PolZeta, being composed of 4 subunits, is the most complex TLSP to date. The fact that Pol31 and Pol32 may be associated with either PolDelta or PolZeta facilitates the switch between these two polymerases when the former encounters a lesion on the template. Well-established evidence connects yeast TLSPs to TLS during DNA replication: Rev1 dCMP insertion is a principal event during bypass of abasic sites in vivo (Gibbs and Lawrence 1995). PolZeta cooperates with Rev1 to accomplish TLS past a broad range of lesions that potentially block replication (Nelson et al. 2000). Rad30 is responsible mainly for bypassing cis-synT-T dimers (Gibbs et al. 2005; Washington et al. 2000). PolZeta and Rev1 physically interact, as shown by co-immunoprecipitation (Hirano and Sugimoto 2006), and this association enhances the efficiency of their TLS functions (Acharya et al. 2006). In mitotically dividing yeast cells, TLSPs are also involved in DNA synthesis associated with DNA double-strand breaks (DSBs) repair by homologous recombination (HR). HO-induced DSBs in mitotic cells were shown to lead to an increase in mutations at sites nearby (Strathern et al. 1995). PolZeta appears to be involved in the repair of at least some of these DSBs, as the frequency of HO-induced base substitution mutations was considerably lower in an isogenic rev3Δ strain (Holbeck and Strathern 1997). Indeed, yeast PolZeta and Rev1 were found to be associated with HO-induced DSBs, as shown by chromatin immunoprecipitation (Hirano and Sugimoto 2006). In mammals, several TLSP genes have been discovered (reviewed by Gan et al. 2008 and Stallons and Mcgregor 2010). These mutagenic proteins are induced during lymphocyte differentiation as part of the immune response (Gearhart and Wood 2001; Poltoratsky et al. 2001). These polymerases are believed to be involved in the repair of induced DSBs in hypermutation sites during B-cell and T-cell receptor maturation, thus amplifying the variability generated during lymphocyte maturation. In addition, several TLSP genes are highly expressed in mouse testis (Aoufouchi et al. 2000; Garcia-Diaz et al. 2000; Gerlach et al. 2000) and in human testis and ovary tissues (Masuda et al. 2001).
Meiosis is a specialized cell division with major events leading to the formation of haploid gametes and increased genetic diversity. Genetic variation is accomplished by recombination, gene conversion, and independent assortment of the aligned chromosomes. In meiosis, two temporally and functionally distinct processes require massive DNA synthesis: genome replication during meiotic S phase and DSBs repair in meiosis prophase I (the latter is also termed meiotic recombination-related DNA synthesis [MRDS]). DSBs are regularly induced throughout the genome at preferred sites during meiosis in budding and fission yeast cells (Cervantes et al. 2000; Keeney 2001; Zenvirth et al. 1992; Zenvirth and Simchen 2000) in mouse spermatocytes (Zenvirth et al. 2003) and presumably in other eukaryotes. In meiosis of S. cerevisiae, about 140–170 DSBs are generated in each cell (Buhler et al. 2007), and the repair of each break involves synthesis of 0.8–1.9 kb of new DNA (Terasawa et al. 2007). The identity of the DNA polymerases that are involved in meiotic DSB repair is largely unknown, although some evidence shows that the replicative PolDelta has an important role (Li et al. 2009; Maloisel et al. 2004). It has been reported that in mitotically dividing human cells, PolEta (Rad30) takes part in extending DNA from D loops of recombination intermediates (Mcilwraith et al. 2005). Rad30 was also involved in DSB repair during chicken IgV diversification (Kawamoto et al. 2005). Is it possible that the yeast TLSPs are also involved in the repair of meiotic DSBs, thus leading to mutation accumulation and adding a new source of genetic variation?
Five decades ago, Magni and Von Borstel (1962) and Magni (1963) reported a marked elevation in mutation frequency during yeast meiosis. They pointed out that this observed hypermutability is associated with nearby recombinational exchanges. Our study suggests that TLSPs are involved in the process that generates mutations during meiosis by taking part in the repair of meiotic DSBs and perhaps, thus, leading to mutations and to increased genetic diversity.
In this study, we showed that the three yeast TLSPs (Rev1, PolZeta, and Rad30) are induced in meiosis at prophase I, clearly after the regular meiotic DNA replication. Expression of TLSPs appears to occur at the same time as meiotic recombination. The absence of all three TLSPs leads to reductions in allelic recombination and ectopic gene conversion events. By using extensive yeast two-hybrid (Y2H) tests, we detected meiosis-specific protein associations between Rev1 and proteins responsible for DSB formation (Spo11 and Mei4), between Rev7 and Rec114, and between Rev1 and Rad30. These results suggest that TLSPs are involved in meiotic DSB processing and in recombination, possibly during recombinational repair, thus perhaps contributing to the elevated frequency of mutations during meiosis.
Materials and Methods
Yeast strains, deletions, and epitope tagging
All strains were of SK1 genetic background. The yeast strains used in this study are listed in Supporting Information, Table S1. Yeast strains were maintained according to standard techniques and media used were YPD, YPA, and SPM, as described previously (Kassir and Simchen 1991). −His and −Ura media are complete media lacking histidine and uracil, respectively; -Trp-Leu is complete medium lacking tryptophan and leucine (Rose et al. 1990). Deletion mutants deleted for full-length open reading frames (ORFs) were generated in our strains by PCR-based gene disruption (using the relevant PCR-amplified cassettes from the kanMX4 disruption strains library [Reid et al. 2002]), and standard molecular biology techniques (Guthrie and Fink 1991). All strain manipulations were verified by PCR. Epitope-tagged TLSPs were constructed as follows. Cassettes containing the 13-Myc or 3-HA sequence were amplified from the pFA6a-13Myc-TRP1 and pFA6a-kanMX6-PGAL1-3HA plasmids, respectively (Longtine et al. 1998). Specific 72-mer primers were designed to link the 13-Myc or the 3-HA sequence to the C terminus of the relevant TLSP gene (and protein). Cassettes were transformed into the haploid MATa strain DAO20-2, and transformants were selected for their ability to grow in the absence of tryptophan (in the case of 13-Myc) or with the addition of G418 (for 3-HA). Yeast strains were generated in which the tags had been integrated into the chromosomal copy of the relevant TLSP gene under the control of its native promoter. For all fusion constructs, integrations at the appropriate genomic loci were confirmed by PCR, and proper expression of the fusion proteins with the expected molecular weights was assayed by immunoblotting with anti-Myc (Roche) or anti-HA (Santa Cruz Biotechnology) antibodies. The tagged strains were then mated to a haploid, MATα, strain (DAO19-1). Following tetrad dissection and mating of progeny, diploid strains homozygous for the tagged fusions were obtained to provide stronger signals.
Protein extraction and Western blot analysis
For Western blot analysis, yeast cells harboring carboxy-terminal epitope-tagged proteins carrying either myc or HA tags (the proteins Cdc5, Aco1, and β-actin did not carry a tag and were detected with appropriate direct antibodies) were grown for 24 hr in liquid YPD-rich medium and, upon saturation, were suspended in sporulation medium (SPM) at a titer of 2 × 107 cells/ml and shaken vigorously. At given times, samples were collected, and denatured whole-cell extracts were prepared using a trichloroacetic acid (TCA) procedure as follows: 5 ml of 2 × 107 cells/ml were washed twice in ice-cold water and suspended to a final volume of 1 ml. Cells were then incubated with 150 µl of suspension buffer (925 µl of 2M NaOH plus 75 µl of 2-mercaptoethanol) on ice for 10 min and lysed in 150 µl of 55% TCA for 15 min on ice. Protein pellets were obtained by centrifugation at 14,000 rpm for 10 min at 4°C. Pellets were resuspended in 60 µl of HU buffer (8M urea, 5% SDS, 200 mM Tris, pH 6.8, 1 mM EDTA, and 1.5% DTT, with bromophenol blue as a coloring agent and pH indicator) plus 3 µl of 2M Tris (pH 8.0). Proteins were brought to denaturation by 10-min incubation at 65°C.
Protein extract samples (∼40 µl) were boiled, separated on sodium dodecyl-sulfate 8% polyacrylamide gels, and transferred to nitrocellulose membranes (BioTrace). Membranes were probed with the relevant primary antibodies in the appropriate dilution, as follows: mouse anti-Myc (MMS-150R; Roche) diluted 1:1000; rabbit anti-HA (SC-805; Santa Cruz Biotechnology) diluted 1:200; rabbit anti-AcoI (made by Dr. Ophry Pines’ laboratory, Hebrew University School of Medicine) diluted 1:200; and rabbit anti-β-actin (1854-1; Epitomics) diluted 1:500. Bound primary antibodies were detected using HRP-conjugated goat anti-mouse IgG (SC-2005; Santa Cruz Biotechnology) diluted 1:10,000 and goat anti-rabbit IgG antibody (111-035-003; Jackson IR Laboratory) diluted 1:10,000. Protein expression profiles were quantified with ImageJ software (Java-based image processing program).
To determine sporulation efficiencies (which were usually ∼80%), a sample was withdrawn from each culture 24 hr after suspension in SPM and examined by light microscopy.
