Abstract
Purpose
Glioblastoma (GBM) demonstrate down-regulated expression of Human Leukocyte Antigen (HLA) Class I, thereby escaping from cytotoxic T cells and limiting the efficacy of immunotherapy. LOH of HLA Class I (6p21) and/or Beta-2 microglobulin (B2m) (15q21) regions represent irreversible down-regulation. In this study, we examined the prevalence of these LOH events and their relations with overall survival in GBM.
Experimental Design
In a cross-sectional analysis on 60 adult GBM patients, DNA from formalin-fixed paraffin-embedded specimens were evaluated for ten microsatellite regions of HLA Class I, B2m, HLA Class II, HLA Class III, and 6q by PCR as well as immunohistochemical evaluation of HLA Class I expression and CD8+ T cell infiltration.
Results
LOH in HLA Class I, B2m, HLA Class II, HLA Class III, and 6q regions were present in 41.4%, 18.2%, 9.4%, 77.8%, and 36.0% of informative cases, respectively. LOH of HLA Class I was associated with shorter overall survival (HR = 4.89, p = 0.0078). HLA Class I was down-regulated in 22 to 43% of cases based on immunohistochemistry. Cases that displayed negative staining were significantly younger. HLA Class I expression correlated with intratumoral CD8+ T cell infiltration.
Conclusion
LOH in the HLA Class I region is frequent in adult GBMs. The association of shorter survival with LOH in this region suggest a crucial role for these genes in immunosurveillance.
Keywords: Loss of Heterozygosity, Human Leukocyte Antigen, Beta-2 Microglobulin, tumor immunology, Glioblastoma
Introduction
GBM is the most lethal adult primary brain tumor with a median life expectancy of 14.6 months. (1) Previous studies have suggestted that the immune system has a significant role in the pathogenesis of glioma. Reports of the decreased risk of developing glioma in individuals with a strong allergic history suggest that the immune system can be effective in opposing the development and progression of gliomas. (2, 3) Indeed, effector T cell infiltration has been found to be associated with longer survival of GBM patients. (4, 5)
The ability of the immune system to recognize and kill cancer cells relies on activated CTLs that recognize tumor antigens presented by the HLA class I-peptide complex. A major mechanism of cancer immune escape appears to involve defects in antigen presentation by HLA Class I and this mechanism negatively impacts the clinical course in some malignancies and the outcome of CTL-based immunotherapy. (6)
The down-regulation or loss of HLA Class I or B2m, an essential component of the complete HLA Class I molecule, can be divided into reversible or irreversible lesions, each representing entirely different mechanisms of down-regulation. (7) Provided that HLA Class I genes, consisting of HLA-A, HLA-B, and HLA-C, are intact, the expression of HLA Class I can be restored in malignant glioma cells by the use of interferon-gamma and -beta. (8) However, loss of heterozygosity (LOH) in 6p21 and 15q21, where HLA Class I and B2m genes are located, respectively, represent irreversible lesions that are not amenable to epigenetic restoration of HLA Class I expression. (9, 10) Although HLA Class I expression is known to be down-regulated in approximately 50% of GBM (11), it is unclear whether LOH in the HLA Class I and B2m regions are prevalent in GBMs, which is possible given the high chromosomal and microsatellite instability in these tumors. (12) Furthermore, it is unknown whether LOH of these genes has any effect on patient survival, based on the rationale that these events could lead to decreased tumor antigen presentation and limited recognition by CTLs.
In this study, we investigated the expression of HLA Class I and the prevalence of LOH in HLA Class I and B2m regions using a PCR-based microsatellite approach in patients who underwent standard treatment, including resection, radiation, and chemotherapy. We hypothesized that LOH in HLA Class I and B2m regions, if prevalent in GBM, would be associated with lower survival. The results in this study support this hypothesis and provide important prognostic insights regarding the frequency and prognostic relevance of LOH in the HLA Class I and B2m regions, as well as having implications for future immunotherapy trials.
Methods
Patient Specimens
Archival formalin-fixed, paraffin-embedded (FFPE) adult GBM samples (n=64) belonging to a total of 60 patients obtained at the time of tumor biopsy or resection were obtained from the Brain Tumor Bank at the University of Pittsburgh and used for this cross-sectional study (see Supplementary Table 1). De-identified patient information, including clinicopathologic data, was accessible only through designated honest brokers (Anatomic Pathology Broker System, Approval # IRB PRO12020252 [PI: Hideho Okada]). Non-stratified patient samples were selected by tissue availability from 2005 to 2011, inclusive, by our honest broker from the database. Out of these 60 patients, 56 patients (48 newly diagnosed and 8 recurrent cases; one specimen per patient) were treated with resection, radiation, and chemotherapy.
Antibodies
The following primary monoclonal antibodies (mAbs) were used at indicated dilutions (see Supplementary Table 2 for allele-specific recognition for each of the mAbs): EMR8-5 (1:3000); HCA2 (1 μg/mL); HC10 (0.25 μg/mL). The quality of HLA Class I staining by each mAb was monitored by using surrounding normal cells, such as endothelial cells, lymphocytes, and microglial cells, as internal positive controls. Negative controls for HLA Class I expression included 5 cases of medulloblastoma, which are known to express little to no HLA Class I. (13) The anti-human CD8-specific mAb clone C8/144B (1:50) (Dako Cytomation, Glostrup, Denmark) was used to identify tumor infiltrating CTLs. Monoclonal rabbit anti-CD11b antibody (1:100) (clone EP1345Y, Abcam, Cambridge, MA, USA) was used to identify myeloid cells. Concentrations of the primary antibodies were optimized to minimize background staining in normal brain tissue.
Immunohistochemistry
Immunohistochemistry on paraffin-embedded tissue sections were performed as described by us recently. (14) Briefly, deparaffinized, antigen-retrieved sections were blocked for endogenous peroxidase and non-specific binding of mAbs before incubated with each of the antigen-specific antibodies at the aforementioned concentrations for 1 hour at room temperature. After washing, slides were then incubated with Dako anti-mouse secondary antibody (Envision+) for 30 minutes at room temperature. Peroxidase labeling was visualized using diaminobenzidine (Dako Cytomation, Glostrup, Denmark). Vector SG (Burlingame, CA, USA) was used for double-staining. The sections were lightly counterstained with Gill’s hematoxylin.
