Abstract
Actin has well-established functions in the cytoplasm, but its roles in the nucleus remain poorly defined. Here, by studying the nuclear actin-containing yeast INO80 chromatin remodeling complex, we provide genetic and biochemical evidence for a role of monomeric actin in INO80 chromatin remodeling. In contrast to cytoplasmic actin, nuclear actin is present as a monomer in the INO80 complex and its barbed end is not accessible for polymerization. An actin mutation affecting in vivo nuclear functions is identified in subdomain 2, which reduces the chromatin remodeling activities of the INO80 complex in vitro. Importantly, the highly conserved subdomain 2 at the pointed end of actin contributes to INO80 interactions with chromatin. Our results establish an evolutionarily conserved function of nuclear actin in its monomeric form and suggest that nuclear actin can utilize a fundamentally distinct mechanism from cytoplasmic actin.
Introduction
The presence and potential functions of nuclear actin have been debated over several decades1–4. Early observations of biochemical co-purifications of actin with nuclear proteins were dismissed as contaminations of cytoplasmic actin, which is a major protein component in the cytosol. Adding to the controversy is the fact that actin cannot be detected in the nucleus by the classical actin stain, phalloidin, which binds to filamentous actin (F-actin) and decorates the extensive actin cytoskeleton. However, a number of studies from various organisms using different experimental approaches continue to suggest that actin is indeed present in the nucleus, and several lines of evidence strongly argue for its presence. First, actin can be detected in the cleanly separated Xenopus oocyte nuclei5. Second, actin can be detected in the nucleus using newly developed monoclonal actin antibodies, which recognizes only the monomeric actin (G-actin)6. Third, nuclear export signals (NES1 and NES2) have been identified for actin, which when mutated, leads to the nuclear accumulation of actin7. Finally, recent studies indicate that actin is a subunit of a number of chromatin modifying complexes, including ATP-dependent chromatin remodeling complexes and histone acetyltransferase complexes, which are all nuclear complexes8–13. These studies demonstrate that at least a fraction of actin can be found in the cell nucleus.
The presence of actin in the nucleus raises the question of its potential function. Early biochemical studies suggested that actin may be involved in transcription by RNA polymerase II14, while more recent evidence has implicated actin in transcription by all the three classes of RNA polymerases15–18. Moreover, actin dynamics also play an important role in the regulation of transcription factors, such as the serum response factor (SRF)19. Furthermore, in the actin-containing BAF chromatin remodeling complex, it has been shown that actin is a stable subunit of the complex13, and that the BAF complex can also interact with actin filaments in vitro20. Despite this emerging evidence, the functions of nuclear actin remained unclear, largely due to the lack of a defined biochemical and genetic system, in which the function of nuclear actin can be unambiguously demonstrated1. For example, the presence of multiple actin isoforms makes genetic studies of actin function in higher organisms difficult and complicates the interpretations of existing studies.
To address the function of nuclear actin in this study, we employed the yeast actin-containing INO80 chromatin-remodeling complex as a model system. We demonstrate that actin is a part of an evolutionarily conserved module, which consists of actin and actin-related proteins (Arps) within the INO80 complex. Taking advantage of the presence of a single actin gene (ACT1) in the yeast genome, we show that a specific mutation in actin causes defects in nuclear functions of actin in vivo and INO80 chromatin remodeling in vitro. Mechanistically, we observed that nuclear actin in the INO80 complex functions as a monomer and contributes to INO80 chromatin remodeling through its subdomain 2. Given the evolutionary conservation of actin and the INO80 complex, these findings reveal that actin in the nucleus has fundamental and conserved functions in addition to its well established functions in the cytoplasm. Intriguingly, nuclear actin may be utilized in mechanistically distinct ways from its cytoplasmic counterpart.
Results
Actin and Arps form a sub-complex within the INO80 complex
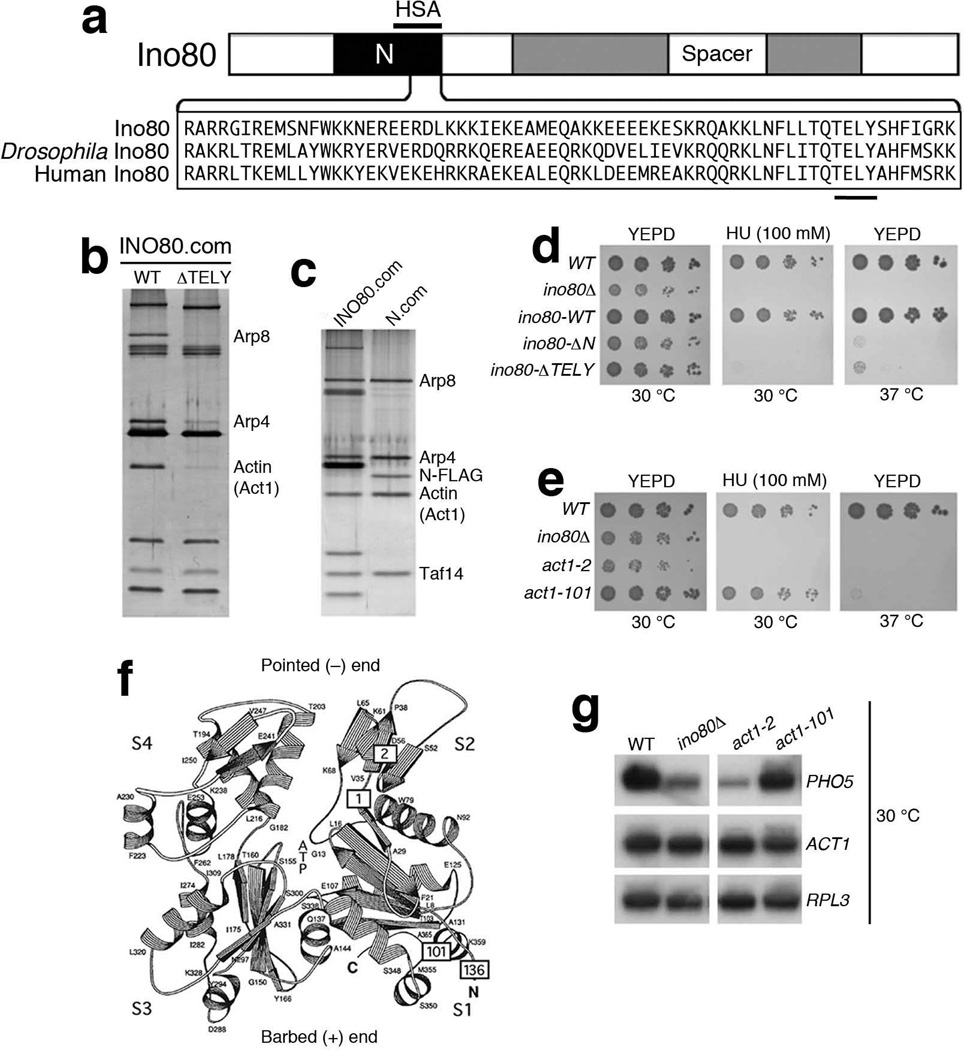
Mammalian BAF, and Drosophila BAP chromatin remodeling complexes were initially identified as containing an actin subunit11,13. In yeast, there are three known actin-containing chromatin modifying complexes - the INO80 and SWR1 chromatin remodeling complexes and the NuA4 histone acetyltransferase complex8,10,12. Using glycerol gradient centrifugation, high stringency purifications, as well as immuno-affinity purification with additional INO80 subunits, we confirmed that actin is a highly stable subunit of the INO80 complex (Supplementary Fig. 1). The N-terminal region of Ino80 core ATPase subunit contains the HSA domain, which was shown to be a platform to recruit actin and Arps in chromatin modifying complexes including INO8021. Curiously, there is a highly conserved TELY motif (amino acids 531 to 598) overlapping with HSA domain within the Ino80 ATPase and its orthologues, such as Drosophila Ino80 and human Ino80 (Fig. 1a and Supplementary Fig. 2a). Deletion of TELY motif from INO80 resulted in a substantial and specific reduction of actin, Arp4 and Arp8 from the INO80 complex, while associations of other subunits remained intact (Fig. 1b), implicating the conserved TELY motif in the recruitment of actin and specific Arps to the INO80 complex. To address whether actin and Arps form a distinct module within the INO80 complex, we tagged the N-terminal region of Ino80 containing TELY motif with the double-FLAG epitope at its C-terminus. Interestingly, FLAG-immuno-affinity purification with the tagged N-terminal region identified a sub-complex consisting of actin, Arp4, Arp8, and Taf14 (Fig. 1c). Moreover, all subunits of this actin-Arp sub-complex co-sediment as a single complex around 27% in a glycerol gradient, consistent with being a module of the INO80 complex, which sediment as a single complex at around 33% (Supplementary Fig. 1a-c). Furthermore, deletions of N-terminal region (∆N) or TELY motif (∆TELY) disrupted INO80 function in vivo (Fig. 1d), indicating that the actin-Arp module is not only an evolutionarily conserved structural entity, but also a functional module within the INO80 complex. Thus, these results established a defined biochemical system for the study of nuclear actin and Arps.
