Abstract
Near-infrared light is favourable for imaging in mammalian tissues due to low absorbance of hemoglobin, melanin, and water. Therefore, fluorescent proteins, biosensors and optogenetic constructs for optimal imaging, optical readout and light manipulation in mammals should have fluorescence and action spectra within the near-infrared window. Interestingly, natural Bacterial Phytochrome Photoreceptors (BphPs) utilize the low molecular weight biliverdin, found in most mammalian tissues, as a photoreactive chromophore. Due to their near-infrared absorbance BphPs are preferred templates for designing optical molecular tools for applications in mammals. Moreover, BphPs spectrally complement existing genetically-encoded probes. Several BphPs were already developed into the near-infrared fluorescent variants. Based on analysis of the photochemistry and structure of BphPs we suggest a variety of possible BphP-based fluorescent proteins, biosensors, and optogenetic tools. Putative design strategies and experimental considerations for such probes are discussed.
Introduction
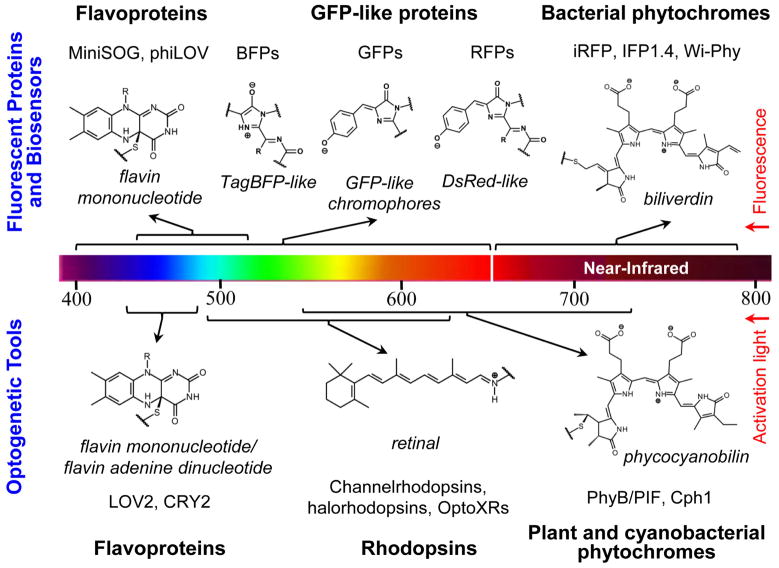
Modern biology is increasingly reliant on optical technologies such as fluorescence imaging, optical detection, and light-induced manipulation. However, the major limitation in this field is the availability of genetically-encoded reagents by which to study processes in vivo. Several types of naturally occurring light-active proteins, such as flavoproteins1, GFP-like proteins2–4, rhodopsins1, and phytochromes5–7 have been successfully employed for engineering of fluorescent proteins (FPs)2–4, 8–12, biosensors13, and optogenetic tools14–19 (Fig. 1). The important component of all light-active holoproteins is a chromophore, typically consisting of a conjugated electron π-system. Chromophore is either autocatalytically derived from amino acid side chains, as in a GFP-like family of proteins3, 4, or incorporated by an apoprotein from the surrounding protein environment1, 5. Spectral properties of light-sensitive proteins are mainly determined by their chromophore structure (Fig. 1) and its immediate protein environment.
Fig. 1.
A diversity of the chromophores in the major groups of currently available fluorescent proteins, fluorescent biosensors, and optogenetic tools developed for biotechnological applications are shown. The upper part of the figure shows the chemical structures of flavin mononucleotide, TagBFP-like, GFP-like, DsRed-like and biliverdin chromophores for the respective fluorescent proteins and biosensors derived from flavoproteins (MiniSOG8, phiLOV9), GFP-like proteins (BFPs, GFPs, RFPs)2, 3, and bacterial phytochromes (iRFP10, IFP1.411, Wi-Phy12). The lower part of the figure shows the chemical structures of flavin mononucleotide, retinal and phycocyanobilin chromophores for the respective optogenetic tools derived from flavoproteins (LOV214, CRY215), rhodopsins (channelrhodopsins16, halorhodopsisns16, OptoXRs17), plant and cyanobacterial phytochromes (PhyB/PIF19, Cph118). The chromophores are shown in their protein-linked forms. A color scale presents the wavelength range of fluorescence emission for the fluorescent proteins and biosensors, and the wavelength range of the activation/de-activation light for the optogenetic tools.
Reduced autofluorescence, low light scattering, and minimal absorbance at longer wavelengths make near-infrared (NIR) FPs superior probes for deep-tissue and whole-body imaging. Phytochromes from fungi, plant, bacteria and cyanobacteria are red/far-red water-soluble photoreceptors utilizing linear tetrapyrrole bilins as chromophores6, 7. However, the subclass of phytochromes found in photosynthetic and non-photosynthetic bacteria20–22, termed Bacteriophytochrome Photoreceptors (BphPs), have certain advantages over other phytochromes such as from plants and cyanobacteria for engineering NIR probes. First, BphPs utilize biliverdin IXα (BV) as a chromophore6, which in contrast to the tetrapyrrole chromophores of other phytochrome types is ubiquitous in mammalian tissues10, 11. This important feature makes BphPs applications in live mammalian cells, tissues and whole mammals as straightforward as conventional GFP-like FPs10, 23. Second, BphPs exhibit red-shifted NIR absorbance and fluorescence relative to other phytochrome types20 and their fluorescent derivatives24–26 and lay within a NIR transparency window of mammalian tissues (650–900 nm) (Fig. 1)27. Third, the domain architecture and pronounced conformational changes upon photoisomerization make BphPs attractive templates for designing optogenetic probes28, 29. Taken together, BphPs are appealing candidates for designing of optical probes for in vivo applications in mammals. Recently, several BphPs have been developed into the first NIR FPs such as IFP1.411, iRFP10, and Wi-Phy12.
Initially in this review, we describe the structure and photochemistry of BphPs as well as conformational changes of the BV chromophore. We then provide a workflow to develop BphPs-based NIR FPs, optical biosensors, and optogenetic tools. Lastly, we indicate possible obstacles in the course of their engineering and suggest potential in vivo applications. We focus on BphPs whereas for phyotochromes from plants and cyanobacteria that bind other than BV tetrapyrroles not found in mammals we refer readers to recent reviews6, 7, 28–30.