The authors (JY and RH) assessing the immunohistochemistry were blinded to the clinical data until grading was completed. When disagreement occurred, further discussion ensued to achieve a consensus. Disagreement of all grading occurred in less than 5% of cases. Results for HLA Class I staining were classified according to the criteria established by the HLA and Cancer component of the 12th International Histocompatibility Workshop. (15) Lesions were scored as positive, heterogeneous, and negative, when the percentage of stained tumor cells in a specimen was >75%, between 75% and 25% inclusive, and <25%, respectively. For statistical comparisons, HLA Class I immunoreactivity was dichotomized into negative and positive, which included heterogenous and homogenous staining. CD8+ positive cells were graded semi-quantitatively by intratumoral and perivascular infiltration patterns on a zero to three scale using a 20X objective. Intratumoral infiltration was graded as 0 (no positive cells in any field), 1 (mild, 0-10 cells in some fields), 2 (moderate, individual positive cells in most fields), and 3 (strong, many individual positive cells in all fields or aggregation/clusters of positive cells in multiple foci). Perivascular infiltration was categorized by 0 (no positive cells around any vessel), 1 (mild, few positive cells around some vessels), 2 (moderate, 10-20 positive cells around some vessels), and 3 (strong, more than 10 positive cells around all vessels).
PCR-based microsatellite analysis
Each FFPE specimen was stained with H&E to ensure that GBM was present. Areas with high density and purity (>90%) of tumor cells were marked for microdissection of adjacent sections to minimize contamination from normal cells and ensure reliable LOH analyses. (16) Overlapping areas in up to 4 adjacent slides were used for microdissection. DNA was extracted from the microdissected tissue using QIAamp DNA Micro Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Ten polymorphic microsatellite markers spanning 6p21 and 15q21 were tagged with FAM at their 5′ ends (Integrated DNA Technologies, Inc. Coralville, IA, USA). The details of the microsatellite markers used in this study are given in Table 1. Relative positions of markers targeting chromosome 6 are displayed in Supplementary Figure 1. An assumption in this study was that a microsatellite marker near a particular gene would serve as a surrogate marker for the allelic status of that gene, such that LOH demonstrated by a marker would indicate an allelic loss of the targeted gene. PCR was performed on GeneAmp® PCR System 2700 thermal cycler (Applied Biosystems, Foster City, CA, USA)) using KOD Xtreme™ Hot Start DNA Polymerase (EMD Millipore, Billerica, MA, USA) and its recommended protocol with the optimized annealing temperature of each primer listed in Table 1. Products of amplification were first visualized by 2% agarose gel electrophoresis to ensure ample amplification and were subsequently subjected to capillary gel electrophoresis that was performed at the Molecular Anatomic Pathology laboratory, University of Pittsburgh Medical Center on ABI3730 (Applied Biosystems, Foster City, CA, USA) according to the protocol. PCR-quality water was used as a negative control in all reactions. The signal waveforms from the microsatellite-based PCR were visually inspected for aberrant signals that may suggest inadequate amplification. The technician performing the capillary electrophoresis analysis was blinded to the results. The relative fluorescence values (peak heights) were obtained for individual alleles and the ratio of peaks was calculated using GeneMapper software v.4.0 (Applied Biosystems, Foster City, CA, USA). LOH for a specific marker was defined as having an allelic ratio fall outside a 99% confidence internal constructed from DNA obtained from peripheral blood of 10 or more normal individuals using the following equation proposed by Slebos et al.:
where X̄ is the average log2(allelic ratio) from normal control samples, tn-1 is the t-score for the sample size available for a specific marker, s is the standard deviation, and n is the number of normal samples available. (17) Retention of heterozygosity (ROH) was defined as the lack of LOH findings with informative allele ratios. Samples with non-informative loci and no LOH for a particular marker are excluded from analysis.
Table 1.
Panel of microsatellite markers targeting chromosome 6 and 15.
| Marker | Specific Location | Size | Primer Sequence | Annealing Temp (°C) |
|---|---|---|---|---|
| D6S105 | Tel.(1500-2500kb)/HLA-A | 116-168 | 5′ GCCCTATAAAATCCTAATTAAC 5′ GAAGGAGAATTGTAATTCCG |
55 |
| D6S265 | Centromeric to HLA-locus A | 122-144 | 5′ ACGTTCGTACCCATTAACCT 5′ ATCGAGGTAAACAGCAGAAA |
58 |
| D6S276 | Tel(6000kb)/HLA-A | 198-230 | 5′ TCAATCAAATCATCCCCAGAAG 5′ GGGTGCAACTTGTTCCTCCT |
64 |
| C.1.2.c/ D6S2800 |
HLA-locus B | 186-286 | 5′ GGATCCTAGGAACTCCCTCCTG 5′ GAGCAGAAGGGAGATGAAATGG |
58 |
| C.1.2.5 | Tel.(62kb)/HLA-B, centr.(19kb)/HLA-C |
178-220 | 5′ CAGTAGTAAGCCAGAAGCTATTAC 5′ AAGTCAAGCATATCTGCCATTTGG |
57 |
| D15S126 | Tel./beta-2-microglobulin | 188-218 | 5′ GTGAGCCAAGATGGCACTAC 5′ GCCAGCAATAATGGGAAGTT |
60 |
| D15S209 | Tel./beta-2-microglobulin | 189-212 | 5′ AAACATAGTGCTCTGGAGGC 5′ GGGCTAACAACAGTGTCTGC |
58 |
| D6S291 | Centromeric to HLA-class II region | 198-210 | 5′ CTCAGAGGATGCCATGTCTAAAATA 5′ GGGGATGACGAATTATTCACTAACT |
57 |
| D6S273 | HLA-class III | 120-140 | 5′ GCAACTTTTCTGTCAATCCA 5′ ACCAAACTTCAAATTTTCGG |
58 |
| D6S311 | 6q | 229-276 | 5′ ATGTCCTCATTGGTGTTGTG 5′ GATTCAGAGCCCAGGAAGAT |
57 |
Statistical analysis
Statistical analyses were performed using STATA Version 12 (College Station, TX, U.S.A.) and TIBCO Spotfire S+® 8.2 for Windows (Somerville, MA, U.S.A). The Mann-Whitney U test was used to compare continuous variables between two groups. Fisher’s Exact test was used to analyze categorical data from two groups. Spearman rank correlation was used to analyze associations in immunoreactivity. Immunohistochemistry data and clinical correlations with LOH status were analyzed using both newly diagnosed and recurrent cases together. Exploratory analyses of LOH events with molecular data from the brain tumor bank, obtained by methods described previously (18-21), were performed using univariate analyses. Samples with non-informative loci and no LOH for a particular marker are excluded from survival analysis. Analyses were performed by excluding non-informative loci due to the assumption that the loci could harbor LOH or balanced alleles. Including non-informative loci may then introduce bias to the results. Differences in overall survival (OS), defined as duration from initial biopsy/resection until death, between groups using only newly diagnosed cases were analyzed using standard proportional hazards regression modeling for censored time-to-event data; inference was based on the likelihood ratio test. Values unless otherwise specified are presented as mean ± standard deviation; p ≤ 0.05 was considered to be significant.