Figure 1. Analysis of the actin subunit in INO80 complex.
(a) Top shows a schematic representation of the Ino80 ATPase with the split ATPase-helicase domain (shaded) and the N-terminal region in black. Bottom shows the amino acid sequence alignment of the TELY motifs within the N-terminal region of the Ino80 ATPases from yeast (Ino80), Drosophila Ino80 and human Ino80. The conserved amino acids TELY is underlined. Region of HSA domain is marked above the N-terminal region (amino acid 496-588 from the N-terminus of Ino80). (b,c) SDS-PAGE and silver staining showing, (b) wild type and ∆TELY INO80 complexes, and (c) wild type INO80 and N-terminal (N.com) complexes. Relevant subunits are shown on the right. (d,e) Phenotypic analysis of (d) ino80 mutants lacking the N-terminal region (∆N) or the TELY motif (∆TELY) on YEPD (30°C), HU (100 mM, 30 °C) and YEPD (37 °C) plates, and (e) actin mutants on YEPD, HU (100 mM) plates at 30 °C and on a YEPD plate at 37 °C. (f) Schematic representation of the structure of actin. The four subdomains of actin are indicated by S1, S2, S3 and S4. The positions of actin mutations described in this study are indicated by boxes labeled with allele numbers: 2 for act1-2 (A58T), 1 for act1-1 (P32A), 101 for act1-101 (D363A E364A) and 136 for act1-136 (D2A). Pointed (−) end region and barbed (+) end region are marked respectively. Adapted from Kabsch et al., (1990) with permission. (g) Northern blot analysis of PHO5 and ACT1 expression in wild type (WT) and mutant strains at 30 °C after 4 hours of induction, using RPL3 as loading control.
Identification of actin mutant defective in nuclear functions
Despite growing evidence that actin is involved in multiple nuclear functions, the in vivo function of nuclear actin remains a mystery, due to the lack of genetic evidence. We reason that if actin is a functional subunit of chromatin modifying complexes, such as the INO80 complex, actin mutants that disrupt actin function in these complexes might exist.
To search for actin mutations that affect the nuclear functions of actin, we used phenotypes such as hypersensitivity to DNA-damaging agents (hydroxyurea, HU), as well as defects in the transcription of PHO5 gene as screening criteria12,22,23. We screened a collection of existing actin temperature sensitive mutants in the S288C background24,25 for defects in nuclear functions and found that only few actin mutants showed hypersensitivity to HU, or defective PHO5 transcription (unpublished observations) at permissive temperature (30 °C). Among these mutants, the act1-2 allele with an A58T mutation24, showed similar defects as an ino80 mutant (Fig. 1e). A58T occurs in subdomain 2 of actin, which is a part of the pointed end of actin (Fig. 1f and Supplementary Fig. 2b). Under permissive temperature, compared to wild type or another temperature sensitive actin mutant act1-101, which contains D363A E364A mutations in subdomain 1 at the barbed end of actin25, act1-2 and ino80 mutants were both hypersensitive to 100 mM of HU (Fig. 1e). Similarly, the activation of PHO5 in both act1-2 and ino80 mutants was markedly reduced (Fig. 1g and Supplementary Fig. 2c). Furthermore, defects of the act1-2 mutant were rescued by a plasmid expressing the wild type actin (Supplementary Fig. 2d). These nuclear defects of act1-2 were prominent at permissive temperature, when the cells continue to grow26. Thus specific nuclear defects of actin can be studied in act1-2 at permissive temperature. Moreover, the allele-specificity of actin mutants in actin-containing chromatin modifying complexes such as INO80 and NuA4 suggests that specific actin mutations may have different effects on distinct complexes (Supplementary Fig. 2e-g).