Structure and photochemistry
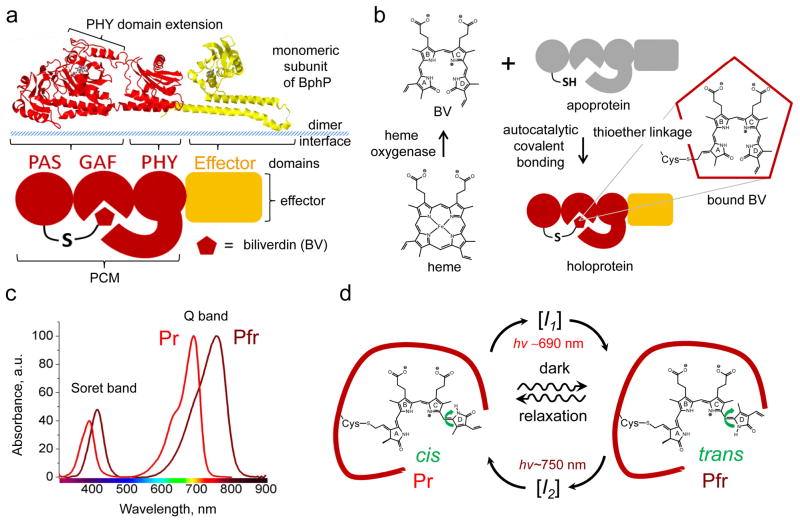
Analysis of the crystal structures and amino acid sequences illustrate that BphPs and their plant and cyanobacterial analogues share a common domain architecture, consisting of a photosensory core module (PCM) and an output effector module, which is typically represented by histidine kinase (HisK) (Fig. 2a)6, 31–34. Besides HisK motifs other effector modules, such as PAS domains that interact with repressors and prevent their binding to DNA35, 36, GGDEF (diguanylate cyclase) and EAL (phosphodiesterase) domains that are involved in second messenger signaling37, have been found in so called non-canonical BphPs20, 21. Biological functions of BphPs are poorly understood, however, some of them may play role in synthesis of light harvesting complexes, in respiration and carotenoid regulation20, 21, 35. The PCM is formed by PAS (Per-ARNT-Sim repeats), GAF (cGMP phosphodiesterase/adenylate cyclase/FhlA transcriptional activator), and PHY (phytochrome-specific) domains connected by α-helix linkers. Despite the low resemblance of their primary structures, PAS, GAF, and PHY domains share a common topology (Fig. 3)30–32. PAS and GAF domains are very distantly related and have been found in other signaling proteins. PHY is a phytochrome-specific GAF domain20. The majority of the chromophore-protein interactions occur at the GAF domain while the PHY domain’s extension serves to shield BV from solvents32, 38. The α-helices of the GAF and effector domains are involved in the formation of head-to-head BphP dimers (Fig. 2a)32, 39.
Fig. 2.
Structure, formation, spectral and photochemical properties of bacterial phytochromes. (a) Structural organization of a monomer subunit of BphP, (b) synthesis of biliverdin IXα (BV) from heme and its incorporation by apoprotein, (c) absorbance spectra of BphPs in the Pr and Pfr states, and (d) photocycle of BV chromophore within the protein environment are shown. (a, top) Structure of the monomer subunit of the BphP photosensory module (PMC) of Pseudomonas aeruginosa in red (PDB accession ID 3C2W) is overlapped with the structure of the effector domain, represented by histidine kinase in yellow (PDB accession ID 2C2A). (a, bottom) Schematic representation of BphP consisting of the PAS, GAF, PHY, and effector domains. A PHY domain’s extension shields BV from solvent and plays a role in BphP photoconversion. Dimer interface is formed by α-helices of the GAF domain and linker between PMC and effector domain. (b) Degradation of heme to BV is catalyzed by heme oxygenase. This reaction proceeds through a common mechanism that leads to formation of BV, which then autocatalytically covalently attaches to conservative Cys residue in the PAS domain of an apoprotein via a thioether linkage, resulting in a haloprotein. (c) Absorbance spectra of the typical Pr and Pfr states presenting the Q and Soret absorbance bands. (d) BV chromophre in the Pr and Pfr states is shown within protein environment of BphP (dark red curve). Transition from the Pr state to the Pfr state and vice versa is induced with 690 nm and 750 nm light, respectively. The transitions result from rotation of D-ring of the BV chromophore around adjacent double bond (green arrow). In the dark the photoconverted state undergoes spontaneous relaxation back to the ground state (waved arrows). The transition from the Pr to Pfr state and vice versa occurs via different intermediate states I1 and I2, respectively.
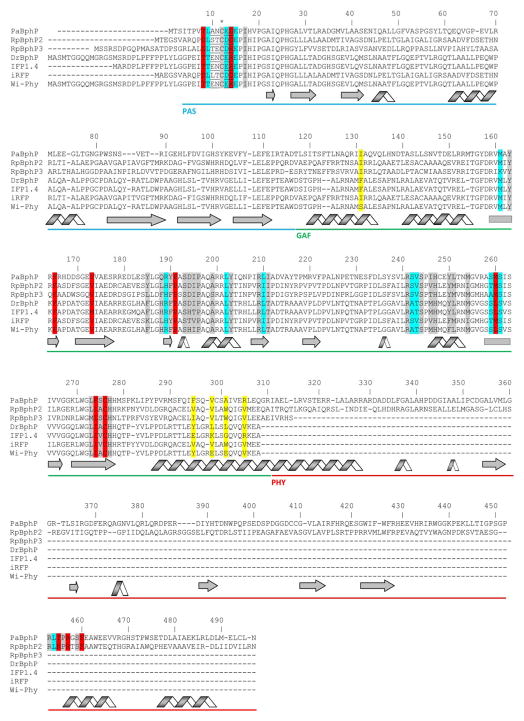
Fig. 3.
Alignment of amino acid sequences of the photosensory modules of the most characterized BphPs. The proteins were chosen based on the availability of the crystal structures (PaBphP, RpBphP3, DrBphP) and those that were developed to the fluorescent proteins (IFP1.4, iRFP, Wi-Phy, and RpBphP2 as the template for iRFP). The numbering of amino acid residues follows that for the PaBphP protein. Cys residue, which is covalently attached to the BV chromophore, is marked with asterisk. The chromophore surrounding residues within 4.5 Å, 4.5–5.5 Å and 5.5–6.5 Å are highlighted with gray, cyan, and red colors, respectively. The residues located in the dimer interface are highlighted with yellow. The residues located in the close proximity to the thioether bond between BV and apoprotein are underlined. The α-helixes and β-sheets demonstrate the secondary structure of BphPs. The PAS, GAF and PHY domains are underlined with the blue, green, and red lines, respectively.
BphPs autocatalytically bind the BV chromophore, which is a product of the oxidative degradation of heme by heme oxygenase (HO) (Fig. 2b)6. Incorporation of BV into the BphP apoprotein likely occurs in two consecutive steps: first, BV is secured to the chromophore-binding pocket in the GAF domain, and second, a thioether bond is formed with a conserved Cys in the PAS domain, which is constrained by adjacent amino acid residues (Fig. 3)40, 41. BphPs can exist in two stable interconvertible forms, termed Pr and Pfr states. The Pr state absorbs “red” light at 690–710 nm while the Pfr state absorbs “far-red” light at 740–760 nm (Fig. 2d). Absorbance bands in the NIR part of spectrum are termed Q bands. Along with absorption at the Q band, each BphP also absorbs at 380–420 nm in the violet range of spectrum, known as the Soret band. In agreement with Kasha’s rule, which states that photon emission occurs in appreciable yield only from the lowest excited state, excitation of either band of the Pr state results in NIR fluorescence41, 42. The Pr state of BphP variants emits at 700–720 nm10–12, while fluorescence of the Pfr state has not been reported yet. The latter is due to the sub-picosecond half-life of the Pfr excited state that results in its negligible quantum yield43. Interestingly, at the acidic pH values BV dimethyl ester exhibits several emission peaks including one at 770 nm that is close to the expected Pfr emission maximum44, 45.