Results
Patient characteristics
The clinical features of patients (n = 56) are shown in Supplementary Table 1. The mean age was 60.0 ± 12.7 years (range 25-88 years). There were 35 males and 21 females. Their median follow-up time was 9.5 months (range 0.7 - 78.5 months).
HLA Class I expression in adult GBM
The results of immunohistochemical staining of 56 FFPE adult GBM with 3 different mAbs (HCA2, EMR8-5, and HC10) varied widely among specimens; representative images are depicted in Figure 1 and Supplementary Figure 2A. Using EMR8-5 mAb, 11 (22%), 13 (27%), 25 (51%) cases displayed negative, heterogenous, and homogeneous expression, respectively. Using HCA2 mAb, 17 (33%), 15 (29%), 20 (38%) cases displayed negative, heterogenous, and homogeneous expression, respectively. Using HC10 mAb, 23 (43%), 5 (9%), 26 (48%) cases displayed negative, heterogenous, and homogeneous expression, respectively.
Figure 1.
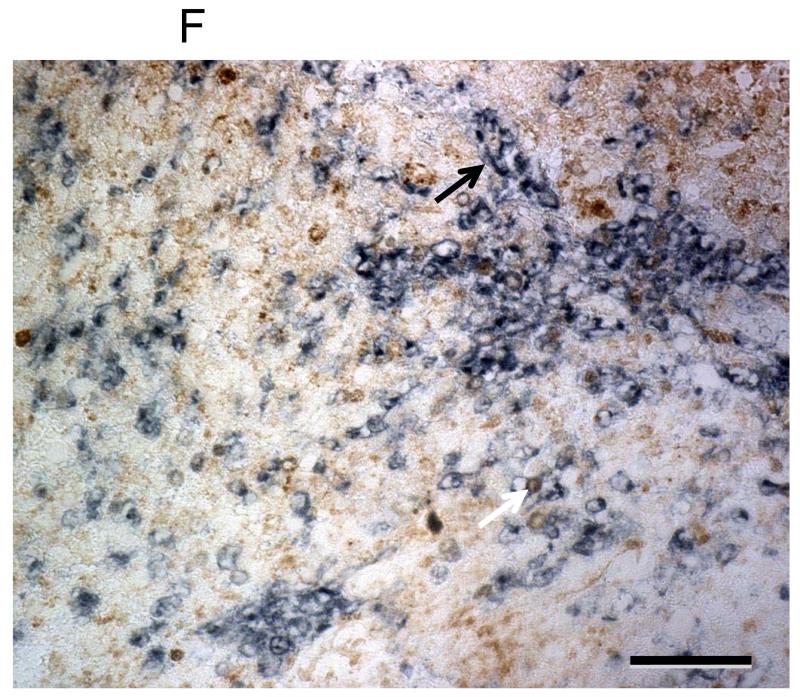
Immunohistochemical staining for HLA Class I expression in adult GBM. Representative images of immunohistochemistry using HCA2 shown at 10X magnification (A-D) and 20X (F). Each scale bar represents 100μm (A-D). Negative control using medulloblastoma, while endothelial cells serve as internal positive control (A). HCA2 staining showing negative (B), heterogeneous (C), and homogeneous (D) expression. Heterogeneous and homogeneous immunoreactivity are considered to be positive HLA I expression. A chart summary of HLA Class I staining (E). While some myeloid cells (CD11b+, dark blue) exhibit HLA Class I (brown) immunoreactivity (white arrow), most infiltrating myeloid cells (CD11b+, dark blue) in GBM are almost negative for HLA Class I (black arrow) (F).
There was a positive correlation between age and HCA2 immunoreactivity (p=0.02), between age and EMR8-5 immunoreactivity (p=.054); however, no correlation between age and HC10 immunoreactivity was found (p = 0.53). Overall survival was not associated with immunoreactivity against EMR8-5, HCA2, or HC10 regardless of whether age was included in the analysis. Myeloid cells (CD11b+ cells) displayed heterogeneous immunoreactivity of HLA Class I (Figure 1F).
CD8+ T cells infiltration
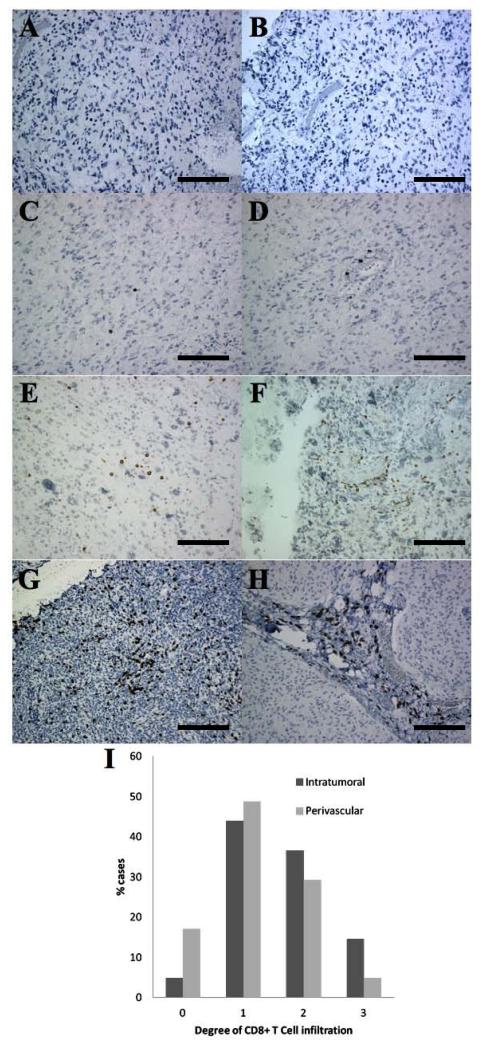
As shown in Figure 2 and Supplementary Figure 2B, of the 41 cases with sufficient tissue for analysis, intratumoral CD8+ T cell infiltration was not observed (grade 0) in 2 cases (4.9%), and was mild (grade 1) in 18 cases (43.9%), moderate (grade 2) in 15 cases (36.6%), and strong (grade 3) in 6 cases (14.6%). Perivascular CD8+ T cell infiltration was not observed (grade 0) in 7 cases (17.0%), and was mild (grade 1) in 20 cases (48.8%), moderate (grade 2) in 12 cases (29.3%), and strong (grade 3) in 2 cases (4.9%). Overall, most cases had some degree of CD8+ T cell infiltration, but rarely did the cases display strong, diffuse infiltration.
Figure 2.