Actin is required for INO80 chromatin remodeling in vitro
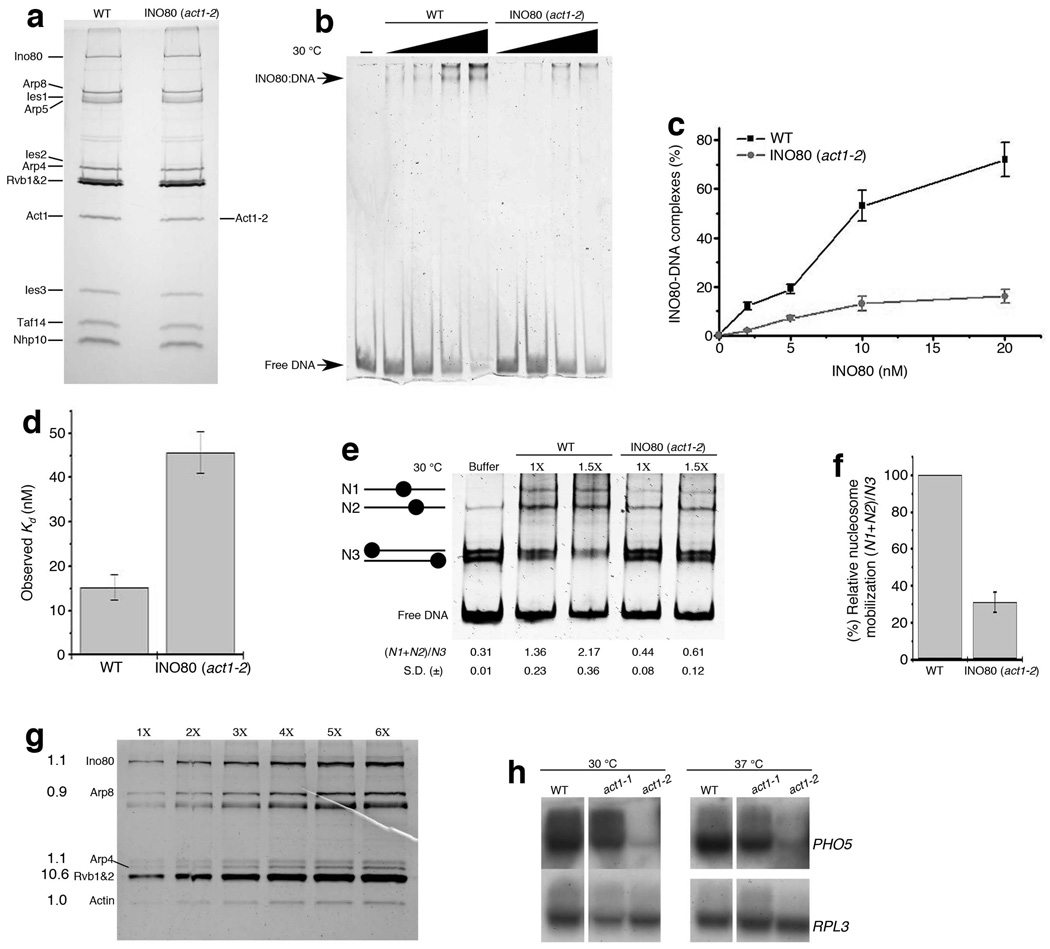
Given that the act1-2 mutation (A58T) in actin markedly affected nuclear functions in vivo and some defects overlapped with ino80 mutant (Fig. 1e), we investigated the potential contribution of actin to INO80 chromatin remodeling in vitro. To this end, we purified the INO80 complex using FLAG-immuno-affinity purification from an act1-2 mutant containing a FLAG-tagged Ino80 ATPase. The purified complex contained all the subunits as a wild type INO80 complex, except wild type actin subunit (Act1) was replaced with the mutant actin (Act1-2, A58T) (Fig. 2a), indicating that the assembly of INO80 complex was not affected by the act1-2 mutation. Therefore, changes in biochemical activities of the INO80 (act1-2) complex compared to the wild type INO80 complex would likely be attributed to the act1-2 (A58T) mutation in actin, rather than defects in other subunits. We compared the biochemical activities of the wild type and mutant INO80 (act1-2) complexes using in vitro assays that were all carried out at 30 °C, at which the nuclear defects of the act1-2 mutant were prominent (Fig. 2). The binding activity of the INO80 (act1-2) complex to free DNA (359 bp INO1 promoter DNA at 5 nM) was substantially reduced compared to that of the wild type INO80 complex (KdINO80= 7.23 nM ± S.D.=1.1 nM, KdINO80 (act1-2)= 21.01 nM ± S.D.=3.4 nM) (Fig. 2b,c), suggesting that actin plays an important role in regulating the DNA-binding activity of the INO80 complex.
Figure 2. Actin contributes to INO80 chromatin remodeling.
(a) SDS-PAGE and silver staining showing wild type (WT) and mutant (act1-2) INO80 complexes. (b) Native PAGE showing 359 bp INO1 DNA (5 nM) in the presence of increasing equimolar concentrations of WT and act1-2 INO80 complexes from 2 nM to 20 nM at 30 °C. (c,d) Graph showing (c) % DNA bound to WT and act1-2 INO80 complexes as shown in (b), data presented is the mean of five independent experiments ± S.D., (d) Kd values for mono-nucleosome binding of WT and act1-2 INO80 complexes using nucleosomes with linker DNA (207 Nuc), assessed using gel shift assay, data presented is the mean of five independent experiments ± S.D. (e) Native PAGE showing nucleosome mobilization by wild type and mutant INO80 complexes (1× equals 5 nM) at 30 °C using the INO1 mono-nucleosome substrate (5.8 nM). Chromatin remodeling by the INO80 complex is indicated by the reduction of N3 band intensity and the increase in N1, N2 band intensity, which is represented by an increase in (N1+N2)/N3 ratios (bottom), ± S.D. from three independent experiments. (f) Graphical representation for mono-nucleosome mobilization as shown in (e), data presented is the mean of three independent experiments ± S.D. (g) SDS-PAGE and deep purple staining of the INO80 complex (1× equals 10 ng actin). Numbers on the left indicate the stoichiometry of labeled subunits with the actin subunit normalized as 1. Note that the Rvb1 and Rvb2 helicases co-migrate as a single band - Rvb1/2. (h) Northern blot analysis of PHO5 expression in wild type (WT) and mutant strains at 30 °C after 3 hours of induction, and at 37 °C after 1.5 hours of induction. RPL3 is a loading control.
To further examine the binding activities to chromatin substrate, we compared the nucleosome binding activities of wild type and mutant INO80 (act1-2) complexes using gel shift assay27. Compared to wild type, mutant INO80 (act1-2) complex showed about 3.0-folds reduction in the nucleosome binding affinity with the nucleosomes containing linker DNA at both sides (207 nucleosomes at 25 nM) (KdINO80= 15.25 nM ± S.E.= 2.8 nM, KdINO80 (act1-2)= 45.68 nM ± S.E.= 4.65 nM) (Fig. 2d and Supplementary Fig. 3a,b), suggesting a important role of actin in association of INO80 complex with nucleosomes. We also validated these results using another method involving fluorescently labeled mono-nucleosomes28, and similarly detected a 3.1-fold decrease in nucleosome affinity between wild type and mutant INO80 (act1-2) complex (Supplementary Fig. 3c and Supplementary table) consistent with a prominent role of actin in regulating chromatin binding.
Moreover, we compared the ATPase activities of the wild type and INO80 (act1-2) complexes. The ATPase activity of INO80 complex is more stimulated by nucleosome core particles than free DNA12. As compared to wild type, INO80 (act1-2) complex showed marked reduction in both the DNA and nucleosome-stimulated ATPase activities (Supplementary Fig. 3d). Since the bulk of ATPase activity of INO80 complex is abrogated by K737A mutation12, the observed reductions can be attributed to effects of the act1-2 (A58T) mutation on the enzymatic activity of Ino80 ATPase. Furthermore, we examined the chromatin remodeling activity of the INO80 (act1-2) complex using mono-nucleosome mobilization assay29,30. INO80 chromatin remodeling activity was revealed by the redistribution of N1-N3 nucleosome species (Fig. 2e). Using equimolar amounts of the wild type and mutant INO80 (act1-2) complexes, we detected a marked reduction, but not a total loss of chromatin remodeling activity for the INO80 (act1-2) complex (Fig. 2e,f). Although the reduction of INO80 activities in the act1-2 mutant was prominent, it may only partially contribute to the in vivo defects since other actin-containing complexes may also contribute to these defects. Nonetheless, these in vitro studies provide evidence that actin itself have a contribution to ATP-dependent chromatin remodeling by the INO80 complex in vitro.
