Abstract
MazF is an mRNA interferase that cleaves mRNAs at a specific RNA sequence. MazF from E. coli (MazF-ec) cleaves RNA at A^CA. To date, a large number of MazF homologues that cleave RNA at specific three- to seven-base sequences have been identified from bacteria to archaea. MazF-ec forms a dimer, in which the interface between the two subunits is known to be the RNA substrate-binding site. Here, we investigated the role of the two loops in MazF-ec, which are closely associated with the interface of the MazF-ec dimer. We examined whether exchanging the loop regions of MazF-ec with those from other MazF homologues, such as MazF from Myxococcus xanthus (MazF-mx) and MazF from Mycobacterium tuberculosis (MazF-mt3), affects RNA cleavage specificity. We found that exchanging loop 2 of MazF-ec with loop 2 regions from either MazF-mx or MazF-mt3 created a new cleavage sequence at (A/U)(A/U)AA^C in addition to the original cleavage site, A^CA, while exchanging loop 1 did not alter cleavage specificity. Intriguingly, exchange of loop 2 with 8 or 12 consecutive Gly residues also resulted in a new RNA cleavage site at (A/U)(A/U)AA^C. The present study suggests a method for expanding the RNA cleavage repertoire of mRNA interferases, which is crucial for potential use in the regulation of specific gene expression and for biotechnological applications.
Keywords: Sequence-specific endoribonucleases, MazE-MazF, TA systems, Chimeric proteins
Introduction
The toxin-antitoxin (TA) systems were originally discovered in low copy number plasmids and stably maintain the plasmid by selectively killing daughter cells which have lost the plasmid 1. In type II TA systems, toxin genes are co-expressed with their cognate antitoxin genes present in the same operons. Toxins and their cognate antitoxins form stable TA complexes in cells under normal growth conditions. Since antitoxins are labile proteins that are readily degraded by stress-induced proteases, the balance between toxin and antitoxin is altered under stress conditions to release free toxins in the cells from the TA complexes. This results in growth arrest and eventual death 2-4.
mRNA interferases are sequence-specific endoribonucleases encoded as toxins in a number of TA systems 5,6. The MazF-ec in Escherichia coli was the first identified mRNA interferase consisting of 111 residues and forms a stable dimer that cleaves RNA specifically at A^CA (^ indicates cleavage site) 7. To date, a large number of MazF homologues have been identified from various bacteria and some species of archaea 8. Staphylococcus aureus contains one MazF homologue, MazF-sa, which has been shown to cleave mRNA at U^ACAU sequences 9. A MazF homologue from Bacillus subtilis (MazF-bs) has 18.3% identity and 40.5% homology to MazF-ec and also cleaves RNA at U^ACAU 10. In addition, the MazF homologue, MazF-hw, was recently identified from a halophilic archaeon, Haloquadratum walsbyi, found in the Sinai Peninsula as a seven-base cutter cleaving RNA at UU^ACUCA sequences 11.
The crystal structure of MazF-ec shows that it forms a stable dimer, and each dimer further forms a heterohexamer complex with a dimer of MazE-ec, the cognate antitoxin for MazF-ec 12. The heterohexamer, MazF2-MazE2-MazF2, is maintained in normally growing cells. However, under stress conditions, stress-induced proteases preferentially eliminate MazE-ec dimers, inducing MazF mRNA interferase activity. This results in cell growth arrest and eventual cell death 13. Each MazF-ec monomer forms a single globular domain consisting of a seven-stranded β-sheet with three α-helices 12, while each MazE-ec molecule contains a long unstructured C-terminal extension, which binds to the RNA substrate binding site created at the interface between two MazF-ec molecules in a MazF-ec dimer 12.
The MazF-ec homodimer has two identical sites for nucleotide substrate binding, to which the C-terminal tail of MazE-ec binds. NMR spectroscopy analysis with the MazF-ec mutant E24A [MazF-ec(E24A)] demonstrated that the nucleotide binding sites on the MazF-ec dimer completely overlaps with MazE-ec binding sites 14. Notably, there are minor differences between the NMR-derived secondary structure of MazF-ec(E24A) 14 and the crystal structure of MazF-ec in the MazE-MazF complex 12: one at β-strand S1, which is four residues longer, and the other at α-helix H3, which is two residues shorter in the NMR structure of MazF-ec(E24A). Putative nucleotide-binding sites in the NMR structure were proposed, as they were perturbed in the presence of an uncleavable substrate 14. Although the nucleotide-binding surface largely overlaps with the MazE interacting regions, there appears to be distinct differences between them. For example, H28 and Y58 are perturbed more strongly with substrate present than with the MazE C-terminal peptide present 14.
