Abstract
The BTD gene codes for production of biotinidase, the enzyme responsible for helping the body reuse and recycle the biotin found in foods. Biotinidase deficiency is an autosomal recessively inherited disorder resulting in the inability to recycle the vitamin biotin and affects approximately 1 in 60,000 newborns. If untreated, the depletion of intracellular biotin leads to impaired activities of the biotin-dependent carboxylases and can result in cutaneous and neurological abnormalities in individuals with the disorder. Mutations in the biotinidase gene (BTD) alter enzymatic function. To date, more than 165 mutations in BTD have been reported. Our group has developed a database that characterizes the known mutations and sequence variants in BTD. (http://arup.utah.edu/database/BTD/BTD_welcome.php). All sequence variants have been verified for their positions within the BTD gene and designated according to standard nomenclature suggested by Human Genome Variation Society (HGVS). In addition, we describe the change in the protein, indicate whether the variant is a known or likely mutation vs. a benign polymorphism, and include the reference that first described the alteration. We also indicate whether the alteration is known to be clinically pathological based on an observation of a known symptomatic individual or predicted to be pathological based on enzymatic activity or putative disruption of the protein structure. We incorporated the published phenotype to help establish genotype-phenotype correlations and facilitate this process for those performing mutation analysis and/or interpreting results. Other features of this database include disease information, relevant links about biotinidase deficiency, reference sequences, ability to query by various criteria, and the process for submitting novel variations. This database is free to the public and will be updated quarterly. This database is a paradigm for formulating databases for other inherited metabolic disorders.
Keywords: biotinidase deficiency, biotinidase, BTD gene, mutation database, polymorphism
Biotin is an essential water-soluble vitamin that is the coenzyme for four carboxylases in humans. The serum concentration of biotin depends on dietary biotin intake and recycling of endogenous biotin. Biotinidase is the enzyme that catalyzes the cleavage of biotin from biocytin or biotinylated peptides and releases biotin and lysine (Wolf et al. 1983a). Biotinidase deficiency (BD) (OMIM no. 609019) is an autosomal recessively inherited disorder caused by a defect in biotinidase resulting in multiple carboxylase deficiency (MCD, OMIM no. 253260). If untreated, BD can lead to neurologic and cutaneous symptoms (Wolf et al. 1983b; Wolf 2011; Pindolia et al. 2011). Profound BD is defined as having less than 10% of mean normal serum enzymatic activity, whereas partial BD is defined as having 10%–30% of mean normal serum enzymatic activity (Wolf 2003). Profound BD can result in seizures, hypotonia, developmental delay, respiratory problems, ataxia, hearing loss, optic atrophy, rash, and alopecia. Partial BD can result in any of the above symptoms, but it is usually milder or only manifests when the individual is stressed or suffers from an illness (Wolf et al. 1985). If a child develops hearing loss, optic atrophy, or developmental delay, the condition is usually irreversible (Wolf et al. 1983a). Essentially all states in the United States and many countries screen for this disorder at birth because early treatment with pharmacological doses of biotin can prevent the disorder from becoming symptomatic in children (Wolf 2002).
The human biotinidase gene is located on chromosome 3p25 and consists of four exons with a total length of 1629 base pairs. To date, there are more than 160 mutations of the BTD gene that have been reported. Four mutations, c.98_104delinsTCC, p.Q456H, p.R538C, and a complex mutation, p.D444H;A171T, are the mutations most frequently observed in individuals with BD and account for approximately 65% of affected individuals (Hymes et al. 2001). The p.D444H mutation alone is a “milder” mutation and in combination with a mutation for profound BD on the other allele results in partial BD.
Diagnosis of BD is usually confirmed by finding deficient biotinidase activity in serum (Cowan et al. 2010). Newborn screening has enabled identification of children with profound and partial BD. However, in some cases, enzymatic activity does not adequately differentiate partial deficiency from heterozygosity for profound deficiency; therefore, mutation analysis is necessary to confirm the diagnosis (Wolf 2010).