Actin in the INO80 complex is a monomer
A key feature of cytoplasmic actin is its ability to polymerize and form F-actin. In the nucleus, whether actin is able to form F-actin remained controversial. In vitro studies with the BAF chromatin-remodeling complex suggest that the actin-containing BAF complex can bind to F-actin in a phosphoinositide (PIP2)-regulated fashion20. To uncover mechanisms of nuclear actin in INO80 chromatin remodeling, we began to investigate if actin polymerization is involved. Based on silver and Coomassie staining of the INO80 complex, actin subunit in the INO80 complex appeared to be monomeric (Supplementary Fig. 1 and data not shown). For more accurate measurement of actin stoichiometry in the INO80 complex, a quantitative method based on fluorescent staining of proteins was used to allow detection of two-fold differences in actin abundance in the complex. From the fluorescence measurements of several INO80 subunits on a SDS-PAGE gel, we estimated that actin in the INO80 complex exists as a monomer as compared to other stoichiometric subunits, such as Ino80, Arp8 and Arp4. In contrast, the Rvb1 and Rvb2 helicases in the INO80 complex together showed a 10.6 to 1 stoichiometry to actin, a value that is close to 12 to 1, as predicted if both Rvb1 and Rvb2 are classical hexameric helicases (Fig. 2g).
Functionally, if actin polymerization is required for INO80 activities, actin mutants that lose the ability to polymerize should be defective in INO80 functions in vivo. The temperature sensitive act1-1 mutant show actin polymerization defects even at permissive temperature26; however, unlike act1-2 mutant, the act1-1 mutant did not showed prominent nuclear defects, as judged by largely normal HU hypersensitivity (Supplementary Fig. 3e) and PHO5 activation at 30°C (Fig. 2h). At non-permissive temperature (37°C), act1-1 mutant loses the ability to polymerize actin within minutes26. Interestingly, although actin polymerization no longer occurs in the act1-1 mutant at 37°C, the activation of PHO5 was relatively normal (Fig. 2h). Given that the act1-2 mutant showed defects in PHO5 activation (Fig. 1g and Fig. 2h), the normal activation of PHO5 in the absence of actin polymerization in act1-1 mutant argues that in PHO5 activation, actin itself is required, but actin polymerization is not required. Moreover, we observed that INO80 complex did not bind actin filaments under the condition when cofilin interacts with actin filaments (Supplementary Fig. 3f). Together, the stoichiometry measurement and the functional analyses suggest that actin polymerization is not required for INO80 chromatin remodeling. However, it is possible that actin polymerization is involved in other nuclear activities, such as the activities of the BAF complex13,20, since the BAF and INO80 complexes are non-orthologous, and are distinct in their composition and the ways they are regulated by inositides (Supplementary Fig. 4a,b). These observations raises the possibility that actin could be utilized in unique ways as a monomer in the nucleus, which is distinct from its cytoplasmic counterpart.
Unique positioning of actin in the INO80 complex
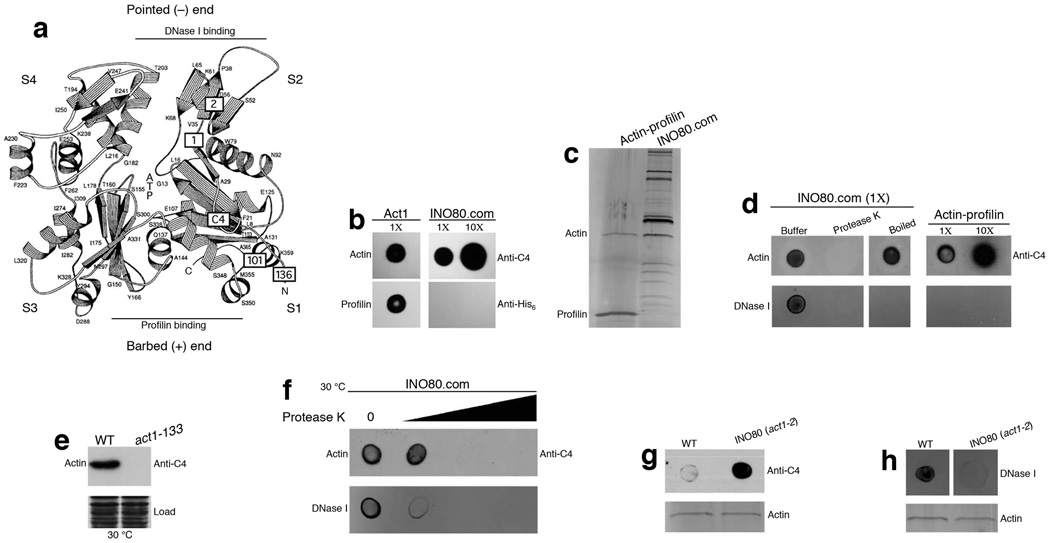
To understand how might actin function as a monomer in INO80, we investigated the microenvironment of monomeric actin in the INO80 complex. Since actin is capable of polymerizing from either the barbed end or the pointed end, we analyzed the accessibility of these two ends (Fig. 3a). Using purified recombinant profilin (with C-terminal His6tag) as a barbed end probe in a blot binding assay, we observed that, in contrast to purified actin which readily binds to profilin, actin in the INO80 complex was unable to interact with profilin (Fig. 3b), suggesting masking of the barbed end region of actin by other INO80 subunits. Since the barbed end region of actin accepts new actin molecule during polymerization31,32, this result suggests that the barbed end of actin in the INO80 complex is unlikely to be available for actin polymerization.
Figure 3. Actin in INO80 complex exists in a unique microenvironment.
(a) Schematic representation of the structure of actin as described in Fig. 1f. The position of the C4 epitope (Asp24 and Asp25) is now indicated in a box labeled with C4. DNase I binding and profilin binding regions are indicated by lines which represents the pointed (−) end region and barbed (+) end region. (b) Dot blot for actin and INO80 complex probed with antibodies as indicated on the right. (c) SDS-PAGE and silver staining of actin-profilin and INO80 complexes purified from cells grown at 30 °C. (d) Dot blot for INO80, and actin-profilin complexes probed with anti-C4 antibodies and fluorescently labeled DNase I as indicated. 1× refers to actin equivalent of the spotted complexes. (e) Western blot analysis of whole cell extracts using the monoclonal anti-C4 actin antibodies. Coomassie staining indicates equal loading of whole cell extracts. (f) Dot blot for INO80 complexes probed with anti-C4 antibodies and fluorescently labeled DNase I as indicated. (g,h) Top panel shows dot blot analysis for INO80 complexes purified from wild type and act1-2 mutant cells probed with anti-C4 antibodies and fluorescently labeled DNase I as indicated. Bottom panel shows equal loading of the complexes in the top panel, as judged by silver staining.
Despite the masking of the barbed end, the pointed end of actin could potentially allow actin polymerization. We probed the pointed end using DNase I. DNase I is known to bind monomeric actin at its subdomain 2 of the pointed end33. Using fluorescently labeled DNase I in a blot binding assay, we observed that actin in the actin-profilin complex (Fig. 3c) (purified by inserting double FLAG-epitope at C-terminus of PFY1 gene in chromosome) failed to bind DNase I due to structural changes in the DNase I binding site induced by the binding of profilin34. By contrast, DNase I was able to bind to the INO80 complex. The binding was due to the protein subunits in the INO80 complex and not due to residual DNA, since Protease K treatment of the INO80 complex abolished DNase I binding. Moreover, binding of DNase I to the INO80 complex was also abolished when the INO80 complex was heat-inactivated (Fig. 3d). These results indicate that, even though the barbed end of actin is masked by other subunits in INO80 complex, subdomain 2 at the pointed end of actin is readily accessible and retain proper conformation for DNase I interaction.