Here, we first propose a computational docking model of MazF-ec complexed with RNA. This model shows that the loops between antiparallel β-strands in the MazF dimer are important for mRNA cleavage specificity. Subsequently, we exchanged two loop regions of MazF-ec with those from MazF-mx and MazF-mt3 as well as poly-glycine sequences to investigate the importance of the two loops on RNA sequence recognition. These approaches revealed that the loop region between the third and the fourth β-sheets of MazF-ec (loop 2) is involved in the RNA recognition since RNA was cleaved at (A/U)(A/U)AA^C in addition to the original A^CA sequences minimally required for MazF-ec recognition.
Materials and Methods
Docking Model
To generate a docked complex of single stranded RNA and MazF-ec, the solution RNA structure (PDB ID: 2K5Z) and the crystal structure of MazF-ec (PDB: 1UB4) were chosen as templates. MazF-ec has missing coordinates for residues 20-26 in the flexible loop, and the loops were reconstituted using ModBase web server 15 to present full atomic coordinates of MazF-ec. An 8-nt RNA fragment (C4UACAUUC11) from the 2K5Z molecule covering the MazF-ec recognition sequence was used to perform rigid-body docking by HADDOCK 16 to obtain the MazF-RNA complex structure. The nucleotides of the RNA fragment C4UACAUUC11 were truncated to C1UACAUUC8 in this present study. Chemical shift perturbations reported in previous NMR studies of MazF-ec 14 were used as primary information for docking restraints. The example docking procedure provided by HADDOCK was adopted to generate 4000 and 1000 models in the first and second iterations, respectively. We assumed that residues 20-26 missing in the crystal structure of MazF (PDB ID: 1UB4) 12 are associated with RNA substrate binding so that the structure of this region can be freely altered during the docking process. Two hundred lowest energy models were further refined in the explicit water system. All water-refined structures are clustered in a single group showing pairwise backbone RMSD constantly smaller than 2 Å. The interactions of RNA and MazF-ec of the 50 lowest energy structures were analyzed using LigPlot 17 and PDBe PISA 18.
Strains and Plasmids
The E. coli DH5α, BW25113 (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) and BW25113 ΔmazEF strains 19 were used for recombinant mutant protein production, for toxicity assay on plate and in liquid, and for in vivo primer extension to identify cleavage sites.
Construction of Mutants Plasmids
The six loop and four poly-glycine mutants were amplified by PCR using pBAD33mazF-ec plasmid as template and primers shown in Table I and then cloned into the pBAD33 vector by using a modified overlap extension technique 20 together with the optimized Shine-Dalgarno (SD) sequence (A−14AGGAGA−8, +1 indicates translation start site).
TABLE I.
Primers used in this study
| Name | Primer sequences (5′ to 3′) | Purpose |
|---|---|---|
| Loop1-mt3 Fw | gttgatttaggcccgccaagtggtagccagccagctaaacgtcgtccaagtgttgtcctg | Cloning |
| Loop1-mt3 Rv | acctgctgcaaatcgaccgaccgatggtgaaccgcccggatttagttgggtttagtctag | Cloning |
| Loop2-mt3 Fw | tgtacaacgtcaaacacagcattagcggccatgccaggaaatgttgttttatccggtcag | Cloning |
| Loop2-mt3 Rv | ttgttgtaaaggaccgtaccggcgattacgacacaaactgcaacatgttccttgtgt | Cloning |
| Loop1-mx Fw | tgatcctgacgattcaggtcccgtgccatcttactctcgtccaagtgttgtcctg | Cloning |
| Loop1-mx Rv | cctgctctcattctaccgtgccctggagaacttagcagtcctagttgggtttagtctag | Cloning |
| Loop2-mx Fw | tgtacaacgtcaaacttacatcgtgcgtccgagggaaatgttgttttatccggtcaggaa | Cloning |
| Loop2-mx Rv | ttttgttgtaaagggagcctgcgtgctacattcaaactgcaacatgttccttgtgt | Cloning |
| Loop1-F1G12 Rv | tcctcctccacctccaccaccacctccacctccaccatcaacccaaatcag | Cloning |
| Loop1-F2G12 Fw | tggaggtggaggaggacgtccagctgttgtc | Cloning |
| Loop2-F2G8 Rv | acctccaccaccacctcctcctcccgttgtacaaggaac | Cloning |
| Loop2-F2G12 Rv | acctccaccaccacctcctcctccacctccacccgttgtacaaggaac | Cloning |
| Loop2-F3 Fw | aggtggtggtggaggtgttgttttatccggt | Cloning |
| R3 | gtttttaccataaacgttgg | Primer extension |
Toxicity Assay of Loop and Poly-glycine MazF Mutants
E. coli BW25113 cells were used for transformation, and the transformants harboring pBAD33mazF-ec plasmids with loop 1, loop 2, or loop 1+2 region from mazF-mt3, –mx, and poly-glycines were streaked onto M9 agar plates in the presence or absence of 0.2% arabinose. Growth curves were measured using E. coli BW25113 cells harboring pBAD33mazF-ec containing loop exchanges at loop 1, loop 2, or loop 1+2 regions from mazF-mt3 or mazF–mx. The cells were grown in LB liquid medium at 37°C in the presence or absence of 0.2% arabinose.