In darkness, most BphPs adopt the Pr state, which typically manifests as the biologically inactive ground or dark relaxed state while some BphPs, designated bathy BphPs, adopt the Pfr state as a ground state22, 32, 46. However, after binding of BV all BphPs initially generate the Pr state and, in the case of bathy BphPs, later spontaneously convert into the Pfr state46. Upon light absorbance, the Pr state photoconverts into the Pfr state, also known as a signaling state. Once generated by red light irradiation, the Pfr state reverts back to the Pr state either relatively slowly and non-photochemically (in a process called dark reversion or thermal relaxation), or rapidly upon irradiation with far-red light (Fig. 2c). The rate of dark reversion, which varies from minutes to hours, can be substantially accelerated or decelerated by introducing point mutations into the GAF and PHY domains, thus affecting the BphP photoperception32, 33, 38, 47.
BphP photoconversion involves a rotation of the D pyrrole ring of BV around a methine bridge between the C and D pyrrole rings7, 30. The photoinduced Pr→Pfr and Pfr→Pr conversions were shown to proceed via distinct pathways involving different metastable intermediates (Fig. 2d), however, similar but inverted proton migration cycles may occur (see reviews for details 7, 30). Deletion of the PHY domain or amino acid residues at the N-terminus of the PAS domain impairs formation of the Pfr state33, 41. Introducing point mutations into the GAF and PHY domains can strongly affect the Bph photochemistry (the rate and efficiency of Pr→Pfr and Pfr→Pr photoconversion, stability of Pr and Pfr states and quantum yield of fluorescence)12, 41–43, 48 as well as non-photochemical transitions (kinetics of dark reversion)32, 33, 38.
The light-driven conformational changes of the BV chromophore are suggested to generate torques about the GAF domain and the C-terminal α-helices, thus propagating a light signal to the output HisK domain and modulating its activity39. The extensive intimate dimerization interface between two BphP monomers is suggested to play an important role in light signal propagation to an output effector domain (see the reviews for details28, 29). It is worth noting that the efficiency of light signal propagation, lifetime of the signaling state and quantum yield of photoconversion are considered to be the significant characteristics in optogenetic tools28, 47.
Fluorescent proteins
Engineering of fluorescent probes based on GFP-like proteins has generated a powerful toolkit for molecular and cell biology2, 4. In addition, several red FPs were developed based on plant and cyanobacterial phytochromes24–26, 49. However, excitation/emission maxima of all these FPs are limited to 660/680 nm. In this respect, BphPs hold great promise for becoming the templates for generation of genetically-encoded NIR probes (Fig. 4). Knowledge of BphPs photochemical properties, their structures, and relevant mutagenesis data makes engineering NIR BphP variants of different spectral phenotypes feasible.
Fig. 4.
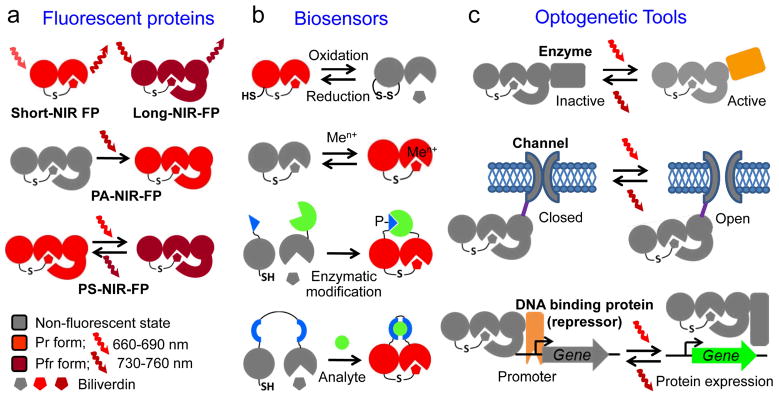
Proposed genetically-encoded near-infrared (NIR) probes based on bacterial phytochromes: (a) versatile two-domain short-NIR and three-domain long-NIR fluorescent proteins (FPs), photoactivatable (PA) and photoswitchable (PS) three-domain NIR fluorescent proteins, (b) two-domain biosensors for redox status and metal ions (Men+), split biosensors for protein interactions resulted from enzymatic modifications, such as phosphorylation (designated as P−), and insertion-based biosensors to detect analytes, and (c) optogenetic tools controlling enzymatic activities, open and closed states of ion channels, and gene expression via regulation of interaction between DNA repressor and gene promoter. The schematic illustration of the structural elements of BphPs corresponds to those shown in Figure 2a. Please see text for more details.
Possible features of NIR FPs based on the PCM of BphPs are shown in Fig. 4a. Compared to GFP-like FPs, the PCM of BphPs has several advantages as well as drawbacks that are summarized in Table 1. Engineering of permanently fluorescent short NIR FPs could involve stabilization of the Pr state of the chromophore, destabilization of the Pfr state, and disruption of the hydrogen bond network between BV and its microenvironment12, 42, 43. This can be achieved by truncating the PHY domain and by introducing specific amino acid substitutions into the chromophore’s immediate environment. This strategy was recently employed to develop IFP1.411, iRFP10, and Wi-Phy12. Furthermore, because the PHY domain plays a crucial role in the stabilization of the Pfr state and BphP photoisomerization, the entire PCM should be used for engineering long NIR FPs, non-fluorescent chromoproteins (CPs) that absorb but do not emit light, photoactivatable (PA) and photoswitchable (PS) NIR FPs. To develop long NIR FPs and CPs, the amino acid positions responsible for stabilization of the Pfr state and disabling Pfr→Pr photoconversion and other Pfr de-excitation pathways43, determined by structural analysis and mutagenesis of PaBphP32, 38, should be the primary targets for site-specific mutagenesis (Table 2). For this, bathy BphPs can be appropriate templates22, 32, 46
Table 1.