Meiosis time course experiments, allelic recombination, and ectopic gene conversion frequencies
The strains used were heteroallelic for the his4-X and his4-B mutations and could yield His+ prototrophs (on –His medium) through allelic recombination. The strains were also homozygous for a deletion of URA3 at its native position on chromosome V (ura3Δ) and for an additional truncated copy of URA3 1400–1500 bp downstream of its original, native position. The truncated insert starts at the 115th bp of URA3 and also contains the adjacent “tail” downstream of URA3, of more than 700 bp (Figure S1). The existence of the truncated copy of URA3 on chromosome V downstream of the original URA3 was verified by pulsed-field gel electrophoresis (clamped homogeneous electrical field [CHEF];Bio-Rad) and by PCR.
Originally our strains were also heterozygous for another, functional copy of URA3 that was present on his4-X-bearing chromosome III. This copy was mutated to contain various point mutations. One of these, ura3-T360G (T to G at position 360), was introduced into all the strains used in these recombination/gene conversion experiments. It conferred uracil auxotrophy (Ura−) on strains that carried it. This phenotype can be reverted to prototrophy (Ura+) by ectopic gene conversion, based on interaction with the truncated copy of URA3 on chromosome V, and can be identified as colonies growing on −Ura medium.
In each time course experiment, cell cultures of two or more strains were each grown overnight in 3 ml of liquid YPD and then resuspended in 300 ml of liquid YPA at a dilution of 1:600 and vigorously shaken at 30°C for ∼20–24 hr to reach a titer of ∼107 cells/ml. Cells were then washed once in water and resuspended in 300 ml of liquid SPM with vigorous shaking. At 2-hr intervals, cell samples were spread (at appropriate dilutions) on YPD plates and on −His and −Ura plates and incubated for 2–3 days at 30°C to obtain colonies, from which the frequencies of allelic and ectopic gene conversions were calculated. The number of colonies appearing on selective plates from time zero (i.e., on −His or −Ura plates) was subtracted from the numbers obtained at each time point during meiosis, as the former reflected events that had occurred in the mitotic divisions prior to meiosis.
Yeast-two-hybrid analysis and plasmids
Yeast-two-hybrid (Y2H proteins were fused to the transcription-activating domain of Gal4 (Gal4AD) or to the DNA-binding domain of bacterial LexA (LexA-BD) protein. These fusions were constructed as previously described (Arora et al. 2004). Spo11, Mei4, Rec104, Rec114, and Rad50 Y2H plasmids were a gift from S. Keeney, Memorial Sloan-Kettering Cancer Center (Arora et al. 2004); Rad51, Rad52, Rad54, and Rad57 Y2H plasmids were a gift from M. E. Dresser, Oklahoma University (Dresser et al. 1997). TLSPs Y2H fusion proteins (Rev1, Rad30, and Rev7) and also a Dmc1 fusion protein were constructed in our laboratory by cloning PCR-generated fragments into either the Gal4AD-bearing plasmid (pACT2) or the LexA-BD-bearing plasmid (pCA1). The Y2H reporter strains 661 and 662, used for the two-hybrid assay, are of the SK1 background and contain Escherichia coli lacZ preceded by two LexA binding sites integrated at the URA3 locus. The strains also carry the ndt80Δ mutation, which causes arrest of cells at prophase of meiosis I (Xu et al. 1995). The Y2H fusion constructs were introduced individually into haploid reporter strains by lithium-acetate transformation (Gietz et al. 1995), and subsequently, the strains were mated in 33 different combinations (thus, every diploid strain contained two different assayed fusion proteins). Cultures for Y2H assays were grown in liquid-selective medium lacking tryptophan and leucine (-Trp-Leu) for 8 hr at 30°C. Cells were then washed and resuspended in either sporulation medium (SPM) or in YPD and incubated for an additional 14 hr at 30°C with vigorous aeration. Cultures were assayed for LacZ expression according to standard protocols (Clontech). Briefly, cells were centrifuged and resuspended in Z buffer, pH 7 (10 mM KCl; 1mM MgSO4; 60 mM Na2HPO4; 40mM NaH2PO4). Cells were divided into two samples of 100 μl each (duplications) and lysed by freezing and thawing cycles. The lysate was mixed with 160 μl of 4 mg/ml ortho-nitrophenyl-β-galactoside (ONPG), used as a substrate for β-galactosidase (β-gal), and incubated at 37°C until development of a yellow color. Once the yellow color appeared, the reaction was stopped by adding 0.4 ml of 1M Na2CO3. Cells were centrifuged, and a 1-ml aliquot was taken to measure absorbance levels at OD420 and OD600. One unit of β-gal hydrolyzes 1 μmol of ONPG per min per OD600. In a typical experiment, two plasmids with fusions to be tested for Y2H interaction were introduced into two haploids of opposite mating types, carrying LexA(op)-LacZ, which were then mated. The resulting diploid was grown for 8 hr in selective liquid medium (-Trp-Leu) until a titer of 2 × 107 cells/ml was reached, and cells were then washed twice in DDW. The culture was then divided into two parallel experiments: half the culture was suspended in liquid SPM and incubated for 14 hr at 30°C with vigorous shaking (the meiotic Y2H interaction experiment). The other half was suspended in fresh selective medium (-Trp-Leu) and grown for 14 hr at 30°C, also shaken vigorously (the mitotic cells Y2H interaction experiment). After 14 hr, ONPG was added to washed cells of the two cultures (see above), and cells were subsequently examined for the color appearance, representing the amount of β-gal activity units.
Control Y2H interactions:
Each diploid was tested together with its two negative controls: one control contained one Gal4AD fusion protein (on a plasmid) and the complementing “empty” plasmid (the LexA-BD vector), and the other control contained the LexA-BD fusion protein together with the first “empty” plasmid (the Gal4AD vector).
Positive controls:
Strong mitotic Y2H interaction was generated by using the same strain harboring plasmids coding for the Mei4 and Rec114 fusion proteins, whereas a strong meiosis-specific Y2H interaction was generated in a diploid harboring plasmids coding for the Spo11 and Rec104 fusion proteins (Arora et al. 2004). Duplicate tests were carried out for each Y2H combination and the two negative controls in every experiment. Every set of Y2H experiments (of a particular combination) was subject to a statistical two-way analysis of variance under the most stringent statistical conditions: the difference between a particular set of Y2H experiments (one Y2H combination) and its two negative controls was tested against their interaction with the 3 or 4 repeated experiments, rather than against the smaller duplicate error; next, the difference between the Y2H value and the higher negative control value was tested, also against the experiment × treatment interaction. The analysis of variance was performed using JMP7 software.
Results
TLSPs are known to help overcome obstacles to replication (Rattray and Strathern 2003). Extensive studies were made of the requirement of TLSPs during DNA replication in mitotically dividing cells (see for example Hirano and Sugimoto 2006). However, much less is known about the involvement of TLSPs in meiosis. In budding yeast, there are three TLSPs, PolZeta (Rev3-Rev7), Rev1, and Rad30, and the role(s) of these proteins in meiosis was hereby studied.
Expression pattern of TLSPs during meiosis
To examine the expression patterns of TLSP genes in meiosis, we used available whole-genome expression microarrays data obtained during yeast sporulation (Chu et al. 1998; Friedlander et al. 2006; Primig et al. 2000). We found that REV3 and REV7 (encoding the catalytic and regulatory subunits of PolZeta, respectively) were significantly induced during meiosis, whereas the transcript levels of REV1 and RAD30 remained low (Figure 1A).