Primer Extension Analysis in vivo
For primer extension analysis of mRNA cleavage sites in vivo, total RNA was extracted from E. coli BW25113 or BW25113 ΔmazEF cells containing pBAD33mazF-ec loop and poly-glycine mutants at different time points: 165 min for loop 2 and loop 1+2, and 210 min for loop 1 after 0.2% arabinose induction according to cell toxicity [Fig. 2(E,G)]. For control reactions without the addition of 0.2% arabinose, cells were collected at 0 hr and at the same time points as above (165 min for loop 2 and loop 1+2 and 210 min for loop 1). After incubation with MazF mutants in vivo, cleaved total RNA was extracted. Primer extension was performed with reverse transcriptase using total cleaved RNA and [γ-32P]ATP-labeled ompA target primers (Table I). The reaction was stopped by the addition of 12 μl of sequencing loading buffer (95% formaldehyde, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol EF), heated at 95 °C for 5 min, and analyzed on a 6% polyacrylamide-containing 8 M urea with a sequence ladder made with the same primer 21.
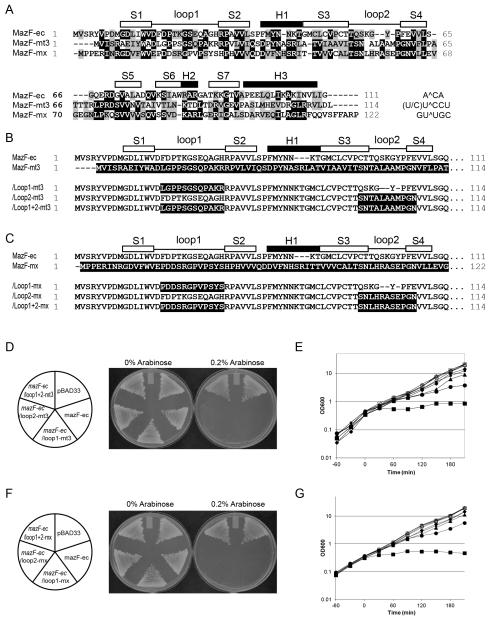
Figure 2. Construction and toxicity of MazF loop mutants.
(A) Sequence alignments of MazF homologues from Escherichia coli (MazF-ec), Mycobacterium tuberculosis (MazF-mt3), and Myxococcus xanthus (MazF-mx). (B and C) Amino acid sequence comparison of loop mutants MazF-ec and MazF-mt3 (B), and MazF-ec and MazF-mx (C). Black shadowed sequences indicate the residues from MazF-mt3 (B) or from MazF-mx (C). White and black boxes indicate β-sheets (S) and α-helices (H), respectively, on the basis of the secondary structure of MazF-ec (PDB ID: 1UB4). (D and F) The transformants harboring pBAD33, mazF-ec, /loop1-mt3, /loop2-mt3, /loop1+2-mt3 plasmids (D), and pBAD33, mazF-ec, /loop1-mx, /loop2-mx, and /loop1+2-mx plasmids (F) were streaked onto M9 plates in the presence or absence of 0.2% arabinose and then incubated at 37°C. (E and G) Growth curves of E. coli BW25113 cells harboring pBAD33 vector (diamond), mazF-ec (rectangle), /loop1-mt3 (circle), /loop2-mt3 (triangle), /loop1+2-mt3 (small rectangle) (E), pBAD33 vector (diamond), mazF-ec (rectangle), /loop1-mx (circle), /loop2-mx (triangle), and /loop1+2-mx (small rectangle) (G). Cells were grown in M9-glucose liquid medium at 37°C in the presence (closed) or absence (open) of 0.2% arabinose.