Comparison of properties of the photosensory module of BphPs and the GFP-like FPs.
| Property | PCM of BphPs | GFP-like FPs | Advantage (+) or Disadvantage (−) of BphPs vs. GFP-like FPs | Ref |
|---|---|---|---|---|
| Overall structure | Consists of two or three domains with common α/β fold topology linked via α-helixes; exists as monomer, dimer or oligomer | Consist of a single domain, rigid β-barrel formed by 11 β-sheets Exist as monomer, dimer, tetramer or oligomer | (+) Domain organization allow diverse strategies for protein engineering (+) Suitable for engineering of optogenetic tools |
4, 7, 28, 29, 32, 33, 39 |
| Size of monomer subunit | PAS-GAF domains: 300–310 a.a. (35–38 kDa) PAS-GAF-PHY domains: 500–530 a.a. (55–60 kDa) | 210–240 a.a. (24–28 kDa) | (−) Potentially may affect proper localization or function of target proteins | |
| Chromophore formation | Apoprotein autocatalytically and covalently incorporates BV as a chromophore | Protein folding followed by autocatalytic chromophore formation in presence of oxygen | (+) Does not require molecular oxygen, therefore, may form in anaerobic conditions (−) Require exogenous BV, whose concentration may vary in different cell types and tissues (−) Presence of HO may improve BV incorporation |
4, 6, 10–12, 50 |
| Absorbance/Emission maxima | 630–750 nm/680–800 nm * | 355–635 nm/425–670 nm | (+) Expands GFP-like fluorescent protein palette into NIR region (+) Optimal for whole-body imaging of mammals |
2, 4, 10, 20, 23 |
| Photo- convertion wavelength and energy | Red (660–690 nm): 0.05–0.1 J/cm2; Far-red (740–760 nm): 0.025–0.1 J/cm2 | Violet-cyan (380–490 nm): up to 180 J/cm2; Orange (560–580 nm): up to 1.6 J/cm2 | (+) Easier photoconversion in deep-tissue samples | 51–53 |
| Quantum yield | Low | High | (−) Low brightness may limit single- molecule imaging applications | 10, 11, 20 |
| Extinction coefficient | High | Moderate | (+) Optimal for optoacoustic imaging (+) Preferable FRET acceptors for red GFP-like FPs |
2, 11, 12, 23 |
The upper value of the emission maxima is estimated based on the BphP absorbance spectra.
Table 2.
The proposed modifications and mutations of the photosensory module of BphPs to achieve specific photochemical effect or biochemical function.
| Phenotype | Template | Modification and Mutations | Effect or Function | Ref |
|---|---|---|---|---|
| Fluorescent proteins and chromoproteins | ||||
| Short NIR | PAS-GAF or PAS- GAF-PHY domains | Truncation of PHY domain; Truncation of up to two amino acids before Cys12; 194A,H,K,L,S; 247A | Stabilization of the chromophore in the Pr state with disabling of Pr→Pfr photoconvertion | 32, 38, 41 |
| 194A,H,K,L,S; 250F; 277Q | Increase in quantum yield | 12, 41, 42 | ||
| 163H, 185L, 195D, 459A, 453A, 277A,Q | Stabilization of the Pr state with limited/reduced photoconversion | 32, 33, 38, 41 | ||
| Long NIR | PAS-GAF-PHY domains of bathy BphPs | 261A | Stabilization of the Pfr state with disabling Pfr→Pr photoconvertion | 38 |
| 163A; 241A; 275A | Stabilization of the Pfr state with reducing Pfr→Pr photoconvertion | 38 | ||
| PS and PA NIR (switching on) | PAS-GAF-PHY domains | 188L; 275A; 190A; 163H; 250F | Decreasing rate of Pr→Pfr dark reversion (from minutes to hours) | 33, 38 |
| 241A; 163A | Increasing rate of Pr →Pfr dark reversion (faster than 3 min) | 38 | ||
| PA NIR (switching off) | PAS-GAF domains | 194A,T,Q; 260A,S | Reversible bleaching of Pr state with no photoconversion to Pfr state | 41 |
| Monomeric | PAS-GAF or PAS- GAF-PHY domains | 131S; 295E; 298D,K; 301D,R; 305R | Disruption of the dimer interface | 11, 12 |
| Biosensors | ||||
| Redox sensor | Optimized BphP- derived FPs | Residues located in close proximity to the thioether linkage between BV and apoprotein | Catalyzing thioether bond formation and influencing its reactivity | 41, 58 |
| Metal sensor | PAS-GAF domains | Truncation of PHY domain | Increasing solvent access to chromophore | 13, 32, 34, 59 |
| Residues within 4.5Å from the chromophore | Improving interactions between metal ion and chromophore | |||
| Split and insertion based sensors * | Optimized BphP- derived FPs | Split/insertion between 112–119 amino acid residues | Unstructured linker between PAS and GAF domains | 32–34, 38, 60, 61 |
| Varying the linkers between PAS domain and sensing moiety, and GAF and sensing moiety | Optimization of PAS and GAF domains collocation for their better interactions | |||
| Optogenetic tools | ||||
| Optogenetic tools with different effector modules | PAS-GAF-PHY domains of BphP and a knowledge-based chosen effector module | Varying the α-helix linker between photosensor and effector modules | Ability of light signal propagation to effector | 18, 62, 63 |
| Point mutations in the α-helix linker and PAS domain | Efficiency of light signal propagation to effector | 64 | ||
| 188L; 275A; 190A; 163H; 250F; 241A; 163A | Optimization of photopreception | 32, 33, 38, 47 | ||
Structure of the PAS-GAF domains contains a 4-crossover knot that may complicate reconstitution of a split protein. Residues at the indicated positions provide the respective phenotype in concerted manner or independently. Residue numbering follows that for PaBphP. See Fig. 3 for the amino acid alignment of several BphPs.
Data on modulation of the rate and efficiency of BphP photoisomerization and/or dark reversion between Pr and Pfr states by amino acid substitutions suggest that it is possible to design reversible PA and PS FPs32, 33, 38, 41. This has recently been demonstrated for a cyanobacteria phytochrome, which was developed into the photoswitchable protein called RGS, although it is not a NIR FP25. Moreover, the ability to independently affect the Pr→Pfr and Pfr→Pr photoconversion rates and the rate of dark reversion may result in different PS FP properties. Amino acid residues affecting quantum yield, Pr→Pfr photoisomerization, and dark reversion can be subjected to random mutagenesis in order to select PA and PS NIR FPs (Table 2). Because of the different chromophore photoconversion mechanisms, the excitation light intensities for photoswitchable BphP-based NIR FPs will likely be substantially lower than those required for the photoswitchable GFP-like FPs. Furthermore, BphP mutants that reversibly decrease (switch off) absorbance in red light without photoisomerization into the Pfr state may be precursors for NIR-to-dark PS FPs (Table 2)11, 41.
Monomerization of BphP-derived FPs may require substitution of a few amino acids11, 12 and could result in NIR FPs for protein tagging (Table 2). BphP-derived CPs exhibiting high extinction coefficients could be useful for photoacoustic imaging23. PA and PS NIR FPs will enable imaging of dynamic processes in whole mammals. These FPs can be turned on in selected locations but otherwise remain undetectable. Photoactivatable fluorescent probes improved the achievable signal-to-background ratio54 and enabled visualization of metastasis originated from cancer cells photoactivated in the primary tumor53, 55. Lastly, the ability of BphPs to emit NIR fluorescence upon excitation in the Soret band makes them attractive templates for probes utilized in stimulated emission depletion (STED)56 microscopy with a single laser for excitation and emission depletion57.
Biosensors
Numerous genetically-encoded fluorescent biosensors, mainly based on GFP-like FPs, have been developed to monitor intracellular environment, enzymatic activities, protein interactions, and intracellular metabolites65. Their excitation and emission wavelengths lay outside of the NIR window, thus, limiting their use deep in mammalian tissues. However, several types of NIR biosensors could be engineered by taking advantage of the multidomain organization of BphPs and the possibility to modulate their spectral properties by altering the BV chromophore directly or by changing the protein tertiary structure. These biosensors include, but are not limited to detection of redox potential or metal ions, as well as protein-protein interactions and analytes using split- or insertion-based design (Fig. 4b). The only BphP-based biosensor available now senses mercury ions13.