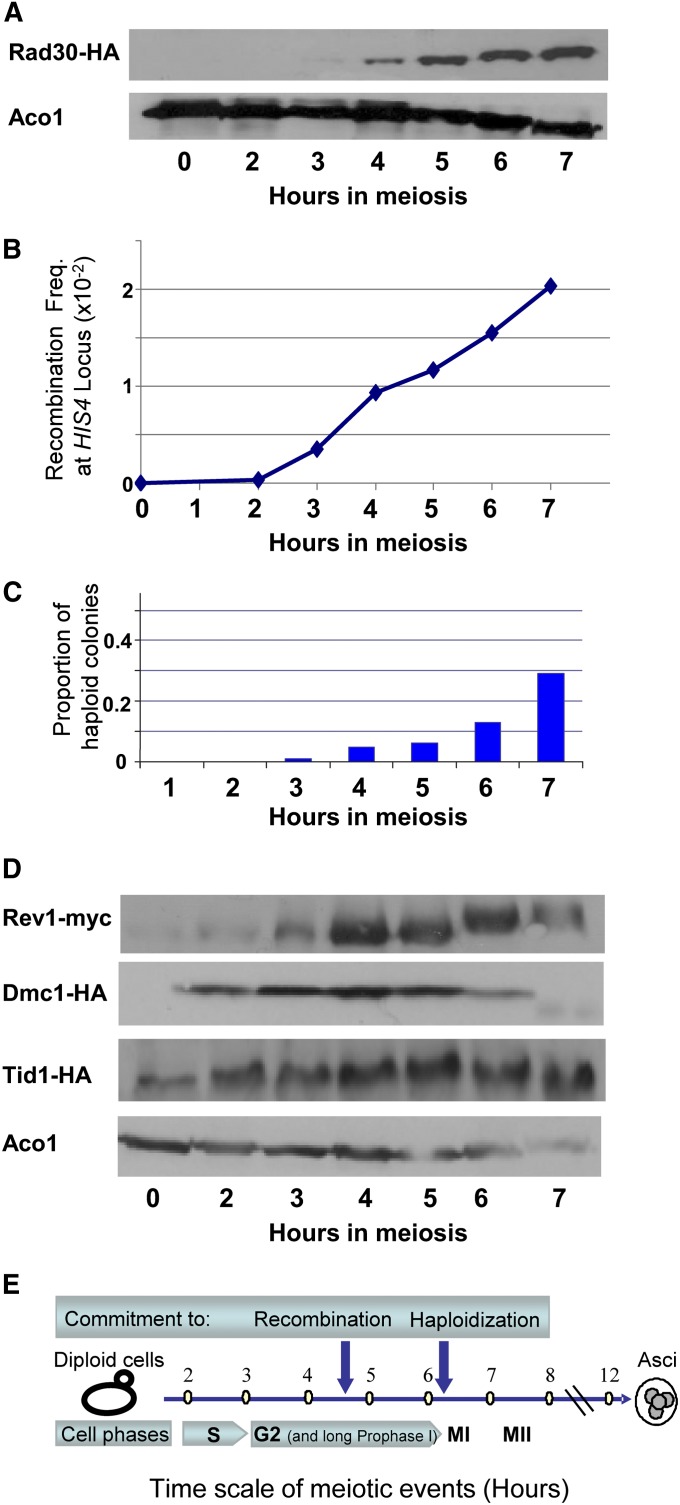
Figure 1 .
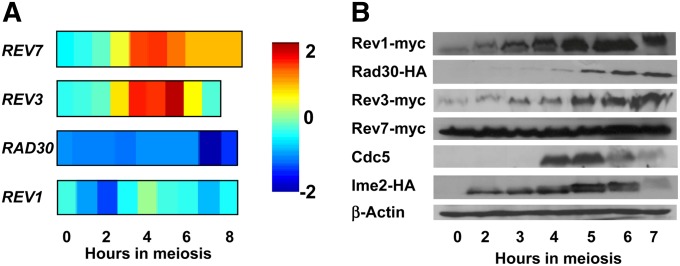
Expression profiles of TLSP genes during yeast meiosis. (A) Levels of RNA of four TLSP genes are shown, based on RNA hybridized to yeast ORF microarrays (Friedlander et al. 2006). Intensity is relative to average abundance levels of the given mRNA. (B) Western blot analysis of TLSP proteins during meiosis. Cells expressing epitope-tagged TLSPs were collected for Western blot analysis at the indicated times after transfer to sporulation conditions. Cdc5 and Ime2 expression peaks mark prophase I and meiotic S phase, respectively (see also Figure 2E). β-Actin was used as a load control.
To further investigate the expression of these genes at the protein level, we constructed strains with their carboxy-terminal epitope-tagged versions, expressed from their native promoters at the endogenous loci. Yeast SK1 cells harboring either Rev3-myc, Rev7-myc, Rev1-myc, or Rad30-HA (strains DAO119, DAO16-1, DAO110, and DAO178, respectively) were examined for expression of the tagged proteins during meiosis by Western blot analysis (Figure 1B). To correlate TLSP expression with meiotic stages, we monitored the levels of the proteins Ime2 and Cdc5 in a closely related strain, DAO212; Ime2 is known to be expressed during the meiotic S phase (Benjamin et al. 2003), and Cdc5 is upregulated in prophase I (Clyne et al. 2003). Sporulation levels were high (∼80%) in all strains tested.
The three trans-lesion DNA polymerases (Rev1, Rev3, and Rad30) were up-regulated during meiosis at the protein level, even though for RAD30 and REV1, we could not identify induction at the RNA level (Figure 1A). The level of Rev1 was approximately threefold increased at 3 hr after transfer to SPM, reached a peak after 4 hr, and decreased after 7 hr. The Rad30 protein was not apparent during early stages of meiosis, and its up-regulation was observed after 5 hr in SPM. This high expression level was maintained during the remaining course of the experiment, ∼14-fold higher than at time zero. Interestingly, the two PolZeta proteins Rev3 and Rev7 showed very different expression patterns. The level of Rev3 increased at 3 hr, and its expression mounted further (∼fourfold relative to that at time zero) between 5 and 7 hr in SPM. Rev7 was maintained at high level during the full course of meiosis, as well as under mitotic conditions (time-0 hr).
All three catalytic TLSP proteins reached maximal levels a few hours after DNA replication (meiotic S), which takes place under these conditions at ∼2 hr in meiosis (at the time of Ime2 induction [Figure 1B and [Benjamin et al. 2003]). Cdc5 expression marks the prophase I stage in yeast meiosis, which occurs in SK1 strains between 4 and 5 hr after transfer to SPM. We found that all three catalytic TLSP proteins showed elevation in their expression levels at this stage. This suggests that these proteins are involved in recombination rather than in the meiotic DNA synthesis phase.
To further investigate the timing and kinetics of TLSPs accumulation, we examined two hallmarks of meiosis, which are easily determined in return-to-growth assays, namely commitment to recombination and commitment to haploidization (Simchen 2009). For this experiment, we used a diploid SK1 strain (DAO178) bearing the Rad30-HA tag (Figure 2, A–C). Recombination commitment was determined at the HIS4 locus. This strain contains a his4::LEU2 insertion (Cao et al. 1990) consisting of prominent DSB sites and is heteroallelic at HIS4. The two nonreversible mutations his4-B and his4-X recombine frequently in meiosis to produce His+ progeny. Haploidization frequencies were measured by assessing the mating types of 100 individual colonies produced on YPD by cells spread at various time points along the time course of meiosis. At the indicated times, cell samples were collected for Western blot analysis to follow Rad30 expression and for plating (on −His and YPD media) to evaluate the frequency of His4+ allelic recombination and haploidy (Figure 2, A, B, and C, respectively). Rad30 up-regulation correlated with the significant rise in the number of recombinant His+ colonies and preceded the appearance of haploid colonies.
Figure 2 .
TLSP protein up-regulation correlates with prophase I events in meiosis. Aliquots of meiotic SK1 cells (strain DAO178) were taken at the indicated times and assayed: (A) Rad30p expression by Western blot analysis; Aco1 was used as a loading control. (B) His+ recombinants. Cells were plated on –His medium to produce colonies resulting from homologous recombination at HIS4. (C) Appropriate dilutions were plated on YPD medium, and colonies were assayed for ploidy by mating to mating-type testers (100 colonies per time point). (D) Meiotic cultures were assayed by Western blotting for levels of Rev1 and Dmc1 (strain DAO137) and Tid1 (strain DAO180) proteins. Western blotting time course analysis of these strains was carried out as described in Materials and Methods. The apparent shift in Rev1 band at 6–7 hr is probably the result of distortion of the gel, as it was not seen in four other meiosis time course experiments. (E) Schematic timeline of the major meiotic events during sporulation in strains of SK1 genetic background (see also Székvolgyi and Nicolas 2010).
As seen above, the expression of all three catalytic TLSPs was elevated considerably later than the time of meiotic DNA replication (represented by Ime2 induction) and occurred before commitment to haploidization (Figure 2E); this time corresponds to prophase I events such as DNA double-strand breakage and recombination (Keeney 2008; Padmore et al. 1991; Szekvolgyi and Nicolas 2010). Proper recombination–repair of DSBs requires many factors, among them the meiosis-specific recombinase Dmc1, which belongs to the RecA/Rad51 superfamily, and the recombinase accessory factor Tid1/Rdh54 (Shinohara et al. 2000). We wished to determine the timing of expression of these DSB repair proteins in relation to TLSP accumulation. Diploid Rev1-Myc tagged SK1strains bearing either Dmc1-HA or Tid1-HA (strains DAO137 and DAO180, respectively) were induced into meiosis. At the indicated intervals, cell samples were taken for protein extraction and blotting (Figure 2D). As we previously found (Figure 1B), here too, significant expression of Rev1 was observed at prophase I. As expected, the meiosis-specific protein Dmc1 was not expressed in mitotic cells (Figure 2D, time 0 in the Dmc1 blot). Its expression was observed 2 hr after transfer of the cells to SPM, reaching maximal levels between 3 and 5 hr in meiosis, at the same time as Rev1 accumulation (Figures 1B and 2D). Afterward, Dmc1 protein levels dropped remarkably and disappeared between 6 and 7 hr when cells were moving from prophase I into the meiotic chromosomal segregation (MI). Tid1 is known to function under mitotic conditions as well as in meiosis (Klein 1997). Indeed, Tid1 was expressed throughout our experiment, showing only a slight elevation at 4–5 hr in meiosis. The overall correlation in timing of protein increase between Rev1 and Dmc1 further supports the possible involvement of TLSPs in recombinational repair during meiotic DSB processing.