We have reported gene variant archives for eight diseases, such as galactosemia (GALT), multiple endocrine neoplasia type 2 (RET), juvenile polyposis syndrome (SMAD4), and X-linked Alport syndrome (COL4A5) (http://arup.utah.edu/). These clinically curated archives uniquely display genotype and associated phenotype information (Calderon et al. 2007; Crockett et al. 2010;Margraf et al. 2009; Wooderchak et al. 2010; Sumner et al. 2011). We now introduce a new disease database for BD and BTD gene variants (http://arup.utah.edu/database/BTD/BTD_welcome.php). This database was developed to serve as a reference for all sequence variations currently reported in the BTD gene as well as to provide the most up-to-date phenotypic information available. The database accepts new mutation submissions. To date, 165 variants of the BTD gene have been described (Hymes et al. 2001; Baykal et al. 2005; Couce et al. 2011; Funghini et al. 2002; Pomponio et al. 1997; Norrgard et al. 1999; Laszlo et al. 2003; Milankovics et al. 2007; Swango et al. 1998; Iqbal et al. 2010), of which 155 have been classified as pathogenic; one mutation is suspected of being pathogenic, and nine are suspected of being benign.
Material and Methods
BTD variant database objectives
The aim of the BTD variant database is to record all known variants in BTD, and their associated phenotypes and/or from abnormal newborn screening results. This database is freely available to the public, and users are encouraged to submit newly discovered variants to the database curator using the “Database Submission” Web page, regardless of publication status. The submission form can also be used to update clinical information or to clarify the classification or phenotype for existing sequence variants. The submission form requests information about the sequence variation; clinical findings, if any; publications; and the submitter’s contact information. Those submitting novel variants to the database are expected to follow their own Institutional Review Board (IRB) protocols or consenting processes according to their institution’s instructions. To maintain the accuracy and utility of the BTD database, content will be updated quarterly by using new sequence variation information from reports in the literature, database submissions, and updates; and routine clinical testing performed at ARUP Laboratories. The date of the latest update is displayed on the BTD “Home” Web page. ARUP’s BTD variant database is disease-specific, including clinical data when available. Each variant in the ARUP BTD database has been verified against the GenBank reference sequence accession numbers NC_000003.10, NM_000060.2, and NP_000051.1 and contains a “Comments” field including haplotype information where available. Previously unpublished submissions are updated to include references once data have been published.
Data sources and limitations
The BTD database was constructed using gene sequence variation data published in the scientific literature since the BTD gene was first identified in individuals with BD, ascertained clinically or by newborn screening. To date, approximately 140 sequence variants have been found by searching PubMed (http://www.ncbi.nlm.nih.gov/sites/entrez) and Google (http://google.com/). Search terms included “BTD,” “biotinidase deficiency,” or “BTD mutation.” Occasionally, molecular sequencing results identify novel gene variants that are also included in this archive. The database was constructed using Human Genome Variation Society (HGVS) and Human Genome Organization (HUGO) Mutation Database Initiative recommendations for essential and optional content (http://www.hgvs.org/mdifaq.html). HGVS nomenclature recommendations for description of sequence variants were used to generate DNA and protein changes for this database. Entries were verified for position and name based on these sequences. All sequence variants in the database, including future updates, are designated based on their position in relation to the coding regions of the cDNA of BTD following recommendation listed in the HGVS (http://www.hgvs.org/mutnomen) and described in den Dunnen and Antonarakis (2000).
Software
SQL tables (MySQL, Inc.) were organized to represent BTD mutation status, associated phenotype, literature references, and any known clinical information. PHP coding was performed in-house for dynamic HTML (hypertext markup language) display to render pages and display SQL tables. Graphics were generated using FusionCharts version 3 software (www.fusioncharts.com/). Web pages are hosted on a Mac OSX Apache server. BTD mutations were added to the database and edited using phpMyAdmin software (www.phpmyadmin.net).