Results and Discussion
Computational Structural Model of the MazF-ec and RNA Complex
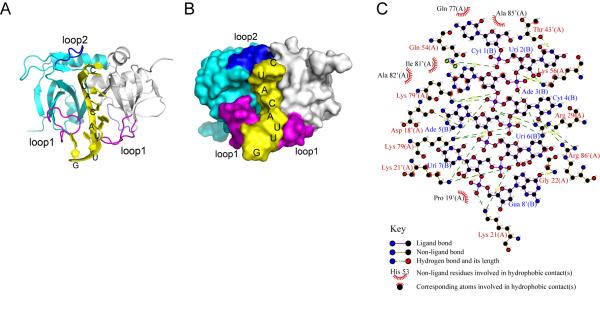
NMR spectroscopy shows that the MazF homodimer contains two identical RNA substrate binding sites 14. It was predicted that one active site loses RNA binding activity when an RNA substrate binds to the other active site in MazF-ec. These sites largely overlap the binding sites for the C-terminal tail of MazE-ec 14. X-ray diffraction determined that the structure of the MazE-MazF complex (PDB ID: 1UB4) does contain electron density at the loop 1 region, presumably due to the loop’s flexibility. Since loop 1 (S1-S2 loop) is suggested to be crucial for RNA binding and cleavage 14, we predicted a structure containing a flexible loop 1 as generated by the ModBase web server 15 using 1UB4 as template. This was used for all molecular docking and structural analyses. Here, we used HADDOCK to construct a docking model for the MazF-ec complex with an 8-nt RNA structure (PDB ID: 2K5Z) and to define the molecular interactions between RNA and MazF-ec. HADDOCK 16 is an information-driven, flexible docking program that uses experimental results, including point mutations, binding, and titration to generate structural models of protein-DNA complexes. The use of HADDOCK has been reported for 47 protein-DNA complexes, and the success rate is higher than 94% 22. The 8-nt RNA fragment contains not only the central cleavable A^CA sequence but also both residues at the 5′- and 3′-ends of the A^CA sequence that interact with MazF-ec. Active and passive residues of MazF-ec used in HADDOCK are obtained from chemical shift perturbations observed in the NMR study of the MazF-ec and single stranded DNA complex 14. Notably, HADDOCK has been successfully used to observe the RNA binding and interaction with Kid, a MazF structural homolog 23. The structural model of the MazF-ec and RNA complex using HADDOCK is shown in Figure 1(A,B). Detailed analysis of molecular interactions between MazF-ec and RNA using LigPlot 17 is shown in Figure 1(C). There are three hydrogen bonds and two hydrophobic contacts between MazF-ec and the 5′ region (C1U2) of the RNA substrate [Fig. 1(C)]. Interestingly, two hydrogen bonds form with Q54 and K56 [cyan in Figure 1(B)], both of which are in the loop 2 region. The other hydrogen bond forms with T43′ [the prime (’) indicates amino acid residues from the second monomer; white in Figure 1(B)]. In U6UG8 of the 8-mer substrate, six hydrogen bonds form with three amino acid residues, K21, K21′, and G22. P19 has hydrophobic contact with the RNA substrate. However, all of the interactions mentioned above may not be involved in specific RNA-sequence recognition since these interactions are observed with extra bases of the RNA substrate at both 5′ (CU at 5′) and 3′ ends (UUG) but not with the A^CA sequence. Differences in the absolute protein structures could not be predicted due to the flexibility of loop regions. Nevertheless, along with A^CA, these extra interactions appear to enhance RNA binding in the concave region located in the interface between two MazF-ec monomers.
Figure 1. A structural model of the MazF-ec and RNA complex.
(A) A ribbon view of the complex of homodimeric MazF-ec with an RNA fragment. In this complex, two monomeric subunits of MazF-ec are colored in cyan and gray ribbons, and the 8-nt RNA fragment (5′-C1UACAUUG8) is presented as a yellow stick. Two important flexible loops, loop 1 (residue 16-28) and loop 2 (residues 53-59) in each subunit involved in RNA binding are colored in purple and blue, respectively. (B) A surface view further displays that the RNA fragment is tightly inserted into the cavity of the dimeric interface, interacting with the two flexible loops. The structure images were prepared using PyMOL. (C) The interactions between RNA and MazF-ec of each model were analyzed using LigPlot 17.
Five hydrogen bonds exist between the phosphate backbone (between A3 and C4) in the RNA cleavage site and MazF-ec residues K79, R29 and K79′; the ribose in A3 forms hydrophobic contacts with MazF-ec [Fig. 1(C)]. The C4 and A5 bases do not have hydrophobic interactions, while these bases form hydrogen interactions with R86′, D18′, and K79. These interactions from our model indicate that loop 1 interacts with the 3′ region downstream of the A^CA sequence in addition to its interaction with the A^CA sequence. The location of the K79, K79′ and R29 residues in MazF-ec correspond structurally to those of catalytic triad residues, R73, D75, and H17 in Kid 23 (Fig. S1). Our model lacks a negatively charged residue corresponding to D75 in Kid, which was predicted to act as the catalytic base in MazF-ec 23. However, D18 and E24 are located close to the catalytic triad region, and alanine substitution of either one of two residues eliminates the MazF-ec toxicity 14. Our data also show that the RNA binding site of MazF-ec is located at the interface between two monomers forming the concave region and that two loop regions, loops 1 and 2, are also involved in RNA binding as demonstrated previously 14. Our computational docking model shows that MazF-ec residues, D18′, R29 (loop 1), K56 (loop 2), and K79, K79′, R86′ (S6) mediate the interaction between MazF-ec and A^CA of RNA substrate. K56 is one of residues in the loop 2 region and hydrogen bonds with U2 and A3 in RNA substrate. This U^ACA site was reported as another cleavage site of MazF-ec in in vitro conditions 24. Both the Kid structure and this docking model suggest that S5-S6 region might contain catalytic residues and/or mediate RNA-sequence recognition region 23. However, K79 and R86 residues in this region are highly homologous among MazF homologues 10, and thus, K79 and R86 are thought to be catalytic sites rather than RNA-sequence recognition sites. Taken together, our data lead us to hypothesize that the flexible loop regions control RNA-sequence recognition specificities for MazF mRNA interferases.