Effects of TLSPs deletion on allelic recombination and on ectopic gene conversion in meiosis
As the three TLSPs were clearly induced during meiosis and the timing of their expression suggested that they may have a role in recombination rather than in meiotic S phase, we further examined various aspects of recombination in strains in which the TLSP-encoding genes were deleted. We constructed diploid strains homozygous for deletions in one of the genes, REV1, REV3, or RAD30 (rev1Δ, rev3Δ, rad30Δ, respectively), double mutants (homozygous for deletions in any two of these TLSP genes), and the triple mutant rev1Δ rev3Δ rad30Δ. The control strain was the wild-type (WT) for the four TLSP-encoding genes. These strains were also heteroallelic for his4-X and his4-B and could yield His+ prototrophs through allelic recombination/gene conversion. Moreover, the strains were homozygous for a deletion of URA3 at its native position on chromosome V (ura3Δ) but contained an additional, truncated copy of URA3 1400–1500 bp downstream of this native position (see Materials and Methods and Figure S1, for more details). The presence of this truncated copy of URA3 enabled us to devise a test for ectopic gene conversion in addition to the allelic gene conversion at HIS4. Our diploid strains were originally also heterozygous for another functional copy of URA3 that was present on his4-X-bearing chromosome III. This copy was mutated to contain various point mutations. One of these, ura3-T360G, was introduced into all the strains used in these recombination/gene conversion experiments. It conferred uracil auxotrophy (Ura−) on strains that carried it, but this phenotype could be reverted to prototrophy (Ura+) by gene conversion resulting from interaction with the truncated copy of URA3 on chromosome V. The frequency of Ura+ prototrophs following meiosis in our WT strain was 6–7 × 10−5 cells, about 100-fold lower than that of His+ prototrophs (6–7 × 10−3 cells). The former represents events of ectopic gene conversion, whereas the latter results from allelic recombination.
Meiosis time course experiments (0–12 hr, starting with mitotically dividing cells that were transferred to SPM liquid sporulation medium) were performed 3–8 times for most strains (only twice for the double-mutant strain rev3Δ rad30Δ). In each experiment, cell cultures of two or more strains were used to obtain cell populations that underwent reasonably synchronous meiosis. The kinetics of appearance of His+ and Ura+ recombinants were assayed every 2 hr by a return-to-growth experiment (see Materials and Methods for details). After 48 hr in SPM, cell samples were examined microscopically to determine sporulation efficiencies.
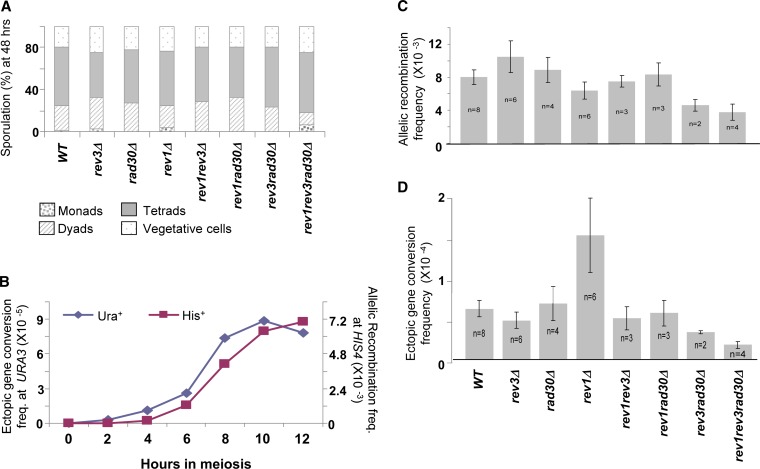
As seen in Figure 3A, sporulation in the TLSP-deleted strains appeared to be normal, and they all showed high sporulation efficiency (around 80% at 48 hr; no significant differences were found between the strains). Several dozen tetrads of the triple-mutant (rev1Δ rev3Δ rad30Δ) and of the WT strain were dissected on YPD medium and ascospore germination was found to be comparably high, 90%–95% in both strains. Thus, the absence of TLSPs does not appear to interfere with chromosome segregation in meiosis.
Figure 3 .
Sporulation, homologous (allelic) recombination, and ectopic gene conversion in strains homozygous for TLSP gene deletions. (A) Sporulation of cultures of isogenic strains deleted for one, two, or all three TLSP genes and their ancestral WT strain (no TLSP deletions). For each strain, 200 cells were examined at 48 hr, using phase-contrast microscopy. No statistically significant differences were found among sporulation frequencies of different strains (chi-square test). (B) Kinetics during meiosis of allelic recombination at HIS4 and ectopic gene conversion of a point mutation in URA3 in the WT strain. (C) Maximal allelic recombination values at HIS4 for the seven TLSP-deletion strains and their ancestral WT, obtained at 10 or 12 hr in sporulation. Each value is the mean of n independent experiments (n = 8, n = 4, and so forth), and the bars denote SEs. (D) Maximal ectopic gene conversion values of a point mutation at URA3 (ura3-T360G), obtained as described in Figure 3C and in Materials and Methods. Columns in C and D represent the same strains, and results were obtained from the same experiments.
Figure 3B represents a typical experiment with the WT strain. It shows that the frequency of cells giving rise to recombinant colonies (His+ and Ura+) increases during meiosis, reaching a maximum at 10–12 hr. Ectopic gene conversion (Ura+) in this experiment may be preceding allelic recombination by 30–60 min, as was the case in most experiments. Similar time course experiments were carried with all the strains, and strains were compared to each other for frequencies of His+ and Ura+ colonies over the whole time course (using the Mann-Whitney non-parametric statistical test). Figures 3, C and D, summarize results of 36 experiments (made with 8 independent strains) for allelic and ectopic gene conversion, respectively, using the maximal values obtained (at 10 or 12 hr). We also tested the events leading to Ura+ and His+ prototrophy in an isogenic strain homozygous for spo11Δ (strain DAO62) to confirm the fact that the events of both allelic and ectopic gene conversion (production of His+ and Ura+ colonies, respectively) are indeed dependent upon DNA double-strand breakage (data not shown): in this Spo11-deficient strain, the frequencies of His+ and Ura+ progeny were very low, virtually zero (and no spores were observed in the “meiotic” culture).
As can be seen in Figures 3, C and D, several of the TLSP-deleted strains show altered frequencies of allelic recombination (in HIS4) and of ectopic gene conversion (in URA3). The most notable result is the impairment of both genetic processes in the strain deleted for all three TLSPs, rev1Δ rev3Δ rad30Δ, which is highly significant over four independent experiments and consistent throughout the entire time course (data not shown; and Printzental 2010). Allelic recombination is more than twofold reduced in the triple-deletion mutant in comparison to that of the WT (Figure 3C), whereas ectopic gene conversion is fourfold reduced (Figure 3D). Interestingly, the double-mutant rev3Δrad30Δ shows reduction that is almost as marked in HIS4 allelic recombination as that in the triple mutant, suggesting that the Rev1 TLSP may have only a secondary role in this process. On the other hand, the strain deleted for REV1 alone (marked rev1Δ) showed more than twofold increase in ectopic gene conversion (Figure 3D) but not a marked effect on allelic recombination (Figure 3C), as if the Rev1 protein has a role of restricting recombination events to allelic sequences, rather than ectopic ones. However, the frequencies of ectopic gene conversion in the two double-mutant strains rev1Δrev3Δ and rev1Δrad30Δ were not significantly different from the rev3Δ or rad30Δ single mutants, or from the ancestral WT, suggesting that the absence of Rev3 or Rad30 is epistatic to the effect of rev1Δ on ectopic gene conversion.
A strain homozygous for rev7Δ (DAO29) was also studied in a similar meiotic time course experiment (data not shown; and Printzental 2010); it did not differ from the WT in allelic and ectopic recombination frequencies or in overall sporulation efficiency.
We conclude from the data reported in this section that the three TLSPs are probably involved in recombinational processes in meiosis, possibly during the repair of meiotic DSBs. The exact role of each of the TLSPs is not clear, however.
Yeast-two-hybrid interactions between TLSPs and some meiotic DSB proteins in cells arrested in meiosis and in cells undergoing mitotic divisions
The expression of TLSPs during meiosis is significantly elevated during meiotic prophase I (Figure 1B). At this stage of meiosis, DSBs are formed and repaired by recombination. Our genetic experiments (above) have shown that in the absence of all three TLSPs, recombination events in meiosis are compromised, both allelic and ectopic gene conversion (at HIS4 and at URA3, respectively). Several distinct protein complexes are known to be involved in meiotic DSB formation and chromosome recombination through DSB repair (Keeney 2008). DSBs are generated by the topoisomerase-like protein Spo11 with the aid of at least four meiotic proteins: Mei4, Rec102, Rec104, and Rec114. The DSB sites are then resected by the MRX complex, composed of Mre11, Rad50, and Xrs2. Recombination is next promoted by the Rad52 epistatic group of proteins, including the recombinase Rad51 and its mediator Rad52, and also Rad54 and the Rad55-Rad57 heterodimer. Dmc1, the meiosis-specific homolog of Rad51 also plays an important role at this stage of recombinational DSBs repair. We therefore inquired whether the TLSP proteins interact physically with any of these protein complexes. We carried out yeast-two-hybrid (Y2H) interaction assays (Fields and Song 1989; Fields and Sternglanz 1994) between Rev1, Rad30, and Rev7 and four representatives of the first group (Spo11, Mei4, Rec104, and Rec114); five proteins from the Rad52 group (Rad51, Dmc1, Rad52, Rad54, Rad57); and, with the Rad50 protein, a component of the MRX complex.