Interestingly, we observed that the epitope of actin monoclonal antibody C4 is located around amino acids Asp24 and Asp25, since mutations of these two amino acids in the act1-133 mutant (D24A D25A) abolished the C4 epitope (Fig. 3e). As such, C4 antibody could be used as a probe for actin subdomain 1 which is a part of the barded end of actin (Fig. 3a). Limited Protease K treatment of the INO80 complex revealed that the loss of DNase I binding precedes the loss of C4 epitope (Fig. 3f), consistent with the distinct microenvironment of actin in the INO80 complex, in which barbed end region is protected while the pointed end region remain exposed (Fig. 3b-f). Interestingly, act1-2 mutation (A58T) enhanced the sensitivity of C4 antibody to detect actin in the native INO80 complex (Fig. 3g). Moreover, act1-2 mutation in actin reduced the DNase I binding ability of the INO80 complex (Fig. 3h). These observations support that actin is the target for DNase I binding, and that the act1-2 mutation affects the conformation of actin in the INO80 complex. Together, these structural probing studies indicate that the monomeric actin in the INO80 complex is uniquely positioned in a microenvironment in which its barbed end is protected and its pointed end, which contains subdomain 2, is exposed.
Actin subdomain 2 contributes to INO80 chromatin remodeling
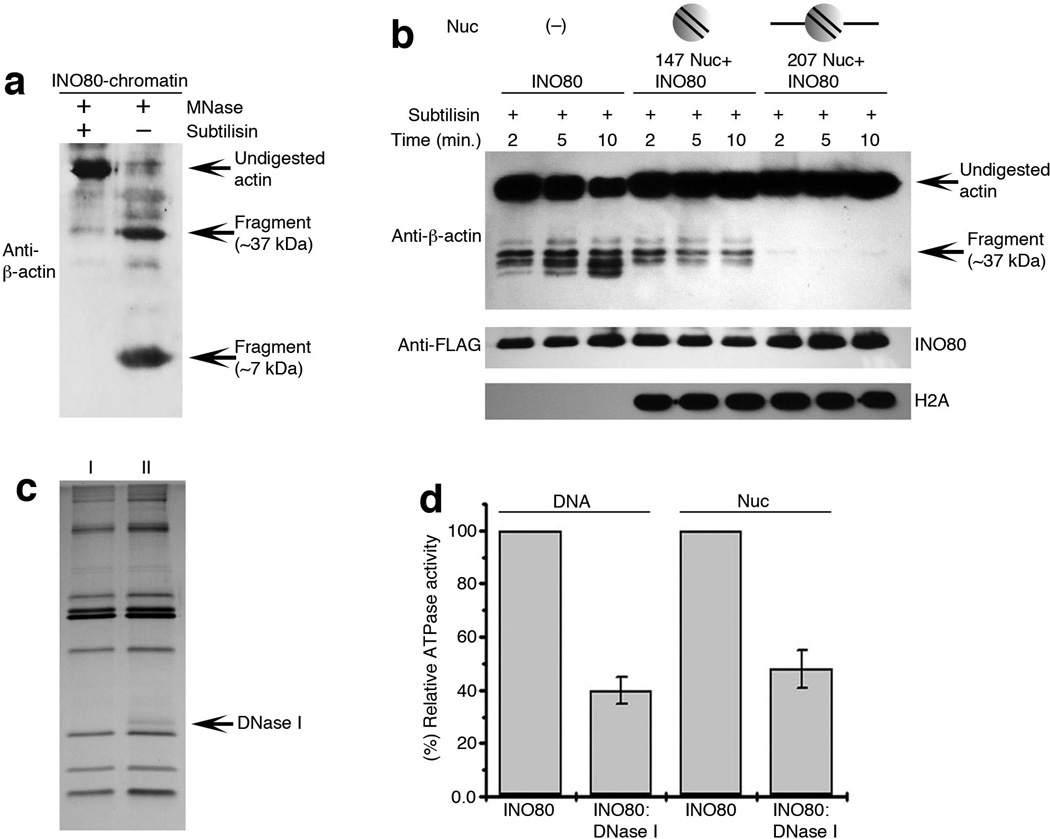
To further reveal the function of the exposed pointed end of actin in INO80 complex, we analyzed the actin subdomains. Curiously, the act1-2 mutation (A58T) that reduced INO80 activity is located in subdomain 2 at the pointed end. Given the reductions of DNA and nucleosome binding activities and chromatin remodeling activity in the INO80 complex containing act1-2 mutation (Fig. 2), we proposed that subdomain 2 at the pointed end of actin in the INO80 complex may be implicated in interaction with chromatin. To test this model, we investigated the interaction between actin in the INO80 complex with its chromatin substrate. We purified the INO80 complex bound to chromatin at lower salt concentration. Under these conditions, INO80 is co-purified with its native chromatin substrate as indicated by the presence of all core histones and DNA10. To probe the accessibility of actin subdomain 2 in the context of chromatin-bound INO80 complex, we utilized subtilisin, a protease from B. licheniformis. When used in 1:1500 (subtilisin: actin) concentration, subtilisin specifically cleaves the subdomain 2 of actin and generate bands around 36 kD and a band at around 7 kD which can be detected by respective actin antibodies35,36. Interestingly, although subdomain 2 in free INO80 complex was accessible, when INO80 was bound to chromatin, subdomain 2 was no longer accessible to subtilisin, suggesting that subdomain 2 at the pointed end of actin is involved in chromatin association. Curiously, after micrococcal nuclease (MNase) treatment of the chromatin which was bound to INO80 complex, the accessibility of subtilisin to subdomain 2 of actin in the INO80 complex reappeared, suggesting that subdomain 2 of actin in INO80 complex may be involved in interaction with MNase-sensitive features of chromatin such as linker DNA (Fig. 4a and Supplementary Fig. 4c-g).
Figure 4. Actin subdomain 2 is crucial for INO80 interaction with chromatin.
(a,b) Western blot analysis of, (a) purified INO80-chromatin complex in presence and absence of MNase after complete subtilisin digestion, probed with polyclonal anti-β-actin antibodies, (b) subtilisin accessibility to actin subdomain 2 in purified INO80 complex without its substrate chromatin, in presence or absence of 147 Nuc or 207 Nuc. (c) SDS-PAGE and silver staining for INO80 complex (lane I), and INO80: DNase I complex (lane II). (d) Graph showing relative DNA or nucleosome (359 bp INO1) dependent ATPase activity of INO80: DNase I compared to INO80 complex using equimolar amount of both the complexes and data presented is the mean of five independent experiments ± S.D.