Analysis of chemical properties of BV and BphPs suggests that the PAS-GAF domains could serve as optical biosensors for redox potential and metal ions. Possible mechanism of redox sensing is based on the two reversible reactions (Fig. 4b). The first reaction is an attachment of BV to an apoprotein. It has been shown that the chromophore binding in phytochromes can be reversible66. The second reaction is the formation of a disulfide bond, which can prevent the chromophore attachment to the apoprotein. In order to engineer redox sensors, amino acid residues surrounding the thioether bond between BV and the apoprotein should be primary targets for mutagenesis in BphP-derived FPs (Table 2). Insertion of an additional Cys into a close proximity to the Cys residue that binds BV may be necessary.
Linear tetrapyrroles can coordinate to some physiologically important metal ions. For example, BV can form stable chelate complexes with Zn(II), Cu(II), Cd(II) and Mn(III) due to the coordination of the metal ion to the doubly NH-deprotonated ligand of the pyrrole rings of the chromophore59. Interaction of the metal ions with BV alter its spectral characteristics and can result in its bright fluorescence67. It has been shown for other linear tetrapyrroles that metal ions can enhance and shift their fluorescence emission68–70. Possibly, formation of metal complexes would occur with BV bound to mutated BphP apoprtotein variants, which exhibit some room in the chromophore-binding pocket for a metal ion. Therefore, non-fluorescent PAS-GAF domains and CPs could be the primary templates because coordination of metal ions typically decreases the flexibility of a chromophore, thus increasing its quantum yield. Truncation of the PHY domain may be required to facilitate an access of the metal ion from solvent to the chromophore (Table 2). Optimization of the sensors to biologically relevant subnanomolar ranges of ions should be performed.
According to structural data32–34, a disordered linker between the PAS and GAF domains might be the preferable location for polypeptide breakage or insertion of sensing moieties to design split- and insertion-based biosensors, respectively (Fig. 4b). It should be noted, however, that all PAS-GAF pairs have a unique 4-crossover knot, which may complicate protein reconstitution. Once the right position to make a split or add an insertion is determined, the next step is the optimization of linkers between the PAS and GAF domains and the fused sensing moieties60, 61. A reversibility of fluorescence resulting from association-dissociation of the sensing moieties in biosensors remains to be studied. It is likely that both monomeric and dimeric versions of BphP-derived FPs are suitable for engineering split and insertion biosensors. Development of BphP-based NIR biosensors will enable in vivo tracking of protein-protein interactions and analyte detection in whole-body imaging.
Optogenetic tools
Optogenetics enables control of biological processes by light in mammalian cells and tissues. Heterologous expression of light-sensitive proteins, such as rhodopsins and flavin-binding proteins (Fig. 1), is used to achieve precise light-controlled stimulation or silencing of neurons16, light activation of enzymes18, and induction of protein heterodimerization19, among many other applications. For example, the activation wavelengths of currently available rhodopsin-based optogenetic tools are limited to ~630 nm16, which is beyond the NIR tissue transparency window. NIR optogentic constructs will allow non-invasive light manipulations of physiology and behavior in animals directly via skin without surgical intervention.
BphPs have not yet been employed as optogenetic tools, however, the PCM possesses all of the necessary features for such a design. An existence in nature of non-canonical BphPs is a good evidence that the typical effector domain HisK can be substituted by others enzymes and motifs. The effector domains are always located at the C-terminus of the PCM. A linker between the PCM and effector domains plays a crucial role in signal transduction and typically consists of an α-helix. A PCM mutagenesis strongly affects signal propagation to the effector domain and photoperception. The latter property is important for optimization of optogenetic constructs due to its strong influence on the lifetime of the effector’s signaling state and its resultant modulation of their light sensitivity.
Several design approaches can be suggested on the basis of the aforementioned properties (Fig. 4c). An overall strategy to engineer optogenetic tools would involve several steps. First, a choice of the appropriate effector domain should be based on the structural and functional mechanisms of its biological activity. Second, the α-helix linker of an optimal length between the PCM and effector domains should be designed with respect to their structures to avoid steric restrictions. Third, an introduction of point mutations into the PCM and the linker can further modulate light sensitivity and effector activity in the ground and signaling states of the chromophore47, 64. For example, in LOV (Light-Oxygen-Voltage) proteins substitutions of residues in chromophore binding site substantially affected the photoadduct lifetimes, thus changing their photoperception47. In plant phytochrome PhyB mutations in the PAS domain interrupted the light signal transfer but did not cause substantial changes in spectral properties and photoperception64. Single-domain enzymes, channels, and DNA binding proteins could be suggested as the putative effector domains (Fig. 4c).
An adaptation of examples in which other phytochromes were successfully utilized in optogenetic tools can facilitate design of the BphP-based constructs. For example, a fusion of PCM of phytochrome from cyanobacteria, Cph1 and bacterial histidine kinase, EnvZ was engineered to achieve gene expression induced by red light18. The light response of the Cph1-EnvZ chimaeras was optimized by varying a linker length between the PCM-Chp1 and EnvZ domains. The Cph1-EnvZ variants exhibited a graded response to increasing light intensity. Another system, based on a red-light regulated interaction between PhyB and PIF (Phytochrome Interaction Factor), was used to control gene expression and translocation of target proteins within a cell19, 71. Fusing the PhyB and PIF to two halves of a protein (or two proteins) via an yeast split ATPase-derived intein enabled the rapid light-activated production of the spliced protein (or the two-protein chimera)72. Activation of WASP (Wiskott Aldrich Syndrome Protein) by Cdc42 GTPase mediated by the PhyB-PIF interaction allowed the light-controlled actin assembly in a cell73. Although yet to be applied in vivo, these examples demonstrate the versatility of phytochromes to design optogenetic tools.
A possible limitation to development of BphP-based optogenetic tools is a relatively low level of HisK activation in phytochromes. Another drawback is a lack of the structural information on the signal transduction from photosensor to effector domain. Regulating biochemical processes with NIR light using various optogenetic constructs will provide new insights into tissue physiology and behavior of mammals.
Experimental considerations
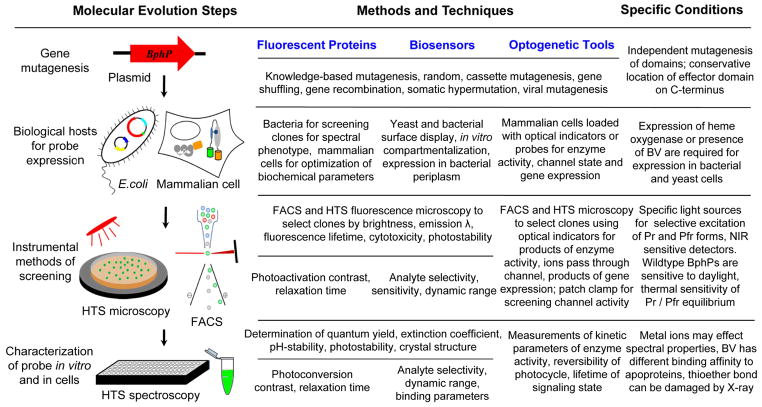
Engineering BphP-based probes with new properties requires advanced methods for directed evolution including generation of libraries of mutants, new hosts for protein expression, and enhanced protein screening and characterization techniques. The molecular evolution approaches used in engineering of advanced GFP-like FPs74 can, to a large extent, be applied to the development of BphP-based probes too. However, several specific properties of BphPs should be considered to design BphP-based FPs, biosensors and optogenetic constructs (Fig. 5).