The two-hybrid analysis was carried out in a diploid strain derived from the high-sporulation SK1 genetic background, strains 661 × 662 (Table S1; see also Arora et al. 2004). The genes REV1, REV7, and RAD30 were fused to the C terminus of the DNA-binding domain of the bacterial LexA protein. Although we attempted it, suitable REV3 fusions could not be obtained, probably due to the large size of the gene, ∼4.5 kb. The reporter construct in these strains is the bacterial gene LacZ, coding for β-gal, fused to the operator of LexA (inserted near URA3 on chromosome V). All meiotic Y2H interaction tests were carried out in cells arrested in meiosis, as a result of being homozygous for the deletion ndt80Δ (Arora et al. 2004). Deletion of the mid-meiosis regulator Ndt80 (ndt80Δ) is known to cause arrest during prophase I after DSB formation but before the first meiotic division (Xu et al. 1995). This meiotic arrest was aimed at capturing and stabilizing transient protein–protein interactions that are unique to this phase and may disappear later on as meiosis proceeds. As we have shown above, TLSPs were intensely up-regulated at the prophase I stage of meiosis (Figure 1B), and DSB-repair proteins were expected to be present (Figure 2D) and available for interactions. Every particular Y2H combination of proteins was tested independently 3–4 times in mitotically dividing cells as well as in cells arrested in meiosis, as described above. In total, 33 combinations were examined, each with its two negative controls: one control contained one fusion protein (on a plasmid) and the complementing “empty” plasmid, and the other control contained the other fusion protein together with the first “empty” plasmid. Two additional positive controls were included in every set of experiments (see Materials and Methods): Mei4 and Rec114 (Y2H interaction in mitotic cells) and Spo11 and Rec104 (meiosis-specific interaction) (Arora et al. 2004).
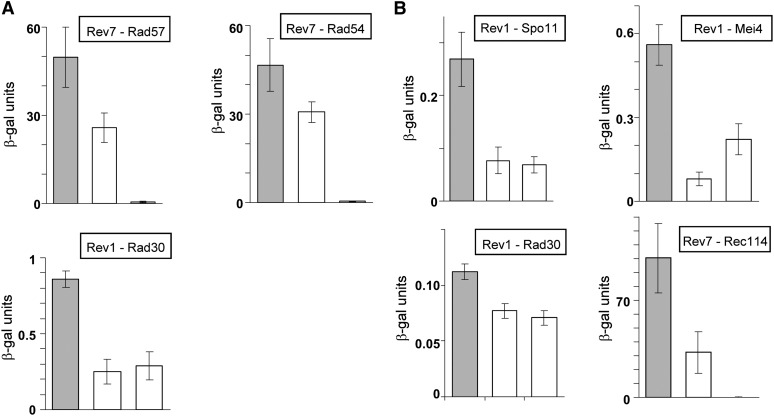
Table 1 summarizes all tested interactions between TLSPs and proteins involved in DSBs formation, resection, and repair during vegetative growth and in meiosis-arrested cells. Table 2 represents corresponding Y2H interactions among the three TLSP proteins. Several statistically significant Y2H interactions were observed, three mitotic interactions and four meiotic interactions. As reported previously (Acharya et al. 2007), we found that Rev1 and Rad30 interacted in vegetative cells (Table 2). Although significant, this interaction was weak (Figure 4A), and the level of β-gal units was considerably lower than the very strong mitotic positive control of Rec114-Mei4, with 50 β-gal units or more. In addition, two new mitotic interactions were found in our analysis: Rev7 showed interaction with Rad54 and with Rad57 (Table 1 and Figure 4A; the Rev7-Rad54 interaction, however, was only of borderline significance in our statistical analysis). We did not observe the reported mitotic interaction between Rev1 and Rev7 (Acharya et al. 2005). Rev1’s interaction with Rev7 was confirmed independently by others using co-immunoprecipitation (D’souza and Walker 2006). It is not clear whether we missed this interaction due to differences in the host strain genetic background, because of minor differences in the LacZ reporter constructs, or differences in orientation of the fusions, or another unrevealed factor(s).
Table 1. Protein-protein interactions between TLSP and DSB proteins.
| Proteins tested for interaction | Mitotically Dividing Cells |
Meiosis-Arrested Cells |
|||||
|---|---|---|---|---|---|---|---|
| Rev1 | Rad30 | Rev7 | Rev1 | Rad30 | Rev7 | ||
| DSB Formation | Spo11 | 0.26 | 0.64 | 0.12 | 4.15 (P = 0.0123) | 0.14 | 0.02 |
| Mei4 | 1.23 | 1.08 | 1.22 | 3.01 (P = 0.0048) | 0.37 | 0.01 | |
| Rec104 | 0.85 | 0.72 | 0.56 | 0.84 | 0.98 | 0.75 | |
| Rec114 | 1.03 | 1.0 | 1.06 | 1.2 | 1.18 | 6.27 (P = 0.0214) | |
| DSB Resection | Rad50 | 0.49 | 0.24 | 0.45 | 0.77 | 0.05 | 0.01 |
| DSB Repair | Dmc1 | 0.45 | 0.17 | 1.33 | 0.83 | 0.48 | 1.23 |
| Rad51 | 0.72 | 0.58 | 1.00 | 0.81 | 0.88 | 1.46 | |
| Rad52 | 0.56 | 0.64 | 1.14 | 0.67 | 0.73 | 1.32 | |
| Rad54 | 1.23 | 0.85 | 1.49 (P = 0.0532) | 1.09 | 0.7 | 1.20 | |
| Rad57 | 0.90 | 1.17 | 2.00 (P = 0.0283) | 1.00 | 0.90 | 1.31 | |
Summaries of Y2H experiments testing interactions between TLSPs and proteins involved in DSB formation, processing, and repair in mitotically dividing cells and in meiosis-arrested cells. LexA-BD and Gal4AD on 2μ vectors were fused to each of the DSB proteins and to TLSPs. Fusions were introduced into Y2H reporter strains and assayed for protein interactions in pair-wise combinations, as described in text. Values indicate mean fold changes in reporter activity between tested interactions and highest negative controls (fold change values were calculated separately for each experiment and then averaged). Every set of Y2H experiments (a particular combination of proteins) was repeated 3–4 times. Each significant (and borderline significant) Y2H interaction is based on four experiments. The given significance level (P value) is based on stringent analysis of variance, as described in Materials and Methods.
Table 2. Protein-protein interactions among TLSPs.
| TLSPs tested | Mitotically Dividing Cells | Meiosis-Arrested Cells |
|---|---|---|
| Rev1-Rad30 | 3.07 (P = 0.0075) | 1.47 (P = 0.002) |
| Rev1-Rev7 | 1.15 | 1.54 |
| Rad30-Rev7 | 0.53 | 0.89 |
Summaries of Y2H experiments testing interactions among the three TLSP proteins in mitotically dividing cells and in meiosis-arrested cells. Rad30 was fused on a 2μ vector to LexA-BD; Rev1 was fused to Gal4AD; and Rev7 was fused to either LexA-BD or Gal4AD to enable all three combinations to take place. Fusions were introduced into Y2H reporter strains as described in the legend to Table 1.
Figure 4 .
Statistically significant Y2H interactions between TLSPs and meiotic DSB proteins. Shown for each case is the level of β-gal units obtained from a diploid strain carrying the two fusion protein plasmids (left column [gray]) and the two corresponding controls (white): one from a strain carrying one plasmid with the Gal4AD fusion protein and an “empty” plasmid (middle column) and the other, a control, with the plasmid harboring the LexA-BD fusion protein and the other “empty” plasmid (right column). All values are means ± SE obtained from four independent experiments. (A) Values obtained from mitotically dividing cells. (B) Values obtained from meiosis-arrested cells.
Four new significant meiotic Y2H interactions were observed: three interactions involving TLSPs and DSB proteins: Rev1-Spo11, Rev1-Mei4, Rev7-Rec114 (Table 1 and Figure 4B); and one interaction involving two TLSPs: Rev1-Rad30 (Table 2 and Figure 4B). It should be noted, however, that only the Rev7-Rec114 interaction was stronger than the meiotic positive control in these experiments, Rec104-Spo11 (Arora et al. 2004), which was normally around 10 β-gal units, whereas the other meiotic interactions were weaker, although statistically significant. The former three interactions shown in Figure 4B were expected to be meiosis-specific because they involved a protein complex that is not expressed during mitotic cell divisions. These meiosis-specific Y2H interactions were indeed absent in the parallel Y2H mitotic experiments, although the relevant proteins were overexpressed (on the 2μ plasmids). These findings may reflect indirect protein interactions that are being established only when a stable and complete meiotic protein complex is formed. Alternatively, this might point toward the existence of meiosis-specific post-translational modifications of these proteins. All interactions presented in Figure 4, although verified in several independent experiments and found to be statistically significant, should be viewed cautiously until confirmed by an independent method, such as co-immunoprecipitation or pulldown experiments. Nevertheless, the Y2H results clearly suggest a physical association in meiosis of TLSPs with DSBs forming proteins rather than with DSBs repair proteins.