BTD Database Content
Database Web site
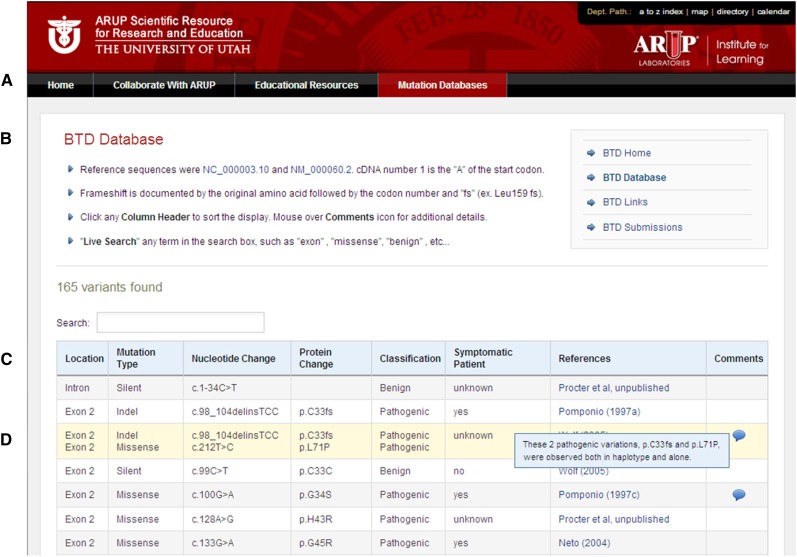
The BTD database can be found under Disease Databases on the ARUP Online Scientific Resource Web page, http://www.arup.utah.edu/, or accessed directly by using the URL http://arup.utah.edu/database/BTD/BTD_welcome.php. Several Web pages can be navigated using the tabs shown in Figure 1A. The BTD “Home” Web page briefly describes profound and partial BD, the database goals, and gene sequencing tests available at ARUP Laboratories. The BTD “Links” Web page has hyperlinks to existing online resource for BD and BTD, including the National Library of Medicine’s Genetics Home Reference and OMIM and UniProt and the Human Protein Reference Database (HPRD) and BTD reference sequences used.
Figure 1 .
Biotinidase database display. (A) Tabs for navigation within the database Website for homepage, links display, search, submission and contact information. (B) Main database Website display. (C) Database column headings where gene variants can be arranged by clicking the sort arrows of select columns in either ascending or descending order. (D) Example of mouse-over function for “Comments” column following the literature reference.
The database can be displayed in its entirety by accessing the “Display Database” Web page shown in Figure 1B. The database contained 165 entries as of January 2013. The default database is displayed according to the location of the sequence variation (5′ to 3′) within BTD. Figure 1 shows the database display columns Location, Mutation Type, Nucleotide Change, Protein Change, Classification, Phenotype, References, and Comments. Information that may further explain the data is displayed for each entry. The database also has search function capabilities such as location, classification, mutation type, literature reference, nucleotide change, or protein change. Searching may be performed from the BTD “Home” or “Search Database” pages.
The “Database Submissions” Web page can be used for a new submission of novel BTD sequence variation to the database or to update information for sequence variations already listed in the database. The “Contact Us” Web page has contact information, including E-mail addresses for the appropriate experts involved in developing and curating the database.
Database display columns
BTD sequence variation location and mutation type:
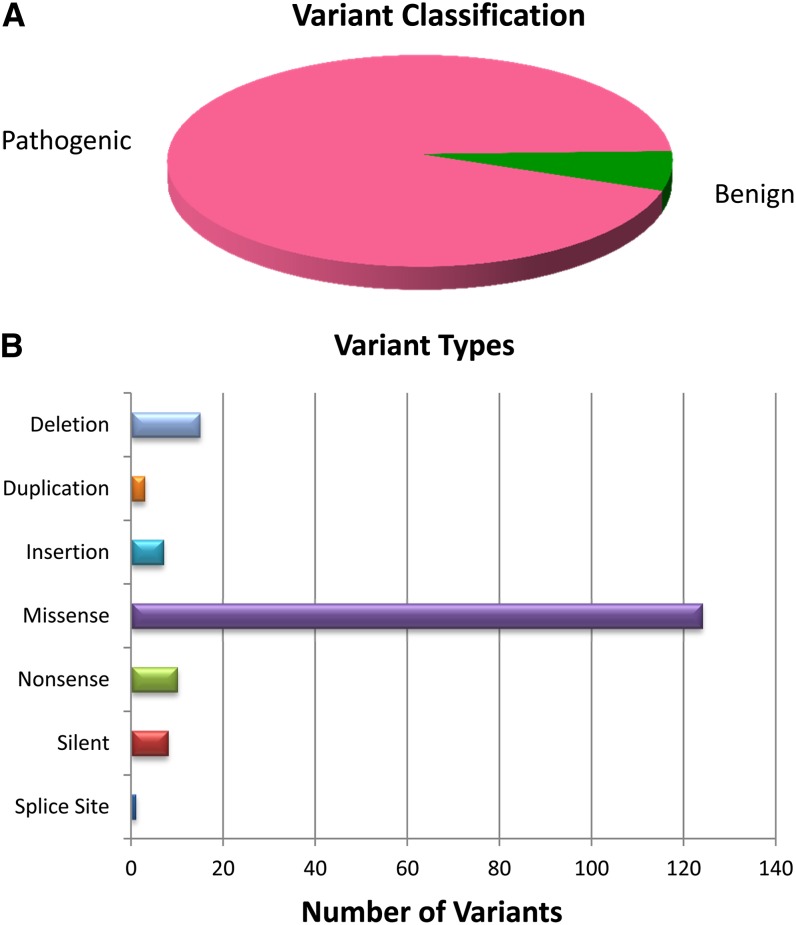
The location column displays the exon (or intron) number for each BTD variant. Mutation type describes the deviation from the reference sequence leading to the change in the DNA. Common types of mutations include small deletions and insertions/duplications, missense, nonsense, splice site, and silent changes. Large deletions and duplications, larger than or equal to one exon, have not yet been reported in individuals with BD. BTD gene variant types are summarized in Figure 2.
Figure 2 .
Summary of (A) variant classification and (B) types of mutations found in the BTD database.
BTD genotype:
The Nucleotide Change column displays the corresponding HGVS nomenclature. For cDNA numbering in the Nucleotide Change column of the BTD database, the first cDNA nucleotide (c.1) is the “A” of the ATG initiation codon.
BTD protein change:
For the Protein Change column, the single amino acid changes are listed in the database display. For splice-site variants, only the genotype is listed.