Exchanging of Loops 1 and 2 of MazF-ec with those from MazF-mx and MazF-mt3
The secondary and tertiary structures of MazF-sa 25, MazF-bs 26 and Kid 23 are highly similar to that of MazF-ec 12,27 because RMSD values of MazF-bs and Kid to MazF-ec (PDB ID: 1NE8, 2C06, and 1UB4, respectively) are high (1.90 and 1.84, respectively). These RMSD values were calculated using PDBeFold 28. The NMR assignment of MazF-sa structure was reported, but the final tertiary structure was not determined 25. We analyzed the chemical shifts of MazF-sa (BMRB accession code: 17288) 25 using TALOS+ 29, and showed that the helix/strand regions of MazF-sa have high similarity to those of MazF-ec (Fig. S2). Although, MazF homologues are not highly similar in terms of primary structure 8, there are a few highly conserved amino acid residues among MazF homologues [Fig. 2(A)]. These conserved residues are most likely involved in the common characteristics of all MazF homologues, such as RNA hydrolysis and protein backbone structure. On the other hand, highly diverse residues may serve for recognition of specific RNA sequences.
As shown in Figures 1 and 2, the loop regions of MazF homologues appear to be important and are highly diverse in their amino acid sequences. Thus, to investigate whether these loops are involved in RNA recognition, we examined the roles of the long loop regions, particularly loop 1 and loop 2 [Fig. 1(A, B)] in MazF. For this purpose, the loop 1 and loop 2 regions of MazF-ec (A^CA specific) were exchanged with the respective loop regions from MazF-mt3 in Mycobacterium tuberculosis [(U/C)U^CCU specific] or from MazF-mx in Myxococcus xanthus [GU^UGC specific]. Sequence similarities between MazF-ec with MazF–mt3 and MazF-mx are 29.8% and 38.1%, respectively. However, the RNA cleavage sequences for the three MazFs are completely distinct. As depicted in Figure 2(B, C), six loop mutations were created, and these chimeric proteins were expressed using the pBAD33 vector in E. coli. The pBAD33 vector has an arabinose responsive promoter; thus, synthesis of the mutant MazF protein was controlled by arabinose, and thus the amount of mutant MazF protein can be manipulated with the amount of arabinose. When BW25113 was used as the host cell, growth on M9 plates with 0.2% arabinose continued to cause toxicity in the individual MazF-ec/loop1 and /loop 2 exchange mutants , while the MazF-ec/loop 1+2 mutants showed less toxicity compared to individual loop mutants [Fig. 2(D, F)]. In a liquid culture, the growth of the cells harboring these mutants was not significantly inhibited after addition of 0.2% arabinose in LB medium, whereas the growth of the cells harboring MazF-ec was strongly inhibited as expected [Fig. 2(E, G)]. All MazF-ec/loop1 mutants [shown as circles in Figure 2(E,G)] showed growth inhibition only after one generation. Inhibition of cell growth by /loop2 (triangles) and /loop1+2 mutants (squares) was found to beless than those of /loop1 mutants [Fig. 2(E, G)]. These results indicate that MazF continues to be active when loop 1 of MazF-ec is exchanged with those from other MazF homologues, while the exchange of loop 2 or both loop 1 and loop 2 severely reduces the endoribonuclease activity. This is likely due to the change in RNA recognition specificities, RNA cleavage activity, and/or reduced expression of MazF mutant proteins.
Determination of Cleavage Sites of Loop MazF mutants by in vivo Primer Extension
Next, in vivo primer extension analysis (in vitro primer extension using mRNA purified in vivo) was carried out to identify RNA cleavage sites recognized by the MazF mutants. The MazF-ec/loop1-mt3 mutant cleaved ompA mRNA at the identical sites (A^CA) that wild type MazF-ec cleaved [Fig. 3(A, B) and Table II]. Interestingly, the MazF-ec/loop2-mt3 mutant also cleaved at new sites, AAAA^C, UUAA^C and UAAA^C, differing from those of MazF-ec (A^CA) and MazF-mt3 (UU^CCU or CU^CCU) [Fig. 3(A, B) and Table II]. We found those cleavages at additional sites were detected when the new cleavage sequences (AAAA^C, UUAA^C and UAAA^C) were followed by any nucleotide, not only A [Fig. 3(A, B), Table (II, S1, and S2)]. We were unable to detect any cleavage site in the RNA when we used MazF-ec/loop1+2-mt3 [Fig. 3(A, B) and Table II]. We found that this protein was highly insoluble, making it impossible to detect activity (Fig. S3). We also identified the cleavage sequences recognized by MazF-ec/loop-mx mutants in vivo. Like MazF-ec/loop1-mt3, MazF-ec/loop1-mxcleaved ompA mRNA at A^CA. Surprisingly, exchanging loop 2 for that of MazF-mx produced the same result as MazF-ec/loop2-mt3, that is cleavage of RNA at (A/U)(A/U)AA^C [Fig. 3(C) and Table (II, S1, and S2)].