Discussion
We found that the three trans-lesion DNA polymerases (TLSPs) of S. cerevisiae are induced in meiosis: the genes REV3 and REV7, encoding the catalytic and regulatory units of PolZeta, respectively, are strongly induced transcriptionally (Figure 1A). All three catalytic TLSP proteins (Rev3, Rev1, and Rad30) increase markedly during meiosis (Figure 1B), whereas Rev7 appears to be abundant throughout meiosis. Rev7’s expression throughout the cell cycle may suggest meiotic functions other than being the accessory subunit of Rev3. Indeed, in human cells, hRev7 (alternatively called Mad2B) was shown to be involved not only in TLS but also in cell cycle regulation (Chen and Fang 2001) and signal transduction (Hong et al. 2009). Regarding Rev1, there also was an earlier report based on a large-scale gene expression screen showing that the protein is induced during meiosis in yeast (when fused to β-gal [Burns et al. 1994]). A meiosis-specific URS1 sequence found in close proximity to the Rev1 coding region presumably enables Rev1’s significant up-regulation during meiosis (Burns et al. 1994).
New DNA synthesis is required during yeast meiosis in two central and distinct events (Figure 2E): in meiotic S, which in strains of SK1 genetic background occurs about 2 hr after transferring the culture to SPM (Figure 1B, Ime2 up-regulation), and during prophase I (Figure 1B, about 4–5 hr, Cdc5 up-regulation), in DSB repair. Our findings are very clear with respect to the timing of expression of TLSPs: The main increase of TLSPs expression during meiosis occurs at 4–5 hr, at a time which coincides with prophase I. At this time, meiotic recombination events take place starting with DSB formation, resection of DSBs, and their repair following homology search, interaction with unbroken homologous chromatids and repair synthesis (Szekvolgyi and Nicolas 2010). Interestingly, we did not observe a marked elevation in TLSP expression at 2 hr at the time of meiotic S phase. Early observations testing REV3 transcription during meiosis support our results by showing only a mild increase in REV3 transcript early in meiosis but an 18-fold increase in REV3 transcription levels later in meiosis (Singhal et al. 1992). Taken together, the time of TLSPs’ expression in meiosis is clearly later than the time of regular meiotic DNA replication (meiotic S), and therefore, we propose that TLSPs are involved in DNA synthesis during recombinational repair of meiotic DSBs.
Two prevalent models have been proposed to elucidate the involvement of TLSPs in DNA damage tolerance during genome replication (i.e., the polymerase switching model and the postreplicative gap-filling model [reviewed by Chen et al. 2011]). One principal difference between the two models accounts for their timing. While the polymerase switching model is coupled to genome replication and can take place only in S phase, the postreplicative gap-filling events in mitotically dividing cells may occur during both S phase and G2 (Daigaku et al. 2010). Occurrence during the latter is supported by the observation that expression of Rev1 peaks in G2/M (Waters and Walker 2006). Our results, showing a significant up-regulation of all three TLSPs at times after genome replication, are in line with these latter findings. Furthermore, it has recently been shown that Rev1 and PolZeta form a complex that is required for efficient HR repair in HeLa cells (Sharma et al. 2012). Hence, the Rev1/PolZeta complex may be needed to operate on resected DSBs, which in meiosis, occur regularly after S, in G2, during prophase I (Keeney 2008; Padmore et al. 1991; Szekvolgyi and Nicolas 2010).
To test whether TLSPs are involved in recombinational repair, we employed two complementary approaches, namely we examined sporulation and meiotic recombination in strains deleted for the TLSP genes, and we examined the physical association during meiosis by Y2H tests between TLSPs and meiotic DSB proteins.
Allelic recombination at HIS4, as well as ectopic gene conversion of a T360G mutation in URA3, were significantly reduced in the triple-deletion mutant rev1Δrev3Δrad30Δ (Figure 3, C and D, respectively), but most of the other deletion strains did not differ significantly from the ancestral WT strain, except the strains deleted for REV1 (rev1Δ) and the double mutant rev3Δrad30Δ. The former strain (rev1Δ) shows a marked increase in ectopic gene conversion compared to that of the WT, which is surprising. When both Rev3 and Rad30 are missing, allelic recombination is also severely compromised, whereas ectopic recombination events are compromised only when all three proteins are absent, as seen in the triple-deletion mutant. The most likely interpretation of these results is that the three TLSPs participate in recombinational events in meiosis that lead to allelic and ectopic gene conversion and that the three enzymes have somewhat interchangeable roles. However, why should the single-deletion mutant rev1Δ show an increase in ectopic gene conversion? Perhaps the answer is that the Rev1 protein restricts gene conversion to allelic sequences (as its effect is seen only at the URA3 locus but not at the HIS4 locus), whereas the other two TLSPs participate in DSB repair regardless of whether it uses allelic or ectopic sequences as templates.
Interestingly, all eight strains (WT and its seven isogenic TLSP-deleted strains) showed comparable sporulation efficiencies in these experiments (i.e., 75%–80% asci after 48 hr) (Figure 2A). This means that the involvement of TLSPs in recombinational repair is not essential to meiosis and that the latter may be completed even in the absence of TLSPs. Germination of spores obtained from the triple-deletion mutant was also very high, comparable to that shown by the isogenic WT strain. The nonessential and fairly mild role of TLSPs in overall meiosis is not surprising, as too much activity of TLSPs during meiosis might lead to high frequency of mutations transmitted to the offspring, which might shift the fine balance from healthy diversity to an unbearable mutation load.
Physical association in meiosis was examined between the three TLSPs and each of 10 meiotic-DSB proteins by Y2H tests in yeast cells arrested in meiotic prophase, as well as in mitotically dividing cells. Of the 30 combinations tested, three new meiosis-specific, statistically significant associations were detected (Table 1; see also Figure 4B), namely Rev1-Spo11, Rev1-Mei4, and Rev7-Rec114. Moreover, a meiotic two-hybrid association was also found between two of the TLSPs, Rev1 and Rad30 (Table 2). Of these four Y2H interactions, only Rev7-Rec114 was stronger than the positive meiotic control, Rec104-Spo11 (established originally by Arora et al. 2004), whereas the three interactions involving Rev1 were much weaker (Figure 4B), although statistically significant. Perhaps the reason why Rev1 was found repeatedly in our meiotic two-hybrid associations is related to its known ability to interact with other proteins. Indeed, in addition to its N-terminal BRCT domain and a central TLS polymerase domain, the Rev1 protein also contains a C-terminal region that has been shown to interact with multiple other TLS polymerases, such as PolZeta (Acharya et al. 2006; Ohashi et al. 2004). These studies suggest an important role for Rev1 as a scaffold protein, perhaps coordinating access of the TLS polymerases to the damaged sites.
In our Y2H analysis, we were able to detect meiotic associations only between TLSPs and proteins of the DSB-generating complex, Spo11, Mei4, and Rec114, and not with DSB repair proteins, which act later in meiotic-DSB processing. The absence of Y2H associations with the latter may be explained in two alternative ways: one possibility is that TLSPs are not involved in the actual recombination-repair process that operates in prophase I, which is Rad51-recombinase-mediated; rather, TLSPs may be associated with the principal complex that generates DSBs and acts immediately on the newly cut DNA by adding a few nucleotides to the 3′-OH ends. DNA ends are then further processed and resected by various nucleases, but because resection acts to generate 3′-end single-stranded DNA, the exonucleolytic events occur primarily on the 5′-P ends in a 5′-to-3′ direction (Mimitou and Symington 2008; Zenvirth et al. 2003) and thus do not affect the new addition(s) made by TLSPs. The resected 3′ end is then ready to invade into homologous templates. We argue that this small addition of nucleotides by TLSPs to the 3′ends may not interfere with strand invasion and homology-based DNA synthesis that occur later in the process, as the DNA stretch needed for invasion extends over hundreds of nucleotides (Zhu et al. 2008). TLSPs are believed to be involved soon after DSB formation in other processes that lead to genetic diversity (Diaz and Casali 2002). Thus, it has been proposed that PolZeta can introduce mutations while filling in 3′ recessed termini during V(D)J recombination, thereby allowing ligation of the two free ends by NHEJ (Zan et al. 2001). Alternatively, the reason for not detecting the Y2H association of TLSPs with any of the five DSB-repair proteins may be related to the low processivity of the TLSPs on the DNA, that may also reflect weak association with the DNA-repair proteins: Being able to synthesize only a few nucleotides at a time, TLSPs promptly dissociate from the site of breakage and hence may fail to show association with the repair proteins recruited to the DSB sites. It is also possible that DSB repair proteins depend on Ndt80, which was absent in our strains. Nevertheless, we suggest that the association of TLSPs with the early, DSB-generating proteins is an important feature of meiosis, by which these polymerases are brought to physical proximity with the DSBs in DNA.