Classification of pathogenicity:
The classification definitions for pathogenicity are: Pathogenic, Suspected Pathogenic, Benign, Suspected Benign, or Uncertain. As is common practice, reduce or complete loss of biotinidase activity is assumed to be pathogenic and the author’s classification for each variant has used enzymatic activity, clinical phenotype and abnormal newborn screening results.
Symptomatic patients:
The status of patients’ clinical manifestations has been included where available. If no symptomatic patient had been observed due to early intervention, the symptomatic status has been stated as “unknown”. In the United States, symptomatic children are relatively rare due to the prevalence of newborn screening to diagnose infants early, as well as the ease of treatment to prevent symptoms.
References and comments:
The final two columns of the database feature links to literature resources and genotype/phenotype findings of each individual variant. As displayed in Figure 1, moving the cursor over comments are a key feature and unique part of this online resource. Comments for each BTD variant often contain additional information or evidence to support the classification designation. Importantly, clinical details, such as age or other medical conditions may be included, but protected information approximately the individual is never listed.
Database contents
Currently, the database displays information and classification of 165 BTD variants. However, this number is actively increasing as the additional variants are described in the literature. In addition, pathogenic mutations that are detected during routine clinical testing of samples at ARUP Laboratories or those submitted by other laboratories will also be added to the database. As mentioned above, the database provides a linked reference to a PubMed abstract for each variant reported in the database. As the classification of each variant is directly based on evidence from the literature, the link to PubMed is a useful feature of this database. However, in cases where a variant has not yet been reported in the literature, the database will still provide the clinical and/or experimental or functional data indicative of its classification. Laboratories and database users may also contact curators via e-mail or telephone regarding database entries, additional references, or clarification of information posted on the site at any time.
Figure 2 summarizes the content of the BTD database by variant classification and mutation types. The database currently has information on 155 pathogenic mutations, one suspected pathogenic mutation, and nine suspected benign variants. A breakdown of the different types of mutations currently contained in the database is also shown in Figure 2B.
Database access and features
The entire database content can be viewed using the display database option. Figure 1 shows an example of the Display Database option for all the sequence variant entries. Database entries may also be queried by certain criteria: variant location, classification (pathogenic, suspected pathogenic, benign, suspected benign and uncertain), mutation type (missense, nonsense, deletions, insertions or duplications, splice-site, or silent changes, nucleotide changes (based on cDNA position), and protein changes (based on codon number), or by the reference in the literature.