Figure 3. Primer extension analyses of the ompA mRNA from the loop mutants.
(A and B) In vivo primer extension analyses of the loop mutants of MazF-ec and MazF-mt3 in BW25113 ΔmazEF (A) and in BW25113 cells (B). (C) In vivo primer extension analyses of the loop mutants of MazF-ec and MazF-mx in wild-type BW25113 cells. A^CA cleavage sites (MazF-ec) are shown as arrows and asterisks to indicate new cleavage sites in loop 2 mutants and as circles in loop 1 mutants.
TABLE II.
Cleavage sites of MazF-ec loop mutants in ompA mRNA
| MazF-ec loop mutants | Cleavage site | Strains |
|---|---|---|
| MazF-ec/loop1-mt3 | A^CA | BW25113 |
| MazF-ec/loop2-mt3 | (A/U)(A/U)AA^C, A^CA | BW25113 |
| MazF-ec/loop1+2-mt3 | n/a | BW25113 |
| MazF-ec/loop1-mt3 | n/a | BW25113 ΔmazEF |
| MazF-ec/loop2-mt3 | (A/U)(A/U)AA^C, A^CA | BW25113 ΔmazEF |
| MazF-ec/loop1+2-mt3 | n/a | BW25113 ΔmazEF |
| MazF-ec/loop1-mx | A^CA | BW25113 |
| MazF-ec/loop2-mx | (A/U)(A/U)AA^C, A^CA | BW25113 |
| MazF-ec/loop1+2-mx | n/a | BW25113 |
| MazF-ec/loop1-G12 | n/a | BW25113 ΔmazEF |
| MazF-ec/loop2-G8 | (A/U)(A/U)AA^C, A^CA | BW25113 ΔmazEF |
| MazF-ec/loop2-G11 | (A/U)(A/U)AA^C, A^CA | BW25113 ΔmazEF |
| MazF-ec/loop1-G12+loop2-G8 | n/a | BW25113 ΔmazEF |
indicates the cleavage site.
n/a indicates no cleavage
When in vivo primer extension was carried out in an mazEF deletion strain, BW25113 ΔmazEF, MazF-ec/loop1-mt3 and MazF-ec/loop1–mx mutants did not show any cleavage activity [Fig. 3(B) and Table II], indicating that the cleavage of RNA at A^CA in BW25113 wild-type resulted from endogenous MazF activity. This endogenous MazF is most likely released from the MazE-MazF complex as MazF-ec/loop1-mt3 or –mx mutant proteins competitively bind to MazE in the endogenous MazE-MazF complex, resulting in the release of free MazF.
Blocking the Function of Loop Regions in MazF-ec by replacement with Poly-glycine
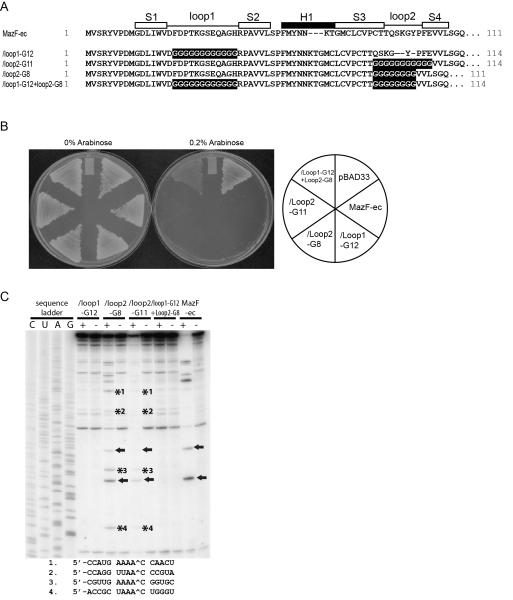
To further investigate how the amino acid residues of loop regions play a role in RNA sequence recognition and MazF catalytic activity, four new loop mutants were constructed in which loop 1 and loop 2 were replaced with poly-Gly sequences. The number of Gly residues in each loop was set to be identical to that in each loop of either MazF-ec or MazF-mt3 [Fig. 4(A)], so that the loop 1 mutant contains 12 Gly residues, while loop 2 mutants were constructed with two different lengths of 8 or 11 Gly residues.
Figure 4. Poly-glycine mutants of MazF-ec.