To test this further, we used a reconstituted nucleosome binding system coupled with protease mapping assay. We analyzed the accessibility of actin to subtilisin in the purified INO80 complex after adding two types of reconstituted mono-nucleosomes: 147 nucleosomes (without linker DNA) and 207 nucleosomes (with linker DNA at both sides) (Supplementary Fig. 5a,b), under the condition in which INO80 complex binds to both 147 and 207 nucleosomes (Supplementary Fig. 5c,d and Supplementary Fig. 3a,b respectively). Subtilisin was accessible to actin in the INO80 complex in the absence of nucleosome substrates, it slightly reduces its accessibility in presence of 147 nucleosome indicating that the binding of linker-less nucleosome was not able to substantially block subdomain 2 of actin in the INO80 complex. By contrast, the addition of nucleosomes with linker DNA (207 nucleosomes) nearly abolished subtilisin accessibility (Fig. 4b), suggesting that the subdomain 2 of actin in INO80 complex is masked by the addition of linker DNA either through direct interaction with linker DNA or potential conformational changes induced by binding to nucleosomes with linker DNA. In contrast, the subdomain 2 of actin in INO80 (act1-2) complex was not markedly blocked by the 207 nucleosomes (with linker DNA) (Supplementary Fig. 5e), consistent with the decreased ability of mutant INO80 (act1-2) complex to bind nucleosomes with linker DNA (Fig. 2d and Supplementary Fig. 3b). As such, although the pointed end of actin in INO80 was exposed, it was involved in regulating the binding of INO80 complex to chromatin. Together, our results suggest a novel mechanism for nuclear actin, in which monomeric actin is contained in a unique microenvironment to regulate chromatin interaction instead of supporting actin polymerization. Although actin alone was unable to bind DNA, INO80 and its actin-Arp module can bind DNA30 (unpublished observation). Thus, nuclear actin in the context of chromatin remodeling complex have gained the unique ability to either directly interact with chromatin or regulate chromatin binding indirectly through conformational changes.
To further analyze the functional importance of subdomain 2 of actin in INO80 activity, we trapped the subdomain 2 of actin in INO80 complex with DNase I and co-purified the INO80: DNase I complex (Fig. 4c). As compared to INO80, DNA as well as nucleosome-dependent ATPase activity of the trapped INO80: DNase I complex was reduced more than 50% (Fig. 4d), confirming a crucial role of subdomain 2 of actin in INO80 activity. Given that the act1-2 (A58T) mutation which reduces INO80 activities is also located in subdomain 2, our combined genetic and biochemical data consistently support for a novel mechanism for nuclear actin, in which the subdomain 2 of actin is implicated in interaction with chromatin.
Discussion
A system to study nuclear actin
Despite increasing evidence suggesting that actin is in the nucleus and may play roles in many nuclear functions, the research on nuclear actin has been stalled by the difficulty of unambiguous demonstrations of actin function in the nucleus both in vivo and in vitro. Such demonstrations require a model system, in which the function of nuclear actin can be cleanly dissected both genetically and biochemically1. Our studies of the yeast INO80 chromatin-remodeling complex establish such a system. The defined stoichiometry and the ability to reduce the INO80 complex into sub-complexes make the INO80 complex a valuable system to demonstrate the function of nuclear actin using biochemical approaches as we have shown in this study. Using the wealth of techniques established to study cytoplasmic actin, we were able to probe the unique microenvironment of actin in the INO80 complex and provide insights into its mechanism. Importantly, our genetic and biochemical analyses have reached the same conclusion on the key function of subdomain 2 of actin in the INO80 complex, thus firmly establishing a novel mechanism for nuclear actin. Taken together, the yeast system serves as an ideal model to study nuclear actin.
An ancient module of nuclear actin and Arp
Our study refines the INO80 complex into a sub-complex containing actin, Arp4, Arp8 and Taf14, and these subunits associate with the N-terminal region of the Ino80 ATPase. Interestingly, all subunits of this actin-Arp sub-complex, including the N-terminal region of the Ino80 ATPase, are evolutionarily conserved, suggesting that this sub-complex represents a unique and ancient module used for INO80 chromatin remodeling. Since actin and Arp4 are consistently present in several chromatin-modifying complexes, such as INO80, SWR1 and NuA48,10,12, and the loss of Arp8 in the INO80 complex results in the loss of actin and Arp430, it can be argued that actin and Arp4 may form a dimer and may represent an even more conserved and basic module involving nuclear actin. This actin-Arp4 module may be used repeatedly in combination with other Arps and proteins in chromatin modifying complexes. For example, actin-Arp4 dimer may associate with Arp8 and the N-terminal region of the Ino80 ATPase, thus forming the observed actin-Arp module in the INO80 complex (Fig. 5). Similarly, actin-Arp4 dimer may form other functional modules in the SWR1 and NuA4 complexes by associating with other proteins. In yeast, this ancient actin-Arp4 module may have also evolved into a less conserved Arp7-Arp9 module found in the SWI/SNF and RSC chromatin-remodeling complexes, where Arp7 and Arp9 has been shown to form a dimer37. Based on these observations, it can be postulated that actin and nuclear Arps in chromatin-modifying complexes may be utilized in a combinatorial fashion to suit specific functions of these complexes.
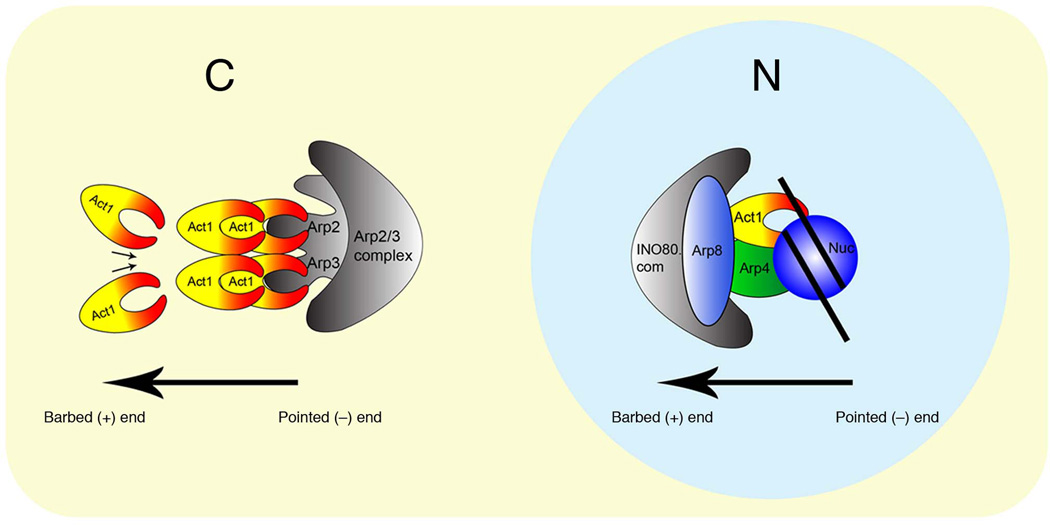
Figure 5. A model for nuclear actin in INO80 complex.
A model showing unique positioning of actin in the INO80 complex in the nucleus compared to the positioning of actin with Arp2/3 complex in the cytoplasm. Whereas the barbed end of actin with Arp2/3 complex is free to polymerize, the barbed end of actin with INO80 complex in the nucleus is masked by other subunits and the pointed end is engaged with chromatin.