Database sort and search functions
The database display can be sorted in ascending or descending order by clicking the arrows found on the Mutation Type, Classification, and References headings (Figure 1). In addition, users can search for variants by location, classification, mutation type, literature reference, nucleotide change, or protein change. The search box feature can be accessed from either the BTD “Home” or “Search Database” Web page. Upon selection of the search criteria, a drop-down list containing possible options appears that allows the user to further narrow search results. A free-text field is also provided which will allow the user to search for a specific nucleotide and/or protein (amino acid). These search features simplify accessibility to the database entries and ease of finding of a particular mutation.
Clinical significance of the database
Clinical and research laboratories that offer full gene analysis for often encountered, rare variants must rely on the published literature or online resources to make informed decisions approximately the classification of the variant when reporting their findings. We developed this database to facilitate this process and assist those testing and interpreting results. The key feature of this database is the ease of access with which it provides pertinent gene variant information and associated genotype-phenotype observations. In addition, the database is updated and curated by individuals with expertise in both molecular genetics, as well as BD, ensuring that all of the information contained in the archive is up-to-date and evaluated with sufficient scientific rigor. Finally, as this database is a publicly available resource, with no login or membership requirements, it serves as an ideal centralized resource for all laboratories offering BTD gene analysis, as well as health professionals treating individuals with possible BD.
Acknowledgments
The authors thank ARUP Institute for Clinical and Experimental Pathology for financial support and the Department of Pathology, University of Utah School of Medicine, for hosting the Web service. We also acknowledge the support of the Safra Research Fund at Henry Ford Hospital Detroit, Michigan 48202 (B.W.).
Footnotes
Communicating editor: J. M. Cherry
Literature Cited
- Baykal T., Gokcay G., Gokdemir Y., Demir F., Seckin Y., et al. , 2005. Asymptomatic adults and older siblings with biotinidase deficiency ascertained by family studies of index cases. J. Inherit. Metab. Dis. 28: 903–912 [DOI] [PubMed] [Google Scholar]
- Calderon F. R., Phansalkar A. R., Crockett D. K., Miller M., Mao R., 2007. Mutation database for the galactose-1-phosphate uridyltransferase (GALT) gene. Hum. Mutat. 28: 939–943 [DOI] [PubMed] [Google Scholar]
- Couce M. L., Perez-Cerda C., Garcia Silva M. T., Garcia Cazorla A., Martin-Hernandez E., et al. , 2011. Clinical and genetic findings in patients with biotinidase deficiency detected through newborn screening or selective screening for hearing loss or inherited metabolic disease. Med. Clin. (Barc.) 137: 500–503 (In Spanish). [DOI] [PubMed] [Google Scholar]
- Cowan T. M., Blitzer M. G., Wolf B., 2010. Technical standards and guidelines for the diagnosis of biotinidase deficiency. Genet. Med. 12: 464–470 [DOI] [PubMed] [Google Scholar]
- Crockett D. K., Pont-Kingdon G., Gedge F., Sumner K., Seamons R., et al. , 2010. The Alport syndrome COL4A5 variant database. Hum. Mutat. 31: E1652–E1657 [DOI] [PubMed] [Google Scholar]
- den Dunnen J. T., Antonarakis S. E., 2000. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 15: 7–12 [DOI] [PubMed] [Google Scholar]
- Funghini S., Donati M. A., Pasquini E., Gasperini S., Ciani F., et al. , 2002. Two new mutations in children affected by partial biotinidase deficiency ascertained by newborn screening. J. Inherit. Metab. Dis. 25: 328–330 [DOI] [PubMed] [Google Scholar]
- Hymes J., Stanley C. M., Wolf B., 2001. Mutations in BTD causing biotinidase deficiency. Hum. Mutat. 18: 375–381 [DOI] [PubMed] [Google Scholar]