CD8+ T cells in adult GBM. Representative images are shown at 20X magnification (A-H). Each scale bar represents 125 μm. Intratumoral (A,C,E,G) and perivascular (B,D,F,H) infiltration. CD8 staining showing negative (A,B), weak (C,D), moderate (E,F), and strong (G,H) infiltration. Results of CD8+ T cell intratumoral and perivascular infiltration in adult GBM (I).
After dichotomizing each form of CD8+ T cell infiltration into negative (grade 0 and 1) and positive infiltration (grade 2 and 3) categories, we found no associations between overall survival and intratumoral or perivascular CD8+ T cell infiltration regardless of whether age was included in the analysis. However, intratumoral CD8+ T cell infiltration significantly correlated with HLA Class I immunoreactivity detected by EMR8-5 (p < 0.01) and HC10 (p < 0.01) but not by HCA2 (p = 0.25). There was not an association found in this study between perivascular CD8+ T cell infiltration and HLA Class I immunoreactivity, regardless of the mAb used.
LOH analysis of 6p21 and 15q21
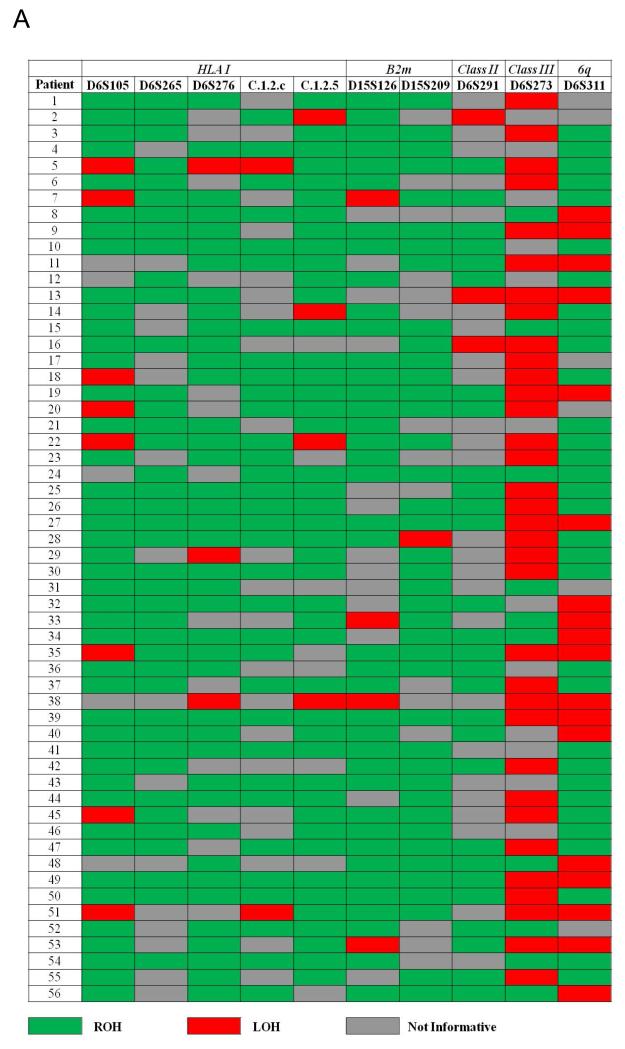
LOH data are illustrated in Figure 3A. Five markers that were used to assess the HLA I region (D6S105, D6S265, D6S276, C.1.2.c, C.1.2.5) demonstrated LOH in 12 of 29 informative cases (41.4%). A marker centromeric to HLA class II region (D6S291) demonstrated LOH in 3 of 32 (9.4%) informative cases. Intriguingly, 35 of 45 (77.8%) informative cases for D6S273, which maps to the HLA class III region, demonstrated LOH. Additionally, using D6S311 (which, if lost in addition to multiple 6p markers, may be inferred to be a marker to assess whole chromosome loss) demonstrated LOH in 18 of 32 (36.0%) informative cases. Two markers mapping to B2m revealed LOH in 6 of 33 (18.2%) informative cases.
Figure 3.
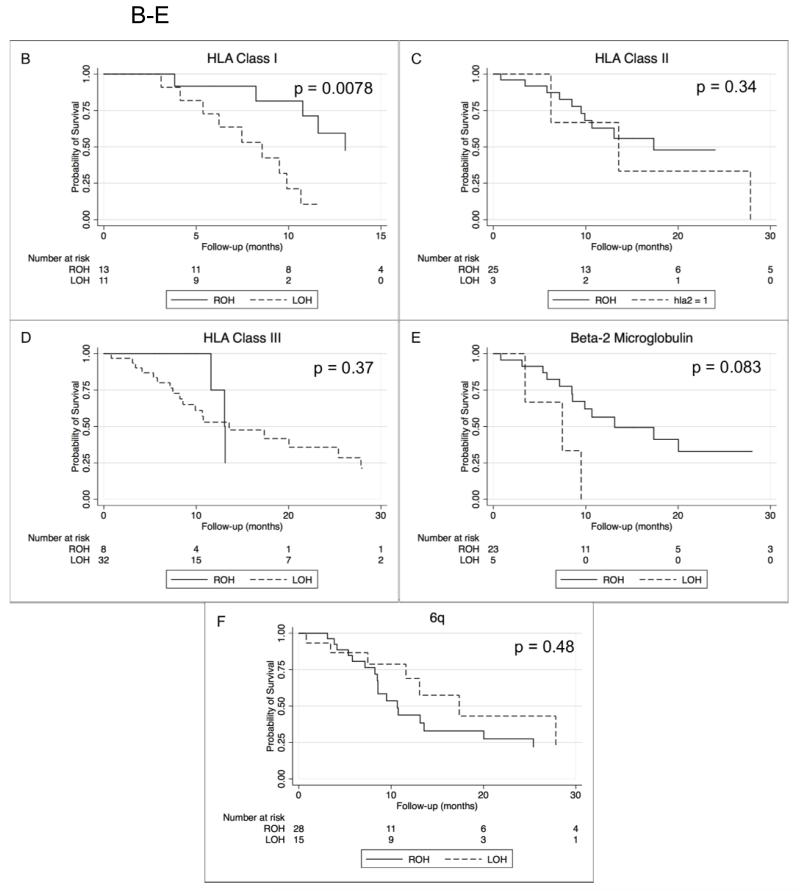
(A) Heat map of LOH in chromosome 6 and 15. (B-F) Kaplan-Meier Survival Curves according to LOH analyses. LOH of HLA Class I region (B) and survival (median 13.1 vs 8.6 months, p = 0.078). LOH of HLA Class II region (C) (p = 0.34) and Class III (D) (p= 0.37). LOH of B2m region (E) and survival (p = 0.083). LOH of 6q (F) and overall survival (p = 0.48). LOH = Loss of Heterozygosity. ROH = Retention of Heterozygosity.