Mechanism of nuclear actin in chromatin remodeling
There are two non-exclusive models concerning the mechanisms of nuclear actin in chromatin modifications. In the first model, nuclear actin undergo dynamic polymerization and depolymerization anchored from actin-containing chromatin modifying complexes3. This model is reminiscent of the actin nucleation or branching mechanism of the Arp2/3 complex38. The in vivo relevance of this model remains to be demonstrated. Our studies established a second model, in which monomeric actin serves as a functional subunit in chromatin-modifying complexes and directly participates in chromatin modifications. This model does not invoke the polymerization of actin in the nucleus. Instead, it requires a novel mechanism of direct interaction between monomeric actin and chromatin. However, it should be noted that the two models are not mutually exclusive. For example, our results do not exclude the possibilities that some aspects of nuclear actin function still require actin polymerization, or that nuclear actin may form unconventional or short filaments.
How might actin participate in chromatin modifications in the second model? It has been shown that Arps, such as Arp4 and Arp8, can bind to histones30,39. Although the precise mechanisms are still vague, during the ATP-dependent chromatin remodeling or histone modifications, the actin-containing chromatin modifying complexes, such as INO80 and NuA4, are likely to directly interact with chromatin (DNA or histones) at various points both temporally and spatially, therefore, the actin-Arp modules may serve as interaction surfaces or chaperones for chromatin. Since actin and Arps form distinct modules as discussed above, it can be postulated that specific combinations of actin and Arps in the chromatin-modifying complexes may be involved in the direct binding to specific features of chromatin, such as DNA, combinations of histones (including histone variants) or histone modifications.
Given that both the Arp2/3 complex and the INO80 complex contain Arps, it was plausible that the actin-Arp subunits could also mimic the function of Arp2/3 dimers in initiating actin polymerization40. However, in the case of cytoplasmic Arp2/3, the Arp2/3 dimer mimics an actin dimer. By contrast, actin in the nuclear INO80 complex is positioned differently (Fig. 5). The barbed end of actin in the INO80 complex is blocked by other INO80 subunits, which prevents actin polymerization. On the other end, although the pointed end is exposed and could potentially be used for actin polymerization, the subdomain 2 of pointed end is utilized to associate with chromatin instead. As such, the unique orientation and microenvironment of actin in different complexes underlies the highly distinct mechanisms between cytoplasmic and nuclear actin (Fig. 5). Given that chromatin is also highly conserved evolutionarily, we propose that one of the previously unrecognized, yet fundamental, function of actin is to directly interact with chromatin in the nucleus. The subdomain 2 of actin has evolved, perhaps together with Arps such as Arp4 and Arp8, to cooperatively interact with chromatin. This mode of actin function, though remarkably distinct from its cytoplasmic counterpart, is likely to be as ancient. We suggest that conventional actin has been utilized to interact with chromatin as one of its fundamental functions since the emergence of eukaryotes.
Online Methods
Yeast manipulations and phenotypic analysis
All S. cerevisiae strains were in the S288C background. A collection of act1 mutants was a gift from David Drubin24,25. Relevant ACT1 mutations were confirmed by PCR and sequencing. To generate strains for protein purification, The INO80 locus in act1-2 mutant was eptiope-tagged with a triple-FLAG sequence at the C-terminus. To generate the N-terminal region expression plasmid, the region coding amino acid 356 to 691 of Ino80 was cloned into a modified pRS416 plasmid with a double-FLAG sequence at the C-terminus, together with native INO80 promoter and terminator sequences. The resulting plasmid, pN-2F, was transformed into an ino80 deletion strain. pACT1 was constructed by cloning a PCR fragment spanning the ACT1 gene from −669 before the start codon to +317 after the stop codon into the pRS416 vector. To purify the actin-profilin complex, the PFY1 gene encoding profilin was epitope-tagged with a double-FLAG tag at the C-terminus in the chromosome.
Standard yeast culture and transformation techniques were followed. Phenotypic analysis was done by plating yeast cells at 5-fold serial dilutions. Plates were incubated at 30 °C for three to five days then scored. For gene expression analysis, yeast strains were grown overnight, then diluted 10- to 20-fold in YEPD. After growth at 30 °C for 4 hours, cells were collected and washed, PHO5 expression was induced in synthetic complete media lacking phosphate at indicted temperature for 1.5 to 4 hours. Total RNA was isolated and northern analysis was performed. The entire ORFs of PHO5, ACT1 and RPL3 were amplified by PCR and used as probes.
Purification of protein complexes
Protein complexes were purified from FLAG-epitope tagged strains as described elsewhere41. All purifications were done using high salt washes (0.5 M KCl) except for INO80 bound chromatin purification where low salt washes were done (0.1 M KCl). For further purification, protein complexes were separated in a 5 ml 17% –35% or 27%–45% glycerol gradient in Buffer H-0.3 (25 mM HEPES-KOH pH 7.6, 1 mM EDTA, 0.02% NP-40, 0.3 M KCl). SDS-PAGE followed by silver staining was done to detect proteins. Quantitative western blotting of the FLAG-tagged Ino80 ATPase was used to normalize complexes used in assays. Deep Purple stain (Amersham) was used to measure the stoichiometry of the INO80 complex using a Typhoon imaging system.
DNA binding and mononucleosome mobilization assays
DNA-binding and mono-nucleosome mobilization assays were performed as previously described30. Briefly, a 359 bp INO1 fragment spanning the INO1 promoter from positions −359 to +1 was used for the DNA-binding assay. The same fragment was used to form mono-nucleosome substrates with recombinant yeast core histones for the mono-nucleosome mobilization assay. Gels from the DNA-binding, mono-nucleosome mobilization assays were stained with SYBR Green I and documented using a Typhoon imaging system.
Quantitative mono-nucleosome binding assays
147 nucleosomes (without linker DNA) and 207 nucleosomes (with linker DNA at both the ends) were prepared as described previously42. Quantitative binding of protein complexes were performed either with unlabelled 147 nucleosomes and 207 nucleosomes by gel shift assays using native PAGE followed by SYBR Green I staining and documented using a Typhoon imaging system, or with fluorescently labeled 147 nucleosomes and 207 nucleosomes using fluorescent measurements as described previously28,42.
Western blot and dot blot assays
Monoclonal anti-C4 antibodies (Chemicon, MAB 1501), in 1:1000 dilutions, as well as polyclonal anti-beta-actin antibodies (Cell signaling, #4967), in 1:2000 dilutions, were used to detect actin in western blots and dot blots. Alexa Fluor-labeled DNase I (Molecular Probes) (5 mg/ml) was used in dot blots to bind purified complexes (10 ng actin equivalents as 1×) spotted on nictrocellulose memeberane, and DNase I binding was detected using a Typhoon imaging system. Global acetylation of histone H4 in whole cell extracts was detected by anti-H4-Penta-Ac antibodies (Upstate Biotechnology, # 06-946), in 1:1000 dilutions, that recognize all five acetylated lysine residues in H4 tail. As a control, under non-permissive temperature (37 °C), a temperature sensitive esa1 mutant was used, esa1-1851 is defective for NuA4 functions and shows a severe reduction in global histone H4 acetylation43, which was detected by the anti-H4-penta-Ac antibodies.