What roles do the TLSPs play in meiosis and why should they be associated with meiotic DSBs and their processing into recombination events? As far as we can judge, the TLSPs are not essential for the completion of meiosis, as strains devoid of all three known enzymes (rev1Δ rev3Δ rad30Δ) undergo normal sporulation, and meiotic chromosome segregation appears to be normal, as spores germinate with high efficiency. One possibility is that there exists in budding yeast another polymerase that functions in meiosis in the absence of the three known TLSPs. Indeed, an additional DNA polymerase, Pol4, is expressed in meiotic cells (Shimizu and Sugino 1993). Pol4-deleted mutants exhibited fivefold increase in meiotic intragenic recombination at HIS4 compared to that of the WT strain (Leem et al. 1994). It is possible that Pol4 fulfils an essential role that the TLSPs have in meiosis, but we have not tested this possibility. Pol32 was recently shown to facilitate the switch from PolDelta to PolZeta when encountering a DNA lesion (Baranovskiy et al. 2012; Johnson et al. 2012; Makarova et al. 2012). Pol32 might also affect the role of TLSPs during meiosis. It would be interesting to test null mutations of the nonessential Pol32 subunit in our system. Alternatively, the role of TLSPs in meiosis is not directly related to chromosome segregation and the mechanics of meiosis. We suggest that TLSPs act mainly outside the principal meiotic DNA replication (meiotic S) in the context of short patches resulting from repair of meiotic DSBs, either by replacing PolDelta in the polymerization of one or two nucleotides (see (Maloisel et al. 2004) for the role of PolDelta in meiotic DSB repair) or earlier, immediately after DSBs have been generated. The outcome of this occasional involvement of TLSPs in DSB processing is a marked increase in the generation of mutations in meiosis compared to the level found during mitotic cell divisions. This may be the explanation for the 6–20-fold increase in mutations during meiosis in yeast found by Magni and Von Borstel (1962) and for their association with recombination events nearby (Magni 1963). However, others (Nishant et al. 2010) did not find convincing evidence for an increase in mutation frequency during meiosis in budding yeast. The role we propose for TLSPs in meiosis is comparable to that of TLSPs in human lymphocytes, which are activated during lymphogenesis and are involved in repair of induced DSBs at Ig hypermutation sites, and thus increase the immunogenic repertoire (Bross et al. 2000; Papavasiliou and Schatz 2000; Poltoratsky et al. 2001; Zan et al. 2001). We therefore suggest that the association of TLSPs with meiotic DSBs leads to an increase in mutations during meiosis and has a long-range evolutionary impact, rather than a direct impact on an individual meiosis and its chromosome mechanics.
Taken together, meiosis reflects a refined balance between the immediate need for genomic stability and the evolutionary requirement for genetic diversity. Meiotic diversity is achieved by chromosome reassortment and recombination, as well as by the newly proposed involvement of TLSPs in meiotic DSB processing.
Supplementary Material
Acknowledgments
We gratefully acknowledge Scott Kenney and Michael Dresser for their gift of Y2H plasmids and strains. We thank Tomer Ravid for advice on Western blot analysis, Jossi Hillel and Nurith Strauss-Liviatan for statistical advice, and Drora Zenvirth for stimulating discussions at various stages of this project. We also thank the reviewers of the manuscript for useful comments that helped us to improve this paper. This work was supported by the Israel Science Foundation (grant 589/07) and U.S.–Israel Binational Science Foundation (grant 2009299). A. A.-E. is supported by Hadassah Academic College, Jerusalem, Israel.
Footnotes
Communicating editor: K. S. McKim
Literature Cited
- Acharya N., Haracska L., Johnson R. E., Unk I., Prakash S., et al. , 2005. Complex formation of yeast Rev1 and Rev7 proteins: a novel role for the polymerase-associated domain. Mol. Cell. Biol. 25: 9734–9740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N., Johnson R. E., Prakash S., Prakash L., 2006. Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase zeta for mismatch extension and for extension opposite from DNA lesions. Mol. Cell. Biol. 26: 9555–9563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N., Haracska L., Prakash S., Prakash L., 2007. Complex formation of yeast Rev1 with DNA polymerase eta. Mol. Cell. Biol. 27: 8401–8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoufouchi S., Flatter E., Dahan A., Faili A., Bertocci B., et al. , 2000. Two novel human and mouse DNA polymerases of the polX family. Nucleic Acids Res. 28: 3684–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora C., Kee K., Maleki S., Keeney S., 2004. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell 13: 549–559 [DOI] [PubMed] [Google Scholar]
- Baranovskiy A. G., Lada A. G., Siebler H. M., Zhang Y., Pavlov Y. I., et al. , 2012. DNA polymerase delta and zeta switch by sharing accessory subunits of DNA polymerase delta. J. Biol. Chem. 287: 17281–17287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin K. R., Zhang C., Shokat K. M., Herskowitz I., 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17: 1524–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bross L., Fukita Y., Mcblane F., Demolliere C., Rajewsky K., et al. , 2000. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity 13: 589–597 [DOI] [PubMed] [Google Scholar]
- Buhler C., Borde V., Lichten M., 2007. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 5: e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns N., Grimwade B., Ross-Macdonald P. B., Choi E. Y., Finberg K., et al. , 1994. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8: 1087–1105 [DOI] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N., 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61: 1089–1101 [DOI] [PubMed] [Google Scholar]
- Cervantes M. D., Farah J. A., Smith G. R., 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5: 883–888 [DOI] [PubMed] [Google Scholar]
- Chen J., Fang G., 2001. MAD2B is an inhibitor of the anaphase-promoting complex. Genes Dev. 15: 1765–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Bozza W., Zhuang Z., 2011. Ubiquitination of PCNA and its essential role in eukaryotic translesion synthesis. Cell Biochem. Biophys. 60: 47–60 [DOI] [PubMed] [Google Scholar]
- Chu S., Derisi J., Eisen M., Mulholland J., Botstein D., et al. , 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705 [DOI] [PubMed] [Google Scholar]
- Clyne R. K., Katis V. L., Jessop L., Benjamin K. R., Herskowitz I., et al. , 2003. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 5: 480–485 [DOI] [PubMed] [Google Scholar]
- Daigaku Y., Davies A. A., Ulrich H. D., 2010. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature 465: 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M., Casali P., 2002. Somatic immunoglobulin hypermutation. Curr. Opin. Immunol. 14: 235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser M. E., Ewing D. J., Conrad M. N., Dominguez A. M., Barstead R., et al. , 1997. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’souza S., Walker G. C., 2006. Novel role for the C terminus of Saccharomyces cerevisiae Rev1 in mediating protein-protein interactions. Mol. Cell. Biol. 26: 8173–8182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Song O., 1989. A novel genetic system to detect protein-protein interactions. Nature 340: 245–246 [DOI] [PubMed] [Google Scholar]
- Fields S., Sternglanz R., 1994. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 10: 286–292 [DOI] [PubMed] [Google Scholar]
- Friedberg E. C., Gerlach V. L., 1999. Novel DNA polymerases offer clues to the molecular basis of mutagenesis. Cell 98: 413–416 [DOI] [PubMed] [Google Scholar]
- Friedberg E. C., Fischhaber P. L., Kisker C., 2001. Error-prone DNA polymerases: novel structures and the benefits of infidelity. Cell 107: 9–12 [DOI] [PubMed] [Google Scholar]
- Friedberg E. C., Wagner R., Radman M., 2002. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296: 1627–1630 [DOI] [PubMed] [Google Scholar]
- Friedlander G., Joseph-Strauss D., Carmi M., Zenvirth D., Simchen G., et al. , 2006. Modulation of the transcription regulatory program in yeast cells committed to sporulation. Genome Biol. 7: R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan G. N., Wittschieben J. P., Wittschieben B. O., Wood R. D., 2008. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res. 18: 174–183 [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M., Dominguez O., Lopez-Fernandez L. A., De Lera L. T., Saniger M. L., et al. , 2000. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol. 301: 851–867 [DOI] [PubMed] [Google Scholar]
- Gearhart P. J., Wood R. D., 2001. Emerging links between hypermutation of antibody genes and DNA polymerases. Nat. Rev. Immunol. 1: 187–192 [DOI] [PubMed] [Google Scholar]
- Gerlach V. L., Feaver W. J., Fischhaber P. L., Richardson J. A., Aravind L., et al. , 2000. Human DNA polymerase kappa: a novel DNA polymerase of unknown biological function encoded by the DINB1 gene. Cold Spring Harb. Symp. Quant. Biol. 65: 41–49 [DOI] [PubMed] [Google Scholar]
- Gibbs P. E., Lawrence C. W., 1995. Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. J. Mol. Biol. 251: 229–236 [DOI] [PubMed] [Google Scholar]
- Gibbs P. E., Mcdonald J., Woodgate R., Lawrence C. W., 2005. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6–4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics 169: 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A., 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360 [DOI] [PubMed] [Google Scholar]