Fig. 5.
Molecular evolution steps, methods and techniques, and specific conditions in the course of development of the BphP-based NIR fluorescent proteins, biosensors, and optogenetic tools. Vertical arrows indicate the typical order of the evolution steps such as gene construction and mutagenesis, biological hosts for protein expression, instrumental methods of screening, protein characterization in vitro and in cells. Methods and techniques proposed for each molecular evolution step are subdivided per the proposed NIR probes. Specific conditions indicate particular qualities of BphPs that should be considered for each directed evolution step. HTS is a high-throughput screening, FACS is a fluorescence-activated cell sorter, and λ is a wavelength. See also Table 2 for details on knowledge-based mutagenesis.
Each BphP domain can be subjected to mutagenesis individually, allowing independent modifications of specific PCM properties (Table 2). Biological hosts for BphP production, such as E. coli and yeast, require co-expression of heme oxygenase for BV synthesis10, 21 (Fig. 5). The internal membrane of E. coli is not permeable to BV, and therefore, heme oxygenase expression is required to synthesize BV in intact bacteria10, 11. The expression systems typically produce a large amount of recombinant BphPs that permits their mutants to be screened in both low- and high-throughput formats10, 11. However, in contrast to BphP-derived FPs, screening for BphP-based biosensors may require modified bacterial and yeast systems. For example, recently reported periplasm targeted expression in E. coli could enable screening of large libraries of BphP biosensors75. The outer membrane of bacteria is easily permeable to metal ions and low molecular weight compounds, thus allowing manipulation of analyte concentration for efficient clone selection. A rapid linker optimization for split and insertion BphP variants can be achieved using a histone methylation-based system adopted for screening in E. coli colonies76.
Although endogenous BV is ubiquitous in mammalian cells at a submicromolar level10, 11, certain applications may demand higher incorporation rates, necessitating artificially raised BV levels. In such cases, BV concentrations may be increased by supplying it exogenously to cell culture as the membranes of mammalian cells are permeable to BV and many other compounds10, 11. The latter property makes mammalian cells advantageous for biosensor screening. For example, the mammalian cell-based system employing printing plasmid DNA arrays and subsequent imaging reversely transfected cells can be applied to optimize BphP-derived biosensors77.
Development of BphP-based optogenetic tools may require expression systems that depend on the origins of effector domains. Moreover, the biological hosts should be compatible with the proposed system for clone selection. Screening systems for directed evolution of BphP-based optogenetic tools remain to be tested, leaving several possible modes of action. Use of colored substrates to report activity of an effector domain fused to the PCM could be one approach. For example, to screen for activity of the Cph1-EnvZ fusion variants18, the S-gal substrate that is converted into black precipitate by LacZ was used. The selection criterion was the black-white contrast between the illuminated and non-illuminated areas of the bacterial film on a Petri dish18. Biological hosts expressing or loaded with optical sensors for enzyme activity and metal ions could facilitate screening of enzyme- and channel-based optogenetic constructs. Screening could employ conventional FP reporters whose expression is controlled by a promoter regulated by light-sensitive DNA binding constructs. Another screening approach could be the phenotypic changes of organism expressing optogenetic constructs under different intensities and wavelengths of light, as it has been shown for hypocotyls elongation and photo-morphogenesis in Arabidopsis26, 64, 78. Lastly, FPs could be fused to optogenetic probes and act as a fluorescence resonance energy transfer (FRET) donor whose fluorescence is modulated upon absorbance changes of the fused probe, corresponding to its activity state79.
Instrumentation and procedures used for directed evolution of GFP-like FPs may require modifications to be suitable for screening of BphP-based NIR probes and optogenetic tools (Fig. 5). Absorbance and emission of BphPs may need specific light sources for selective excitation of Pr and Pfr forms as well as detectors sensitive to NIR fluorescence. Light-emitting diodes, which are currently available in a wide range of wavelengths and output powers80, are good alternatives to traditional light sources based on arc lamps, which often provide insufficient power above 700 nm due to infrared cut-off filters in the output light path (http://zeiss-campus.magnet.fsu.edu/articles/lightsources/). Applications of light-emitting diodes with narrow emission spectra enable selective excitation and allow the omission of excitation filters for screening of mutant clones. It is also advisable to use CCD cameras with high sensitivity in the NIR range or remove the infrared cut-off filter frequently installed in scientific CCD cameras to detect fluorescence.
Natural sensitivity of BphPs to daylight is an important variable in screening BphP-based probes and constructs38. Experiments should be performed using a blue-green safelight (460–560 nm) or in the dark to ascertain ground (dark relaxed) and photoconverted states22, 81. Since the Pr↔Pfr equilibrium is sensitive to temperature the spectral properties and biological activities of the BphP-derived constructs may vary substantially at different temperatures81. It is also important to avoid artifacts during protein purification and characterization. First, in commonly used in metal-affinity purification procedures imidazole can compete with BV for binding to apoproteins66. Second, certain metal ions can affect BphP brightness13 and spectral properties59, 67. Third, a Cys24 SH-group responsible for BV attachment can be easily oxidized and thereby lose its ability to form a thioester bond. Fourth, the thioether bond is typically sensitive to radiation; thus, gentle X-ray data collection from BphP crystals may prevent artifacts in determination of the crystal structures12, 34. Lastly, the BphP apoproteins have different BV binding affinities10, 11, which can strongly affect values of their extinction coefficient determined at various BV concentrations. It should also be mentioned that the BphP apoproteins can efficiently bind BV added in pure form to solution12, 22, 40, 46, thus, demonstrating the versatility of BphPs. This property allows the determination of the kinetic and thermodynamic parameters of the BV-apoprotein interaction in vitro.
Conclusions
Use of BphPs as templates will allow the development of FPs, biosensors, and optogenetic elements that emit or are activated in NIR and utilize the BV chromophore, abundant in mammalian tissues. These probes will avoid autofluorescence in live cells, but more importantly also in vivo, due to tissue transparency in NIR. NIR FPs and biosensors will extend the methods developed for conventional microscopy into a deep-tissue in vivo “macroscopy” including multicolor cell and tissue labeling, FRET, cell photoactivation and tracking, and detection of enzymatic activities and metabolites in tissues. The NIR optogenetic tools will allow noninvasive light-manipulation of biochemistry and physiology of a living mammal directly through the skin.