- Iqbal F., Item C. B., Vilaseca M. A., Jalan A., Muhl A., et al. , 2010. The identification of novel mutations in the biotinidase gene using denaturing high pressure liquid chromatography (dHPLC). Mol. Genet. Metab. 100: 42–45 [DOI] [PubMed] [Google Scholar]
- Laszlo A., Schuler E. A., Sallay E., Endreffy E., Somogyi C., et al. , 2003. Neonatal screening for biotinidase deficiency in Hungary: clinical, biochemical and molecular studies. J. Inherit. Metab. Dis. 26: 693–698 [DOI] [PubMed] [Google Scholar]
- Margraf R. L., Crockett D. K., Krautscheid P. M., Seamons R., Calderon F. R., et al. , 2009. Multiple endocrine neoplasia type 2 RET protooncogene database: repository of MEN2-associated RET sequence variation and reference for genotype/phenotype correlations. Hum. Mutat. 30: 548–556 [DOI] [PubMed] [Google Scholar]
- Milankovics I., Kamory E., Csokay B., Fodor F., Somogyi C., et al. , 2007. Mutations causing biotinidase deficiency in children ascertained by newborn screening in Western Hungary. Mol. Genet. Metab. 90: 345–348 [DOI] [PubMed] [Google Scholar]
- Norrgard K. J., Pomponio R. J., Hymes J., Wolf B., 1999. Mutations causing profound biotinidase deficiency in children ascertained by newborn screening in the United States occur at different frequencies than in symptomatic children. Pediatr. Res. 46: 20–27 [DOI] [PubMed] [Google Scholar]
- Pindolia K., Jordan M., Guo C., Matthews N., Mock D. M., et al. , 2011. Development and characterization of a mouse with profound biotinidase deficiency: a biotin-responsive neurocutaneous disorder. Mol. Genet. Metab. 102: 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomponio R. J., Hymes J., Reynolds T. R., Meyers G. A., Fleischhauer K., et al. , 1997. Mutations in the human biotinidase gene that cause profound biotinidase deficiency in symptomatic children: molecular, biochemical, and clinical analysis. Pediatr. Res. 42: 840–848 [DOI] [PubMed] [Google Scholar]
- Sumner K., Crockett D. K., Muram T., Mallempati K., Best H., et al. , 2011. The SPRED1 Variants Repository for Legius Syndrome. G3: Genes, Genomes, Genetics 1: 451–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swango K. L., Demirkol M., Huner G., Pronicka E., Sykut-Cegielska J., et al. , 1998. Partial biotinidase deficiency is usually due to the D444H mutation in the biotinidase gene. Hum. Genet. 102: 571–575 [DOI] [PubMed] [Google Scholar]
- Wolf B., 2002. Children with profound biotinidase deficiency should be treated with biotin regardless of their residual enzyme activity or genotype. Eur. J. Pediatr. 161: 167–168 (Letter to the Editor: author reply 169). [DOI] [PubMed] [Google Scholar]
- Wolf B., 2003. Biotinidase deficiency: new directions and practical concerns. Curr. Treat. Options Neurol. 5: 321–328 [DOI] [PubMed] [Google Scholar]
- Wolf B., 2010. Clinical issues and frequent questions approximately biotinidase deficiency. Mol. Genet. Metab. 100: 6–13 [DOI] [PubMed] [Google Scholar]
- Wolf B., 2011. The neurology of biotinidase deficiency. Mol. Genet. Metab. 104: 27–34 [DOI] [PubMed] [Google Scholar]
- Wolf B., Grier R. E., Allen R. J., Goodman S. I., Kien C. L., 1983a Biotinidase deficiency: the enzymatic defect in late-onset multiple carboxylase deficiency. Clin. Chim. Acta 131: 273–281 [DOI] [PubMed] [Google Scholar]
- Wolf B., Grier R. E., Allen R. J., Goodman S. I., Kien C. L., et al. , 1983b Phenotypic variation in biotinidase deficiency. J. Pediatr. 103: 233–237 [DOI] [PubMed] [Google Scholar]
- Wolf B., Heard G. S., Weissbecker K. A., McVoy J. R., Grier R. E., et al. , 1985. Biotinidase deficiency: initial clinical features and rapid diagnosis. Ann. Neurol. 18: 614–617 [DOI] [PubMed] [Google Scholar]
- Wooderchak W. L., Spencer Z., Crockett D. K., McDonald J., Bayrak-Todemir P., 2010. Repository of SMAD4 mutations: reference for genotype/phenotype correlation. J. Data Mining in Genom. and Proteomics 1: 101. [Google Scholar]