Overall, the LOH status in the HLA Class I region was not associated with HLA Class I immunoreactivity using EMR8-5, HCA2, or HC10. Furthermore, LOH in the HLA Class I region was not associated with intratumoral or perivascular CD8+ T Cell infiltration. The associations between LOH events with available clinico-molecular data are summarized in Table 2. Of note, patients with LOH in the region of HLA Class III were significantly younger (57.3 ± 12.8 vs 67.2 ± 12.1 years, p = 0.04). Secondary GBM are characterized by a high incidence of p53 mutation. (22) In our study, 28.6% of cases demonstrated p53 aberration, as detected by immunohistochemistry. In univariate analyses, we found no association between p53 with LOH in the HLA Class I and B2m regions.
Table 2. Exploratory analyses: association of LOH events with available clinico-molecular data.
MIB1, p53 expression, were assessed using immunohistochemistry. EGFR amplification, chromosome 7 hyperploidy, and % cells with p16 deletion were assessed using Fluorescence In Situ Hybridization (FISH). LOH of 1p and 19q were determined by using both FISH and PCR. LOH of 9p, 10q, and 17p were determined using PCR only. Overall, loss of any 1 marker was considered as LOH for this study. MGMT promoter methylation was assessed using methylation-specific PCR. The above analyses were performed and quantified as described previously. (18-21)
| P-value | |||||
|---|---|---|---|---|---|
| LOH Class I | LOH Class II | LOH Class III | LOH B2m | LOH 6q | |
| Age * | 0.82 | 1.00 | 0.04 | 0.13 | 0.69 |
| Sex ** | 0.15 | 0.22 | 0.15 | 0.38 | 1.00 |
| MIB1 (immunohistochemistry) * | 0.85 | 0.13 | 0.68 | 0.98 | 0.78 |
| EGFR (FISH) ** | 0.06 | 1.00 | 0.72 | 1.00 | 1.00 |
|
p53 expression
(immunohistochemistry) ** |
0.42 | 0.18 | 0.70 | 0.15 | 0.06 |
|
% cells with hyperdiploidy
(Chromosome 7) * |
0.44 | 0.71 | 0.63 | 0.30 | 0.18 |
| LOH 1p (FISH) ** | 0.70 | 0.12 | 1.00 | 0.30 | 0.16 |
| LOH 1p (PCR) ** | 1.00 | 1.00 | 1.00 | 0.36 | 1.00 |
| LOH 19q (FISH) ** | 0.72 | 0.58 | 0.30 | 1.00 | 0.13 |
| LOH 19q (PCR) ** | 1.00 | 1.00 | 1.00 | 0.55 | 0.33 |
| LOH 9p (PCR) ** | 0.43 | 0.28 | 0.07 | 0.19 | 1.00 |
| LOH 10q (PCR) ** | 0.43 | 0.16 | 0.69 | 0.30 | 0.08 |
| LOH 17p (PCR) ** | 0.70 | 0.58 | 0.30 | 1.00 | 0.13 |
| % cells with p16 deletion (FISH) * | 0.10 | 0.82 | 0.55 | 0.47 | 0.41 |
| MGMT promoter methylation ** | 0.49 | 1.00 | 1.00 | 1.00 | 1.00 |
Mann–Whitney U test
Fisher’s exact test
Kaplan-Meier survival curves are shown in Figure 3 (B-F). Strikingly, survival analyses revealed that LOH in the HLA Class I region was significantly associated with shorter survival in newly diagnosed GBM (median 13.1 vs 8.6 months). (Figure 3B), whereas LOH in the regions of HLA Class II and Class III were not (Figure 3C-3D). In univariate analysis using standard proportional hazards regression, the unadjusted hazard ratio (HR) for LOH in the HLA Class I region was 4.89 (p = 0.0078). In univariate analysis using standard proportional hazards regression, LOH in the B2m region showed a strong trend towards shorter survival (HR = 3.83, p = 0.083). (Figure 3E). Previous studies have found that young age is associated with longer survival. (23, 24) In our study, when we dichotomized age about its median, we found a very weak association (p = 0.30) of old age with longer survival (HR = 0.7). We therefore decided not to include age as a covariate in the proportional hazards regressions. There was no association found between LOH of 6q and survival (Figure 3F).
Discussion
Although it is known that decreased HLA Class I expression is frequent in GBM (11), it was unclear whether LOH in the region of HLA Class I, an irreversible mechanism of down-regulation, is common in GBM and has any impact on the clinical course. Our results provide evidence of the frequency of this genetic aberration in GBM and show a significant association between LOH of these regions and shorter survival.
LOH of HLA Class I genes, consisting of HLA-A, HLA-B, and HLA-C, has been suggested to be the most common irreversible mechanism underlying HLA Class I down-regulation. (10) Our reported prevalence of 41.4% in the current study is comparable to other studies in colorectal carcinoma (40%), melanoma (23%), bladder carcinoma (35%), laryngeal carcinoma (53%), and head and neck squamous cell carcinoma (49%). (10, 25-28) No previous investigations reported any association of LOH of the HLA Class I and B2m regions with survival. Strikingly, our data demonstrated a significant association of LOH in these two regions with poorer survival in GBM patients. Surprisingly, we did not find any association between LOH and HLA Class I expression, even with the use of 3 different mAb that target different allotypic components of the molecule. Similar results have been reported in head and neck squamous cell carcinomas in which, with all 6p21-23 loci considered together, only LOH of D6S291 was found to be associated with HLA Class I expression. (28)
Multiple targets involving the other intact HLA Class I allele and components of the antigen presenting machinery, such as transporter associated with antigen processing and low molecular weight polypeptides proteins, (29) may act in concert to result in detectable down-regulation of HLA Class I, analogous to Knudson’s multiple hit hypothesis. (30) The latter hypothesis would help to consolidate our findings of 1) the lack of association between LOH of HLA Class I region and protein expression, 2) lack of association between LOH of HLA Class I region and CD8+ T cell infiltration seen in this group, and 3) the significant survival difference between LOH and ROH of the HLA Class I region. These temporal events may not be captured by analyzing tissue samples obtained at the time of diagnosis. Although haploinsufficiency of HLA Class I protein due to LOH is an obvious mechanistic explanation for shorter overall survival, mechanisms unrelated to immune escape may also be involved. It was postulated that loss of HLA class I surface expression on human melanoma cells grown in immunodeficient mice with no autologous immune response could generate more oncogenic tumor cells in vivo. (31) Furthermore, HLA Class I may have effects on cell cycle gene expression, invasion and intrinsic tumorigenicity that are not amenable to detection by immunohistochemistry and accounted for by extrinsic immunologic interactions. (32) Precise mechanisms for these observations, in which no interaction with the immune system is needed, remain to be elucidated.