Availability of the BphP-derived probes will further stimulate the development of novel in vivo detection and light-manipulation technologies. Once BphP-based tools are available, future efforts will include optimization of strategies for gene delivery to specific cells and tissues in vivo, design of targeted noninvasive illumination, and refining optical readouts. Overall this will result in a wide range of noninvasive studies of chemical and metabolic status, as well as molecular and cellular interactions in intact tissues and whole living mammals.
Acknowledgments
This work was supported by grants GM073913, CA164468, and EB013571 from the US National Institutes of Health.
Major abbreviations used
- BphP
bacterial phytochrome photoreceptor
- BV
biliverdin IXα
- CP
chromoprotein
- FP
fluorescent protein
- FRET
fluorescence resonance energy transfer
- GAF
cGMP phosphodiesterase/adenylate cyclase/FhlA transcriptional activator
- HisK
histidine kinase
- HO
heme oxygenase
- NIR
near-infrared
- PA
photoactivatable
- PAS
Per-ARNT-Sim repeats
- PCM
photosensory core module
- PHY
phytochrome-specific domain
- PS
photoswitchable
References
- 1.van der Horst MA, Hellingwerf KJ. Acc Chem Res. 2004;37:13–20. doi: 10.1021/ar020219d. [DOI] [PubMed] [Google Scholar]
- 2.Wu B, Piatkevich KD, Lionnet T, Singer RH, Verkhusha VV. Curr Opin Cell Biol. 2011;23:310–317. doi: 10.1016/j.ceb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Subach FV, Verkhusha VV. Chem Rev. 2012;112:4308–4327. doi: 10.1021/cr2001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piatkevich KD, Verkhusha VV. Curr Opin Chem Biol. 2010;14:23–29. doi: 10.1016/j.cbpa.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomelsky M, Hoff WD. Trends Microbiol. 2011;19:441–448. doi: 10.1016/j.tim.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Rockwell NC, Lagarias JC. Chemphyschem. 2010;11:1172–1180. doi: 10.1002/cphc.200900894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulijasz AT, Vierstra RD. Curr Opin Plant Biol. 2011;14:498–506. doi: 10.1016/j.pbi.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, Tsien RY. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie JM, Hitomi K, Arvai AS, Hartfield KA, Mettlen M, Pratt AJ, Tainer JA, Getzoff ED. J Biol Chem. 2012;287:22295–22304. doi: 10.1074/jbc.M111.318881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha VV. Nat Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auldridge ME, Satyshur KA, Anstrom DM, Forest KT. J Biol Chem. 2012;287:7000–7009. doi: 10.1074/jbc.M111.295121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu Z, Zhao M, Sheng Y, Bentolila LA, Tang Y. Anal Chem. 2011;83:2324–2329. doi: 10.1021/ac103236g. [DOI] [PubMed] [Google Scholar]
- 14.Strickland D, Yao X, Gawlak G, Rosen MK, Gardner KH, Sosnick TR. Nat Methods. 2010;7:623–626. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Nat Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattis J, Tye KM, Ferenczi EA, Ramakrishnan C, O’Shea DJ, Prakash R, Gunaydin LA, Hyun M, Fenno LE, Gradinaru V, Yizhar O, Deisseroth K. Nat Methods. 2012;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 18.Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, Voigt CA. Nature. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- 19.Levskaya A, Weiner OD, Lim WA, Voigt CA. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auldridge ME, Forest KT. Crit Rev Biochem Mol Biol. 2011;46:67–88. doi: 10.3109/10409238.2010.546389. [DOI] [PubMed] [Google Scholar]
- 21.Giraud E, Vermeglio A. Photosynth Res. 2008;97:141–153. doi: 10.1007/s11120-008-9323-0. [DOI] [PubMed] [Google Scholar]
- 22.Rottwinkel G, Oberpichler I, Lamparter T. J Bacteriol. 2010;192:5124–5133. doi: 10.1128/JB.00672-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filonov GS, Krumholz A, Xia J, Yao J, Wang LV, Verkhusha VV. Angew Chem Int Ed Engl. 2011 doi: 10.1002/anie.201107026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer AJ, Rockwell NC, Jang AY, Ernst LA, Waggoner AS, Duan Y, Lei H, Lagarias JC. Biochemistry. 2005;44:15203–15215. doi: 10.1021/bi051633z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Wu XJ, Wang ZB, Chen Y, Wang X, Zhou M, Scheer H, Zhao KH. Angew Chem Int Ed Engl. 2010;49:5456–5458. doi: 10.1002/anie.201001094. [DOI] [PubMed] [Google Scholar]
- 26.Su YS, Lagarias JC. Plant Cell. 2007;19:2124–2139. doi: 10.1105/tpc.107.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konig K. Journal of microscopy. 2000;200:83–104. doi: 10.1046/j.1365-2818.2000.00738.x. [DOI] [PubMed] [Google Scholar]
- 28.Moglich A, Moffat K. Photochem Photobiol Sci. 2010;9:1286–1300. doi: 10.1039/c0pp00167h. [DOI] [PubMed] [Google Scholar]
- 29.Vierstra RD, Zhang J. Trends Plant Sci. 2011;16:417–426. doi: 10.1016/j.tplants.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Nagatani A. Curr Opin Plant Biol. 2010;13:565–570. doi: 10.1016/j.pbi.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Sharrock RA. Genome Biol. 2008;9:230. doi: 10.1186/gb-2008-9-8-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang X, Kuk J, Moffat K. Proc Natl Acad Sci U S A. 2008;105:14715–14720. doi: 10.1073/pnas.0806718105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X, Stojkovic EA, Kuk J, Moffat K. Proc Natl Acad Sci U S A. 2007;104:12571–12576. doi: 10.1073/pnas.0701737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner JR, Zhang J, Brunzelle JS, Vierstra RD, Forest KT. J Biol Chem. 2007;282:12298–12309. doi: 10.1074/jbc.M611824200. [DOI] [PubMed] [Google Scholar]
- 35.Giraud E, Fardoux J, Fourrier N, Hannibal L, Genty B, Bouyer P, Dreyfus B, Vermeglio A. Nature. 2002;417:202–205. doi: 10.1038/417202a. [DOI] [PubMed] [Google Scholar]
- 36.Bellini D, Papiz MZ. Structure. 2012;20:1436–1446. doi: 10.1016/j.str.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Tarutina M, Ryjenkov DA, Gomelsky M. J Biol Chem. 2006;281:34751–34758. doi: 10.1074/jbc.M604819200. [DOI] [PubMed] [Google Scholar]
- 38.Yang X, Kuk J, Moffat K. Proc Natl Acad Sci U S A. 2009;106:15639–15644. doi: 10.1073/pnas.0902178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Zhang J, Vierstra RD. Proc Natl Acad Sci U S A. 2010;107:10872–10877. doi: 10.1073/pnas.1001908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Murphy JT, Lagarias JC. Biochemistry. 1995;34:7923–7930. doi: 10.1021/bi00024a017. [DOI] [PubMed] [Google Scholar]