ATP hydrolysis assays
The ATPase assays were performed with 359 bp INO1 promoter DNA as well as with mono-nucleosomes prepared from same DNA using thin-layer chromatography (TLC) in 0.75 M KH2PO4 with (γ-32P) ATP, and signals were quantified on the Typhoon imaging system as described elsewhere12.
Limited proteolysis assays
Protease K and Subtilisin digestions were performed using 10 µl of purified complexes (10 ng actin equivalents), amount of protease used were determined empirically. The reaction was stopped by addition of two volumes of SDS sample buffer and immediate incubation at 100 °C for 5 min. Samples were separated by SDS-PAGE (12 or 15%) followed by either silver staining or western blot analysis.
Supplementary Material
Acknowledgements
We thank C. Wu, Laboratory of Biochemistry and Molecular Biology, National Cancer Institute, Bethesda, MD, USA, in whose lab we initiated early studies on actin; D. Drubin, Department of Molecular and Cell Biology, University of California, Berkeley, CA, USA, for providing a collection of actin mutants; C. Boone, Donnelly Center for Cellular and Biomolecular Research, University of Toronto, Canada, on sharing genetic data on actin and J. Highland, T. Wehr and J. Xiao for technical assistance. Funding to P.K. is provided by the Odyssey postdoctoral program and the Theodore N. Law Endowment for Scientific Achievement at The University of Texas MD Anderson Cancer Center. X.S. is supported by funds and grants from US National Institute of Health NCI (K22CA100017), NIGMS (R01GM093104), and Center for Cancer Epigenetics at MD Anderson Cancer Center.
Footnotes
Author Contributions
P.K., M.C. and X.S. designed the experiments. P.K., M.C. conducted the experiments. D.D.W. and K.L. provided the purified 147 & 207 nucleosomes and performed the quantitative nucleosome binding assay using fluorescently labeled 207 nucleosomes. P.K., M.C. and X.S. analyzed the data and wrote the paper.
References
- 1.Bettinger BT, Gilbert DM, Amberg DC. Actin up in the nucleus. Nat Rev Mol Cell Biol. 2004;5:410–415. doi: 10.1038/nrm1370. [DOI] [PubMed] [Google Scholar]
- 2.Blessing CA, Ugrinova GT, Goodson HV. Actin and ARPs: action in the nucleus. Trends Cell Biol. 2004;14:435–442. doi: 10.1016/j.tcb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem. 2002;71:755–781. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- 4.Shumaker DK, Kuczmarski ER, Goldman RD. The nucleoskeleton: lamins and actin are major players in essential nuclear functions. Curr Opin Cell Biol. 2003;15:358–366. doi: 10.1016/s0955-0674(03)00050-4. [DOI] [PubMed] [Google Scholar]
- 5.Clark TG, Merriam RW. Diffusible and bound actin nuclei of Xenopus laevis oocytes. Cell. 1977;12:883–891. doi: 10.1016/0092-8674(77)90152-0. [DOI] [PubMed] [Google Scholar]
- 6.Gonsior SM, et al. Conformational difference between nuclear and cytoplasmic actin as detected by a monoclonal antibody. J Cell Sci. 1999;112(Pt 6):797–809. doi: 10.1242/jcs.112.6.797. [DOI] [PubMed] [Google Scholar]
- 7.Wada A, Fukuda M, Mishima M, Nishida E. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J. 1998;17:1635–1641. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galarneau L, et al. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell. 2000;5:927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- 9.Ikura T, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 10.Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 11.Papoulas O, et al. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 12.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 13.Zhao K, et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 14.Egly JM, Miyamoto NG, Moncollin V, Chambon P. Is actin a transcription initiation factor for RNA polymerase B? EMBO J. 1984;3:2363–2371. doi: 10.1002/j.1460-2075.1984.tb02141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann WA, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6:1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 16.Hu P, Wu S, Hernandez N. A role for beta-actin in RNA polymerase III transcription. Genes Dev. 2004;18:3010–3015. doi: 10.1101/gad.1250804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Percipalle P, et al. An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc Natl Acad Sci U S A. 2003;100:6475–6480. doi: 10.1073/pnas.1131933100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Philimonenko VV, et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol. 2004;6:1165–1172. doi: 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- 19.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 20.Rando OJ, Zhao K, Janmey P, Crabtree GR. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc Natl Acad Sci U S A. 2002;99:2824–2829. doi: 10.1073/pnas.032662899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szerlong H, et al. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol. 2008;15:469–476. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebbert R, Birkmann A, Schuller HJ. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol Microbiol. 1999;32:741–751. doi: 10.1046/j.1365-2958.1999.01390.x. [DOI] [PubMed] [Google Scholar]
- 23.Steger DJ, Haswell ES, Miller AL, Wente SR, O'Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shortle D, Novick P, Botstein D. Construction and genetic characterization of temperature-sensitive mutant alleles of the yeast actin gene. Proc Natl Acad Sci U S A. 1984;81:4889–4893. doi: 10.1073/pnas.81.15.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wertman KF, Drubin DG, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- 27.Udugama M, Sabri A, Bartholomew B. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol Cell Biol. 2011;31:662–673. doi: 10.1128/MCB.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winkler DD, Luger K, Hieb AR. Quantifying Chromatin-Associated Interactions: The HI-FI System. Methods Enzymol. 2012;512:243–274. doi: 10.1016/B978-0-12-391940-3.00011-1. [DOI] [PubMed] [Google Scholar]
- 29.Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 30.Shen X, Ranallo R, Choi E, Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- 31.Bugyi B, Carlier MF. Control of actin filament treadmilling in cell motility. Annu Rev Biophys. 2010;39:449–470. doi: 10.1146/annurev-biophys-051309-103849. [DOI] [PubMed] [Google Scholar]
- 32.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 34.Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 35.Schwyter DH, Kron SJ, Toyoshima YY, Spudich JA, Reisler E. Subtilisin cleavage of actin inhibits in vitro sliding movement of actin filaments over myosin. J Cell Biol. 1990;111:465–470. doi: 10.1083/jcb.111.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapoor P, et al. Leishmania actin binds and nicks kDNA as well as inhibits decatenation activity of type II topoisomerase. Nucleic Acids Res. 2010;38:3308–3317. doi: 10.1093/nar/gkq051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szerlong H, Saha A, Cairns BR. The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J. 2003;22:3175–3187. doi: 10.1093/emboj/cdg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schafer DA, Schroer TA. Actin-related proteins. Annu Rev Cell Dev Biol. 1999;15:341–363. doi: 10.1146/annurev.cellbio.15.1.341. [DOI] [PubMed] [Google Scholar]
- 39.Harata M, et al. The nuclear actin-related protein of Saccharomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol Biol Cell. 1999;10:2595–2605. doi: 10.1091/mbc.10.8.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
Methods-only references
- 41.Shen X. Preparation and analysis of the INO80 complex. Methods Enzymol. 2004;377:401–412. doi: 10.1016/S0076-6879(03)77026-8. [DOI] [PubMed] [Google Scholar]
- 42.Winkler DD, Muthurajan UM, Hieb AR, Luger K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem. 2011;286:41883–41892. doi: 10.1074/jbc.M111.301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.