Survival analyses aimed at HLA Class I immunoreactivity and linking it with LOH events are complicated due to the various antibodies available, such that contradicting results have been demonstrated even in the same cancer type. (33) Facoetti et al. were not able to demonstrate any survival difference by HLA Class I expression, which the authors attributed to insufficient sample size. (11) That would be particularly true in this study as our sample size is slightly more than half of theirs. Furthermore, mAbs used for the current study are not allotype-specific. Although we included the use of EMR8-5, which has been purported as a true pan-HLA Class I antibody based on its reactivity against recombinant alleles,(34) the high number of serotypes for each HLA-A/B/C precludes the existence of a true pan-HLA Class I mAb. In light of that, the strong association of LOH of HLA Class I with overall survival presents a potential biomarker for GBM without the variability inherent in immunohistochemistry.
Intriguingly, CD8+ T cell infiltration has been repeatedly shown to correlate with survival in GBM patients, where the rationale involves proper antigen presentation by tumor cells. (4, 5) Although we were not able to reproduce these findings possibly due to limited amounts of available tissues, our results have demonstrated for the first time, to our knowledge, a positive correlation between HLA Class I expression and the degree of intratumoral, but not perivascular, CD8+ T cell infiltration in adult GBM. These two patterns of CD8+ T cell accumulation in melanoma specimens have been shown to have a significant association with patient survival. (35) Surprisingly, not many CD11b+ cells are convincingly positive for HLA Class I, suggesting that these CD11b-positive cells may be immature myeloid cells. Overall, CD8+ T cell infiltration was observed to be related to the HLA Class I expression of tumor cells, especially since areas with high-density of tumor cells were selected for this study.
Our results demonstrating a statistically significant younger age for specimens displaying negative HLA Class I immunoreactivity present a paradoxical scenario. It is established that GBM patients who are younger at diagnosis have a better prognosis than those who are older. (36) In younger patients, it is possible that immune cells, capable of effectively targeting and killing adequately presented tumor cells, negatively select for tumor cells that lack proper antigen presentation. We did not find an association between age and overall survival, possibly due to our sample size.
A surprisingly high number of GBM specimens in our study demonstrated LOH in the region of HLA Class III. HLA Class III region encodes many proteins with immune functions, including components of the complement system, cytokines (Tumor Necrosis Factor-α/(TNF-α), Lymphotoxin A, Lymphotoxin B), heat shock proteins, and many genes with no obvious immune-related functions (37). Of immunological interest, TNF-α has been shown to evoke robust antitumor functions by increasing cytotoxic T cell activity (38), enhancing monocyte, granulocyte, and natural killer cell cytotoxicity (39), and activating downstream pro-inflammatory cytokines. (40) Our data showing a high prevalence of LOH in the HLA Class III region are consistent with the above aspects of immunosuppression when the anti-tumor effects are lost. Our survival analysis was limited by the small number of patients with ROH.
Frequent allelic losses were detected using D6S311. Our finding of LOH of 6q in 36% of cases is in close agreement with 25% (tumor grade not defined) previously reported by Miyakawa et al. (41) The region of 6q has been implicated to harbor tumor suppressor genes and represent more aggressive tumors and lower survival in other cancers. (42, 43) (44) In our study, however, we did not find an association between LOH of D6S311 with overall survival.
An obvious limitation in the present study is the cross-sectional design, which does not allow us to discern the temporal relationships and causality of LOH in the region of HLA Class I and survival. However, there is strong biological plausibility concerning the role of HLA Class I in tumor recognition by the immune system. Another concern is the lack of normal, autologous tissue from each patient to perform traditional comparisons in our LOH analyses. However, we have chosen to employ an artificially constructed confidence interval derived from healthy donor DNA to define a stringent criteria for LOH and avoid false positive findings. Any false positive would inherently bias our results towards the null hypothesis, thereby making our current survival finding more convincing. The gain of alleles might produce a false positive LOH finding or “pseudo-LOH” but abnormalities of chromosome 6 are almost always deletions. (45, 46). In addition, any contamianting non-tumor cells would bias towards the null hypothesis and add confidence to our findings. Lastly, the lack of association between age and OS in this selected population was likely due to sampling variance and the effects of age as a function of LOH of the HLA Class I region on OS should be further studied with a larger population.
In conclusion, our findings could have potential prognostic value for adult GBM patients and provide new insight into the GBM immunogenetics.
Supplementary Material
Translational Relevance.
Glioblastoma (GBM) demonstrates down-regulated expression of Human Leukocyte Antigen (HLA) Class I to evade the recognition by CD8+ cytotoxic T cells. In this investigation, we found, for the first time, that loss of heterozygosity (LOH) in the HLA Class I gene is associated with shorter overall survival (OS) in newly diagnosed GBM patients. This sheds new light onto the allelic status of these well-characterized genes and its importance in GBM immunosurveillance. These findings provide greater understanding of how GBM evade the immune system and allow further prognostication for GBM patients.
Acknowledgements
Justin Murphy (Molecular Anatomic Pathology Lab at the University of Pittsburgh) for performing the capillary electrophoresis. Stephanie Bortoluzzi (Division of Neuropathology at the University of Pittsburgh) for compiling patient data.