- 41.Wagner JR, Zhang J, von Stetten D, Gunther M, Murgida DH, Mroginski MA, Walker JM, Forest KT, Hildebrandt P, Vierstra RD. J Biol Chem. 2008;283:12212–12226. doi: 10.1074/jbc.M709355200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toh KC, Stojkovic EA, van Stokkum IH, Moffat K, Kennis JT. Phys Chem Chem Phys. 2011;13:11985–11997. doi: 10.1039/c1cp00050k. [DOI] [PubMed] [Google Scholar]
- 43.Toh KC, Stojkovic EA, van Stokkum IH, Moffat K, Kennis JT. Proc Natl Acad Sci U S A. 2010;107:9170–9175. doi: 10.1073/pnas.0911535107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margulies L, Stockburger M. Journal of the American Chemical Society. 1979;101:743–744. [Google Scholar]
- 45.Braslavsky SE, Holzwarth AR, Lehner H, Schaffner K. Helvetica Chimica Acta. 1978;61:2219–2222. [Google Scholar]
- 46.Karniol B, Vierstra RD. Proc Natl Acad Sci U S A. 2003;100:2807–2812. doi: 10.1073/pnas.0437914100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zoltowski BD, Vaccaro B, Crane BR. Nat Chem Biol. 2009;5:827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Stetten D, Seibeck S, Michael N, Scheerer P, Mroginski MA, Murgida DH, Krauss N, Heyn MP, Hildebrandt P, Borucki B, Lamparter T. J Biol Chem. 2007;282:2116–2123. doi: 10.1074/jbc.M608878200. [DOI] [PubMed] [Google Scholar]
- 49.Fischer AJ, Lagarias JC. Proc Natl Acad Sci U S A. 2004;101:17334–17339. doi: 10.1073/pnas.0407645101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah R, Schwach J, Frankenberg-Dinkel N, Gartner W. Photochem Photobiol Sci. 2012;11:1026–1031. doi: 10.1039/c2pp05374h. [DOI] [PubMed] [Google Scholar]
- 51.Subach FV, Patterson GH, Renz M, Lippincott-Schwartz J, Verkhusha VV. Journal of the American Chemical Society. 2010;132:6481–6491. doi: 10.1021/ja100906g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subach FV, Zhang L, Gadella TW, Gurskaya NG, Lukyanov KA, Verkhusha VV. Chemistry & biology. 2010;17:745–755. doi: 10.1016/j.chembiol.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subach OM, Patterson GH, Ting LM, Wang Y, Condeelis JS, Verkhusha VV. Nat Methods. 2011;8:771–777. doi: 10.1038/nmeth.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marriott G, Mao S, Sakata T, Ran J, Jackson DK, Petchprayoon C, Gomez TJ, Warp E, Tulyathan O, Aaron HL, 5YIsacoff E, Yan Y. Proc Natl Acad Sci U S A. 2008;105:17789–17794. doi: 10.1073/pnas.0808882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kedrin D, Gligorijevic B, Wyckoff J, Verkhusha VV, Condeelis J, Segall JE, van Rheenen J. Nat Methods. 2008;5:1019–1021. doi: 10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wildanger D, Medda R, Kastrup L, Hell SW. J Microsc. 2009;236:35–43. doi: 10.1111/j.1365-2818.2009.03188.x. [DOI] [PubMed] [Google Scholar]
- 57.Bianchini P, Harke B, Galiani S, Vicidomini G, Diaspro A. Proc Natl Acad Sci U S A. 2012;109:6390–6393. doi: 10.1073/pnas.1119129109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stevens JM, Daltrop O, Allen JW, Ferguson SJ. Acc Chem Res. 2004;37:999–1007. doi: 10.1021/ar030266l. [DOI] [PubMed] [Google Scholar]
- 59.Goncharova I, Urbanova M. Anal Biochem. 2009;392:28–36. doi: 10.1016/j.ab.2009.05.040. [DOI] [PubMed] [Google Scholar]
- 60.Kerppola TK. Nat Protoc. 2006;1:1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Souslova EA, Belousov VV, Lock JG, Stromblad S, Kasparov S, Bolshakov AP, Pinelis VG, Labas YA, Lukyanov S, Mayr LM, Chudakov DM. BMC Biotechnol. 2007;7:37. doi: 10.1186/1472-6750-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krauss U, Lee J, Benkovic SJ, Jaeger KE. Microb Biotechnol. 2010;3:15–23. doi: 10.1111/j.1751-7915.2009.00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohlendorf R, Vidavski RR, Eldar A, Moffat K, Moglich A. J Mol Biol. 2012 doi: 10.1016/j.jmb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 64.Oka Y, Matsushita T, Mochizuki N, Quail PH, Nagatani A. PLoS Genet. 2008;4:e1000158. doi: 10.1371/journal.pgen.1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Physiol Rev. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 66.Quest B, Gartner W. Eur J Biochem. 2004;271:1117–1126. doi: 10.1111/j.1432-1033.2004.04015.x. [DOI] [PubMed] [Google Scholar]
- 67.Petrier C, Dupuy C, Jardon P, Gautron R. Photochemistry and Photobiology. 1979;29:389–392. [Google Scholar]
- 68.Chowdhury MH, Ray K, Aslan K, Lakowicz JR, Geddes CD. Journal of Physical Chemistry C. 2007;111:18856–18863. doi: 10.1021/jp0731250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berkelman TR, Lagarias JC. Analytical Biochemistry. 1986;156:194–201. doi: 10.1016/0003-2697(86)90173-9. [DOI] [PubMed] [Google Scholar]
- 70.Van Norman JD, Yatsko ET. Bioinorganic Chemistry. 1978;9:349–353. [Google Scholar]
- 71.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. Nature Biotechnology. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 72.Tyszkiewicz AB, Muir TW. Nature Methods. 2008 doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]
- 73.Leung DW, Otomo C, Chory J, Rosen MK. Proceedings of the National Academy of Sciences. 2008;105:12797–12802. doi: 10.1073/pnas.0801232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Subach FV, Piatkevich KD, Verkhusha VV. Nat Methods. 2011;8:1019–1026. doi: 10.1038/nmeth.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, Campbell RE. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ibraheem A, Yap H, Ding Y, Campbell RE. BMC Biotechnol. 2011;11:105. doi: 10.1186/1472-6750-11-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Piljic A, de Diego I, Wilmanns M, Schultz C. ACS Chem Biol. 2011;6:685–691. doi: 10.1021/cb100402n. [DOI] [PubMed] [Google Scholar]
- 78.Wang X, Roig-Villanova I, Khan S, Shanahan H, Quail PH, Martinez-Garcia JF, Devlin PF. J Exp Bot. 2011;62:2973–2987. doi: 10.1093/jxb/err062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bayraktar H, Fields AP, Kralj JM, Spudich JL, Rothschild KJ, Cohen AE. Photochemistry and Photobiology. 2012;88:90–97. doi: 10.1111/j.1751-1097.2011.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wessels JT, Pliquett U, Wouters FS. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2012;81:188–197. doi: 10.1002/cyto.a.22023. [DOI] [PubMed] [Google Scholar]
- 81.Njimona I, Lamparter T. PLoS One. 2011;6:e25977. doi: 10.1371/journal.pone.0025977. [DOI] [PMC free article] [PubMed] [Google Scholar]