- Goodman M. F., 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 71: 17–50 [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R., 1991. Guide to Yeast Genetics and Molecular Biology. Methods in Enzymology. Academic Press, San Diego, CA: [PubMed] [Google Scholar]
- Harfe B. D., Jinks-Robertson S., 2000. DNA polymerase zeta introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol. Cell 6: 1491–1499 [DOI] [PubMed] [Google Scholar]
- Hirano Y., Sugimoto K., 2006. ATR homolog Mec1 controls association of DNA polymerase zeta-Rev1 complex with regions near a double-strand break. Curr. Biol. 16: 586–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbeck S. L., Strathern J. N., 1997. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics 147: 1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C. F., Chou Y. T., Lin Y. S., Wu C. W., 2009. MAD2B, a novel TCF4-binding protein, modulates TCF4-mediated epithelial-mesenchymal transdifferentiation. J. Biol. Chem. 284: 19613–19622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. E., Prakash L., Prakash S., 2012. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc. Natl. Acad. Sci. USA 109: 12455–12460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir Y., Simchen G., 1991. Monitoring meiosis and sporulation in Saccharomyces cerevisiae. Methods Enzymol. 194: 94–110 [DOI] [PubMed] [Google Scholar]
- Kawamoto T., Araki K., Sonoda E., Yamashita Y. M., Harada K., et al. , 2005. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol. Cell 20: 793–799 [DOI] [PubMed] [Google Scholar]
- Keeney S., 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1–53 [DOI] [PubMed] [Google Scholar]
- Keeney S., 2008. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn. Stab. 2: 81–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H. L., 1997. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics 147: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem S. H., Ropp P. A., Sugino A., 1994. The yeast Saccharomyces cerevisiae DNA polymerase IV: possible involvement in double strand break DNA repair. Nucleic Acids Res. 22: 3011–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Stith C. M., Burgers P. M., Heyer W. D., 2009. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase delta. Mol. Cell 36: 704–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., Mckenzie A., III, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Magni G. E., 1963. The origin of spontaneous mutations during meiosis. Proc. Natl. Acad. Sci. U S A 50: 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magni G. E., Von Borstel R. C., 1962. Different rates of spontaneous mutation during mitosis and meiosis in yeast. Genetics 47: 1097–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova A. V., Stodola J. L., Burgers P. M., 2012. A four-subunit DNA polymerase zeta complex containing Pol delta accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 40: 11618–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloisel L., Bhargava J., Roeder G. S., 2004. A role for DNA polymerase delta in gene conversion and crossing over during meiosis in Saccharomyces cerevisiae. Genetics 167: 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y., Takahashi M., Tsunekuni N., Minami T., Sumii M., et al. , 2001. Deoxycytidyl transferase activity of the human REV1 protein is closely associated with the conserved polymerase domain. J. Biol. Chem. 276: 15051–15058 [DOI] [PubMed] [Google Scholar]
- Matsuda T., Bebenek K., Masutani C., Hanaoka F., Kunkel T. A., 2000. Low fidelity DNA synthesis by human DNA polymerase-eta. Nature 404: 1011–1013 [DOI] [PubMed] [Google Scholar]
- Matsuda T., Bebenek K., Masutani C., Rogozin I. B., Hanaoka F., et al. , 2001. Error rate and specificity of human and murine DNA polymerase eta. J. Mol. Biol. 312: 335–346 [DOI] [PubMed] [Google Scholar]
- Mcilwraith M. J., Vaisman A., Liu Y., Fanning E., Woodgate R., et al. , 2005. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell 20: 783–792 [DOI] [PubMed] [Google Scholar]
- Mimitou E. P., Symington L. S., 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. R., Gibbs P. E., Nowicka A. M., Hinkle D. C., Lawrence C. W., 2000. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol. 37: 549–554 [DOI] [PubMed] [Google Scholar]
- Nishant K. T., Wei W., Mancera E., Argueso J. L., Schlattl A., et al. , 2010. The baker’s yeast diploid genome is remarkably stable in vegetative growth and meiosis. PLoS Genet. pii: e1001109; doi: 10.1371/journal.pgen.1001109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi E., Murakumo Y., Kanjo N., Akagi J., Masutani C., et al. , 2004. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells 9: 523–531 [DOI] [PubMed] [Google Scholar]
- Ohmori H., Friedberg E. C., Fuchs R. P., Goodman M. F., Hanaoka F., et al. , 2001. The Y-family of DNA polymerases. Mol. Cell 8: 7–8 [DOI] [PubMed] [Google Scholar]
- Padmore R., Cao L., Kleckner N., 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66: 1239–1256 [DOI] [PubMed] [Google Scholar]
- Papavasiliou F. N., Schatz D. G., 2000. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature 408: 216–221 [DOI] [PubMed] [Google Scholar]
- Poltoratsky V., Woo C. J., Tippin B., Martin A., Goodman M. F., et al. , 2001. Expression of error-prone polymerases in BL2 cells activated for Ig somatic hypermutation. Proc. Natl. Acad. Sci. USA 98: 7976–7981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig M., Williams R. M., Winzeler E. A., Tevzadze G. G., Conway A. R., et al. , 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26: 415–423 [DOI] [PubMed] [Google Scholar]
- Printzental, O., 2010 Involvement of Error-Prone DNA Polymerases in Yeast Meiosis: Recombination and Enhanced Mutagenesis M.Sc. Thesis, The Hebrew University of Jerusalem, Israel. [Google Scholar]
- Rattray A. J., Strathern J. N., 2003. Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu. Rev. Genet. 37: 31–66 [DOI] [PubMed] [Google Scholar]
- Reid R. J., Sunjevaric I., Keddache M., Rothstein R., 2002. Efficient PCR-based gene disruption in Saccharomyces strains using intergenic primers. Yeast 19: 319–328 [DOI] [PubMed] [Google Scholar]
- Rose M., Winston F., Hieter P., 1990. Methods Yeast Genetics—A Laboratory Course Manual. Cold Spring Harbor Press, New York [Google Scholar]
- Sharma S., Hicks J. K., Chute C. L., Brennan J. R., Ahn J. Y., et al. , 2012. REV1 and polymerase zeta facilitate homologous recombination repair. Nucleic Acids Res. 40: 682–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K., Sugino A., 1993. Purification and characterization of DNA helicase III from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 268: 9578–9584 [PubMed] [Google Scholar]
- Shinohara M., Gasior S. L., Bishop D. K., Shinohara A., 2000. Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. USA 97: 10814–10819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simchen G., 2009. Commitment to meiosis: what determines the mode of division in budding yeast? Bioessays 31: 169–177 [DOI] [PubMed] [Google Scholar]
- Singhal R. K., Hinkle D. C., Lawrence C. W., 1992. The REV3 gene of Saccharomyces cerevisiae is transcriptionally regulated more like a repair gene than one encoding a DNA polymerase. Mol. Gen. Genet. 236: 17–24 [DOI] [PubMed] [Google Scholar]
- Stallons L. J., Mcgregor W. G., 2010. Translesion synthesis polymerases in the prevention and promotion of carcinogenesis. J. Nucleic Acids pii: 643857; doi: 10.4061/2010/643857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J. N., Shafer B. K., Mcgill C. B., 1995. DNA synthesis errors associated with double-strand-break repair. Genetics 140: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekvolgyi L., Nicolas A., 2010. From meiosis to postmeiotic events: homologous recombination is obligatory but flexible. FEBS J. 277: 571–589 [DOI] [PubMed] [Google Scholar]
- Terasawa M., Ogawa H., Tsukamoto Y., Shinohara M., Shirahige K., et al. , 2007. Meiotic recombination-related DNA synthesis and its implications for cross-over and non-cross-over recombinant formation. Proc. Natl. Acad. Sci. U S A 104: 5965–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington M. T., Johnson R. E., Prakash S., Prakash L., 2000. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase eta. Proc. Natl. Acad. Sci. U S A 97: 3094–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L. S., Walker G. C., 2006. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc. Natl. Acad. Sci. U S A 103: 8971–8976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Ajimura M., Padmore R., Klein C., Kleckner N., 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 6572–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H., Komori A., Li Z., Cerutti A., Schaffer A., et al. , 2001. The translesion DNA polymerase zeta plays a major role in Ig and bcl-6 somatic hypermutation. Immunity 14: 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenvirth D., Simchen G., 2000. Meiotic double-strand breaks in Schizosaccharomyces pombe. Curr. Genet. 38: 33–38 [DOI] [PubMed] [Google Scholar]
- Zenvirth D., Arbel T., Sherman A., Goldway M., Klein S., et al. , 1992. Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae. EMBO J. 11: 3441–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenvirth D., Richler C., Bardhan A., Baudat F., Barzilai A., et al. , 2003. Mammalian meiosis involves DNA double-strand breaks with 3′ overhangs. Chromosoma 111: 369–376 [DOI] [PubMed] [Google Scholar]
- Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G., 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.