Grant support
Doris Duke Clinical Research Fellowship (JTY), Musella Foundation for Brain Tumor Research and Information (HO), the National Institutes of Health [2R01 NS055140 and 1P01 CA132714 (HO)]. This project used UPCI shared resources (Biostatistics Facility) that are supported in part by NIH P30CA047904.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: J.Yeung, R. Hamilton, I. Pollack, H. Okada
Development of methodology: J.Yeung, R. Hamilton, Pollack, H. Okada
Acquisition of data: J.Yeung, R. Hamilton, K. Ohnishi, M. Ikeura, D. Potter
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): J.Yeung, R. Hamilton, D. Potter, R. Jakacki, I. Pollack, H. Okada
Writing, review, and/or revision of the manuscript: J.Yeung, R. Hamilton, M. Nikiforova, S. Ferrone, R. Jakacki, I. Pollack, H. Okada
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): J.Yeung, R. Hamilton, M. Nikiforova, S. Ferrone, R. Jakacki, I. Pollack, H. Okada
Study supervision: R. Hamilton, S. Ferrone, R. Jakacki, I. Pollack, H. Okada
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Wiemels JL, Wiencke JK, Sison JD, Miike R, McMillan A, Wrensch M. History of allergies among adults with glioma and controls. International journal of cancer Journal international du cancer. 2002;98:609–15. doi: 10.1002/ijc.10239. [DOI] [PubMed] [Google Scholar]
- 3.Schwartzbaum J, Ding B, Johannesen TB, Osnes LT, Karavodin L, Ahlbom A, et al. Association between prediagnostic IgE levels and risk of glioma. Journal of the National Cancer Institute. 2012;104:1251–9. doi: 10.1093/jnci/djs315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang I, Tihan T, Han SJ, Wrensch MR, Wiencke J, Sughrue ME, et al. CD8+ T-cell infiltrate in newly diagnosed glioblastoma is associated with long-term survival. Journal of clinical neuroscience: official journal of the Neurosurgical Society of Australasia. 2010;17:1381–5. doi: 10.1016/j.jocn.2010.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lohr J, Ratliff T, Huppertz A, Ge Y, Dictus C, Ahmadi R, et al. Effector T-cell infiltration positively impacts survival of glioblastoma patients and is impaired by tumor-derived TGF-beta. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:4296–308. doi: 10.1158/1078-0432.CCR-10-2557. [DOI] [PubMed] [Google Scholar]
- 6.Marincola FM, Jaffee EM, Hicklin DJ, Ferrone S. Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Advances in immunology. 2000;74:181–273. doi: 10.1016/s0065-2776(08)60911-6. [DOI] [PubMed] [Google Scholar]
- 7.Garrido F, Algarra I, Garcia-Lora AM. The escape of cancer from T lymphocytes: immunoselection of MHC class I loss variants harboring structural-irreversible “hard” lesions. Cancer immunology, immunotherapy: CII. 2010;59:1601–6. doi: 10.1007/s00262-010-0893-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satoh E, Mabuchi T, Satoh H, Asahara T, Nukui H, Naganuma H. Reduced expression of the transporter associated with antigen processing 1 molecule in malignant glioma cells, and its restoration by interferon-gamma and -beta. Journal of neurosurgery. 2006;104:264–71. doi: 10.3171/jns.2006.104.2.264. [DOI] [PubMed] [Google Scholar]
- 9.Paschen A, Mendez RM, Jimenez P, Sucker A, Ruiz-Cabello F, Song M, et al. Complete loss of HLA class I antigen expression on melanoma cells: a result of successive mutational events. International journal of cancer Journal international du cancer. 2003;103:759–67. doi: 10.1002/ijc.10906. [DOI] [PubMed] [Google Scholar]
- 10.Maleno I, Lopez-Nevot MA, Cabrera T, Salinero J, Garrido F. Multiple mechanisms generate HLA class I altered phenotypes in laryngeal carcinomas: high frequency of HLA haplotype loss associated with loss of heterozygosity in chromosome region 6p21. Cancer immunology, immunotherapy: CII. 2002;51:389–96. doi: 10.1007/s00262-002-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Facoetti A, Nano R, Zelini P, Morbini P, Benericetti E, Ceroni M, et al. Human leukocyte antigen and antigen processing machinery component defects in astrocytic tumors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11:8304–11. doi: 10.1158/1078-0432.CCR-04-2588. [DOI] [PubMed] [Google Scholar]
- 12.Martinez R, Schackert HK, Plaschke J, Baretton G, Appelt H, Schackert G. Molecular mechanisms associated with chromosomal and microsatellite instability in sporadic glioblastoma multiforme. Oncology. 2004;66:395–403. doi: 10.1159/000079488. [DOI] [PubMed] [Google Scholar]
- 13.Raffaghello L, Nozza P, Morandi F, Camoriano M, Wang X, Garre ML, et al. Expression and functional analysis of human leukocyte antigen class I antigen-processing machinery in medulloblastoma. Cancer research. 2007;67:5471–8. doi: 10.1158/0008-5472.CAN-06-4735. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Komohara Y, Domenick N, Ohno M, Ikeura M, Hamilton RL, et al. Expression of antigen processing and presenting molecules in brain metastasis of breast cancer. Cancer immunology, immunotherapy: CII. 2012;61:789–801. doi: 10.1007/s00262-011-1137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrido FC, Accolla RS, et al. Genetic diversity of HLA: functional and medical implications. Hum Immunol; 12th International Histocompatibility Conference; Paris, France. 1996; Jun 9-12, 1996. pp. 1–184. Abstracts. [PubMed] [Google Scholar]
- 16.Hatanpaa KJ, Burger PC, Eshleman JR, Murphy KM, Berg KD. Molecular diagnosis of oligodendroglioma in paraffin sections. Laboratory investigation; a journal of technical methods and pathology. 2003;83:419–28. doi: 10.1097/01.lab.0000059948.67795.ef. [DOI] [PubMed] [Google Scholar]
- 17.Slebos RJ, Umbach DM, Sommer CA, Horner GA, Choi JY, Taylor JA. Analytical and statistical methods to evaluate microsatellite allelic imbalance in small amounts of DNA. Laboratory investigation; a journal of technical methods and pathology. 2004;84:649–57. doi: 10.1038/labinvest.3700076. [DOI] [PubMed] [Google Scholar]
- 18.Nikiforova MN, Hamilton RL. Molecular diagnostics of gliomas. Archives of pathology & laboratory medicine. 2011;135:558–68. doi: 10.5858/2010-0649-RAIR.1. [DOI] [PubMed] [Google Scholar]
- 19.Horbinski C, Nikiforova MN, Hagenkord JM, Hamilton RL, Pollack IF. Interplay among BRAF, p16, p53, and MIB1 in pediatric low-grade gliomas. Neuro-oncology. 2012;14:777–89. doi: 10.1093/neuonc/nos077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hobbs J, Nikiforova MN, Fardo DW, Bortoluzzi S, Cieply K, Hamilton RL, et al. Paradoxical Relationship Between the Degree of EGFR Amplification and Outcome in Glioblastomas. The American journal of surgical pathology. 2012;36:1186–93. doi: 10.1097/PAS.0b013e3182518e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horbinski C, Hamilton RL, Nikiforov Y, Pollack IF. Association of molecular alterations, including BRAF, with biology and outcome in pilocytic astrocytomas. Acta neuropathologica. 2010;119:641–9. doi: 10.1007/s00401-009-0634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe K, Tachibana O, Sata K, Yonekawa Y, Kleihues P, Ohgaki H. Overexpression of the EGF receptor and p53 mutations are mutually exclusive in the evolution of primary and secondary glioblastomas. Brain Pathol. 1996;6:217–23. doi: 10.1111/j.1750-3639.1996.tb00848.x. discussion 23-4. [DOI] [PubMed] [Google Scholar]