(A) Amino acid sequences of poly-glycine mutants. White and black boxes indicate β-sheets (S) and α-helices (H), respectively, on the basis of the secondary structure of MazF-ec (PDB ID: 1UB4). (B) Toxicity of the poly-glycine mutants of MazF-ec. Transformants harboring pBAD33 vector, mazF-ec, /loop1-G12, /Loop2-G8, /Loop2-G11, and /loop1-G12+loop2-G8 plasmids were streaked onto M9 plates in the presence or the absence of 0.2% arabinose and then incubated at 37°C. (C) In vivo primer extension analyses of the ompA mRNA from BW25113 ΔmazEF. A^CA cleavage sites (MazF-ec) are indicated by arrows, and asterisks indicate new cleavage sites in the loop 2 poly-glycine mutants.
To measure the toxicity of the mutants, BW25113 cells harboring four poly-Gly mutants were grown at 37°C. As shown in Figure 4(B), all mutants except mazF-ec/loop1-G12+loop2-G8 showed toxicity on M9 plates in the presence of 0.2% arabinose at 37°C. Next, to determine RNA cleavage sites, in vivo primer extension analysis was carried out in BW25113 ΔmazEF to eliminate endogenous MazF-ec. Surprisingly, although loop regions in MazF were replaced with poly-Gly sequences, all cleavage sites of MazF-ec/loop1-G12, MazF-ec/loop2-G8, MazF-ec/loop2-G11, and MazF-ec/loop1-G12+loop2-G8 were identical to those of the loop mutants described above (Table II). MazF-ec/loop1-G12 did not show any cleavage activity, while MazF-ec/loop2-G8 and MazF-ec/loop2-G11 cleaved RNA at AAAA^C, UUAA^C or UAAA^C [(A/U)(A/U)AA^C; Fig. 4(C)]. This consensus sequence is identical to the consensus sequence of the loop 2 exchange mutants of MazF-ec with MazF-mt3 and MazF-mx, indicating that the loop 2 sequence of MazF-ec is unique for recognition and cleavage of the A^CA site.
Conclusion
To date, NMR structural studies have been carried out to determine the interaction of mRNA interferases with RNA substrates using Kid 20 and MazF-ec 14,23. In the RNA-Kid complex, the hydrogen bonds and hydrophobic contacts between Kid and RNA have been characterized, and D75, R73, and H17 residues were proposed to be part of the catalytic residues of Kid 23. For MazF-ec, the Kd for substrate binding was estimated to be in the sub-μM range30. In the present paper, we constructed a computational structural model based on two previously proposed models 14,23 and we hypothesized that two loop regions, loop 1 (residue 16-28) and loop 2 (residues 53-59), in MazF-ec play important roles in RNA recognition and in catalytic activity. In this model, loop 1 binds to and the downstream of the ACA sequence and is involved in catalytic activity in a similar manner to the loop 1 region of Kid 23. In our model, loop 2 alsointeracts with the 5′ upstream region of the ACA sequence. Therefore, RNA recognition specificities may be altered by exchanging these loop regions with those of other MazF homologues. Our results reveal that exchange of loop 1 to either loop 1 of MazF-mt3 or MazF-mx resulted in complete loss of the activity of MazF-ec (Fig. 2). On the other hand, exchanging loop 2 resulted in a new cleavage site recognized by MazF, AAAA^C, UUAA^C or UAAA^C in addition to the original A^CA, regardless of the MazF homologue (MazF-mt3 or MazF-mx) used for the exchange. These new cleavage sequences do not reflect the cleavage sites of wild-type MazF-mt3 and MazF-mx, which cleave RNA at UU^CCU or CU^CCU and GU^UGC, respectively. To our surprise, the exchange of the loop 2 of MazF-ec with poly-Gly (either Gly8 or Gly11) resulted in identical RNA cleavage specificity to that of the loop 2 exchange mutants with MazF-mt3 and MazF-mx, indicating that RNA cleavage specificity is not directly associated with the amino acid sequence of the loop 2 region. At present, it is not known how the loop 2 sequence of MazF-ec specifically recognizes the ACA sequence.