- 23.Helseth R, Helseth E, Johannesen TB, Langberg CW, Lote K, Ronning P, et al. Overall survival, prognostic factors, and repeated surgery in a consecutive series of 516 patients with glioblastoma multiforme. Acta Neurol Scand. 2010;122:159–67. doi: 10.1111/j.1600-0404.2010.01350.x. [DOI] [PubMed] [Google Scholar]
- 24.Lacroix M, Abi-Said D, Fourney DR, Gokaslan ZL, Shi W, DeMonte F, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. Journal of neurosurgery. 2001;95:190–8. doi: 10.3171/jns.2001.95.2.0190. [DOI] [PubMed] [Google Scholar]
- 25.Maleno I, Cabrera CM, Cabrera T, Paco L, Lopez-Nevot MA, Collado A, et al. Distribution of HLA class I altered phenotypes in colorectal carcinomas: high frequency of HLA haplotype loss associated with loss of heterozygosity in chromosome region 6p21. Immunogenetics. 2004;56:244–53. doi: 10.1007/s00251-004-0692-z. [DOI] [PubMed] [Google Scholar]
- 26.Maleno I, Aptsiauri N, Cabrera T, Gallego A, Paschen A, Lopez-Nevot MA, et al. Frequent loss of heterozygosity in the beta2-microglobulin region of chromosome 15 in primary human tumors. Immunogenetics. 2011;63:65–71. doi: 10.1007/s00251-010-0494-4. [DOI] [PubMed] [Google Scholar]
- 27.Maleno I, Romero JM, Cabrera T, Paco L, Aptsiauri N, Cozar JM, et al. LOH at 6p21.3 region and HLA class I altered phenotypes in bladder carcinomas. Immunogenetics. 2006;58:503–10. doi: 10.1007/s00251-006-0111-8. [DOI] [PubMed] [Google Scholar]
- 28.Feenstra M, Veltkamp M, van Kuik J, Wiertsema S, Slootweg P, van den Tweel J, et al. HLA class I expression and chromosomal deletions at 6p and 15q in head and neck squamous cell carcinomas. Tissue antigens. 1999;54:235–45. doi: 10.1034/j.1399-0039.1999.540304.x. [DOI] [PubMed] [Google Scholar]
- 29.Kloetzel PM. The proteasome and MHC class I antigen processing. Biochimica et biophysica acta. 2004;1695:225–33. doi: 10.1016/j.bbamcr.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 30.Knudson AG., Jr. Mutation and cancer: statistical study of retinoblastoma. Proceedings of the National Academy of Sciences of the United States of America. 1971;68:820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garrido C, Algarra I, Maleno I, Stefanski J, Collado A, Garrido F, et al. Alterations of HLA class I expression in human melanoma xenografts in immunodeficient mice occur frequently and are associated with higher tumorigenicity. Cancer immunology, immunotherapy: CII. 2010;59:13–26. doi: 10.1007/s00262-009-0716-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrido C, Paco L, Romero I, Berruguilla E, Stefansky J, Collado A, et al. MHC class I molecules act as tumor suppressor genes regulating the cell cycle gene expression, invasion and intrinsic tumorigenicity of melanoma cells. Carcinogenesis. 2012;33:687–93. doi: 10.1093/carcin/bgr318. [DOI] [PubMed] [Google Scholar]
- 33.Powell AG, Horgan PG, Edwards J. The bodies fight against cancer: is human leucocyte antigen (HLA) class 1 the key? Journal of cancer research and clinical oncology. 2012;138:723–8. doi: 10.1007/s00432-012-1192-4. [DOI] [PubMed] [Google Scholar]
- 34.Sato N, Hirohashi Y, Tsukahara T, Kikuchi T, Sahara H, Kamiguchi K, et al. Molecular pathological approaches to human tumor immunology. Pathology international. 2009;59:205–17. doi: 10.1111/j.1440-1827.2009.02353.x. [DOI] [PubMed] [Google Scholar]
- 35.Erdag G, Schaefer JT, Smolkin ME, Deacon DH, Shea SM, Dengel LT, et al. Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer research. 2012;72:1070–80. doi: 10.1158/0008-5472.CAN-11-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamborn KR, Chang SM, Prados MD. Prognostic factors for survival of patients with glioblastoma: recursive partitioning analysis. Neuro-oncology. 2004;6:227–35. doi: 10.1215/S1152851703000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. Journal of human genetics. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- 38.La Riviere G, Klein Gebbinck JW, Schipper CA, Roos E. Tumour necrosis factor-alpha stimulates invasiveness of T-cell hybridomas and cytotoxic T-cell clones by a pertussis toxin-insensitive mechanism. Immunology. 1992;75:269–74. [PMC free article] [PubMed] [Google Scholar]
- 39.Roth W, Weller M. Chemotherapy and immunotherapy of malignant glioma: molecular mechanisms and clinical perspectives. Cellular and molecular life sciences: CMLS. 1999;56:481–506. doi: 10.1007/s000180050447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanabe K, Matsushima-Nishiwaki R, Yamaguchi S, Iida H, Dohi S, Kozawa O. Mechanisms of tumor necrosis factor-alpha-induced interleukin-6 synthesis in glioma cells. Journal of neuroinflammation. 2010;7:16. doi: 10.1186/1742-2094-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyakawa A, Ichimura K, Schmidt EE, Varmeh-Ziaie S, Collins VP. Multiple deleted regions on the long arm of chromosome 6 in astrocytic tumours. British journal of cancer. 2000;82:543–9. doi: 10.1054/bjoc.1999.0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Healy E, Belgaid C, Takata M, Harrison D, Zhu NW, Burd DA, et al. Prognostic significance of allelic losses in primary melanoma. Oncogene. 1998;16:2213–8. doi: 10.1038/sj.onc.1200203. [DOI] [PubMed] [Google Scholar]
- 43.Chuaqui R, Silva M, Emmert-Buck M. Allelic deletion mapping on chromosome 6q and X chromosome inactivation clonality patterns in cervical intraepithelial neoplasia and invasive carcinoma. Gynecologic oncology. 2001;80:364–71. doi: 10.1006/gyno.2000.6087. [DOI] [PubMed] [Google Scholar]
- 44.Rodabaugh KJ, Blanchard G, Welch WR, Bell DA, Berkowitz RS, Mok SC. Detailed deletion mapping of chromosome 6q in borderline epithelial ovarian tumors. Cancer research. 1995;55:2169–72. [PubMed] [Google Scholar]
- 45.Mohapatra G, Kim DH, Feuerstein BG. Detection of multiple gains and losses of genetic material in ten glioma cell lines by comparative genomic hybridization. Genes, chromosomes & cancer. 1995;13:86–93. doi: 10.1002/gcc.2870130203. [DOI] [PubMed] [Google Scholar]
- 46.Weber RG, Sabel M, Reifenberger J, Sommer C, Oberstrass J, Reifenberger G, et al. Characterization of genomic alterations associated with glioma progression by comparative genomic hybridization. Oncogene. 1996;13:983–94. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.