It is interesting to note that in the loop 2 exchange mutants, RNA recognition specificity is altered in the 5′ region of the RNA substrate. This result is consistent with our model in which Q54 and K56 in loop 2 of MazF-ec form three hydrogen bonds and two hydrophobic contacts at the 5′ region (C1U2) of the RNA substrate [Fig. 1(C)]. Apparently, loop 2 mutants do not directly affect the catalytic activity of MazF-ec, as all loop 2 mutants retained the A^CA cleavage activity (the original cleavage site of MazF-ec) in both the wild-type and ΔmazEF BW25113 cells in addition to the new (A/U)(A/U)AA^C cleavage specificity (Figs. 3 and 4). In this new cleavage site, the cleavage specificity of MazF-ec shifted from the 5′ side to the 3′ side of the RNA substrate. In summary, we propose that the loop 1 region of MazF-ec participates in the catalytic activity of MazF-ec, while the loop 2 region is involved in RNA recognition at the 5′ region of RNA substrates as well as the specificity of RNA recognition. Therefore, both loops 1 and 2 play crucial but separate roles at the MazF-ec mRNA interferase. This paper reports the generation of new RNA recognition sequences and RNA cleavage sites for mRNA interferases for the first time. Since there are a number of MazF homologues with different RNA cleavage specificities 31, the exchange of loop 2 or other regions of these MazF homologues with those of other homologues is also likely to generate many new RNA recognition sequences. This approach not only leads to deciphering the exact roles of loop 2 in MazF mRNA interferases but also opens a new avenue for generating a large amount of homogeneous RNA of a certain size for other biological applications. In particular, by using sequence-specific mRNA interferases, both the 5′ and 3′ ends of homogeneous RNA can be uniformly produced, which has not been previously achieved. The approaches described in the present paper may be useful for expansion of RNA cleavage repertoire for mRNA interferases, which is crucial for their potential use for the regulation of specific gene expression in cells and for biotechnological applications, such as high production of homogeneous RNA of a specific sequence.
Supplementary Material
Figure S1. Sequence alignments based on overlaid tertiary structures between MazF-ec and Kid
Sequence alignments were carried out based on the overlaid tertiary structures between MazF-ec (PDB ID: 1UB4) and Kid from Escherichia coli (PDB ID: 1M1F). White and black boxes indicate β-sheets (S) and α-helices (H), respectively, on the basis of the secondary structures of MazF-ec (PDB ID: 1UB4).
Figure S2. The helix/strand regions of MazF-ec and MazF-sa based on NMR assignments
Sequence alignments were carried out using ClustalW2. The helix/strand regions of MazF-ec and MazF-sa were analyzed by NMR assignments. White and black boxes indicate β-sheets (S) and α-helices (H), respectively.
Figure S3. Characterization of protein solubility of MazF-ec/loop1+2-mt3.
mazF-ec/loop1+2-mt3 was cloned into peT28a with a C-terminal His-tag and was introduced into E. coli BL21(DE3). Protein expression was induced by the addition of 1M isopropyl-β-□-1-thiogalactoside (IPTG) for 5 h (lane 3 to 7) at 37°C in M9 media including 0.4% glucose. The cells were disrupted by sonication in 500μl buffer (20 mM Tris-HCl pH 8.0). The supernatants and pellet were separated by centrifugation at 18,000 × g for 10 min. Lane1, whole-cell lysate before induction; lane 2, whole-cell lysate after incubation with IPTG for 3 h at 37°C; lane 3, whole-cell lysate after incubation with IPTG for 5 h at 37°C; lane 4, supernatants (sup) without NaCl added; lane 5, pellet (pel) without NaCl added; lane 6, sup with 0.5M NaCl added to extract protein; lane 7, pel with 0.5M NaCl added to extract protein; lane 8, molecular mass markers. Arrow indicates the band size of MazF-ec/loop1+2-mt3.
ACKNOWLEDGMENTS
We thank Dr. Bonnie Firestein for manuscript suggestions; Dr. Sangita Phadtare, Ana Rodriguez and Dr. Munjin Kwon for the critical reading of this manuscript. This work was supported by the National Institutes of Health grant, [1RO1GM081567] and [3RO1GM081567-02S1] and partly by the KRIBB Research Initiative Program.
REFERENCES
- 1.Gerdes K. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. Journal of bacteriology. 2000;182(3):561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends in biochemical sciences. 2005;30(12):672–679. doi: 10.1016/j.tibs.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Engelberg-Kulka H, Sat B, Reches M, Amitai S, Hazan R. Bacterial programmed cell death systems as targets for antibiotics. Trends in microbiology. 2004;12(2):66–71. doi: 10.1016/j.tim.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nature reviews Microbiology. 2005;3(5):371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 5.Inouye M. The discovery of mRNA interferases: implication in bacterial physiology and application to biotechnology. Journal of cellular physiology. 2006;209(3):670–676. doi: 10.1002/jcp.20801. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi Y, Inouye M. mRNA interferases, sequence-specific endoribonucleases from the toxin-antitoxin systems. Prog Mol Biol Transl Sci. 2009;85:467–500. doi: 10.1016/S0079-6603(08)00812-X. [DOI] [PubMed] [Google Scholar]
- 7.Zhang JJ, Zhang YL, Inouye M. Characterization of the interactions within the mazEF addiction module of Escherichia coli. Journal of Biological Chemistry. 2003;278(34):32300–32306. doi: 10.1074/jbc.M304767200. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annual review of genetics. 2011;45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 9.Zhu L, Inoue K, Yoshizumi S, Kobayashi H, Zhang Y, Ouyang M, Kato F, Sugai M, Inouye M. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. Journal of bacteriology. 2009;191(10):3248–3255. doi: 10.1128/JB.01815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JH, Yamaguchi Y, Inouye M. Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS letters. 2011;585(15):2526–2532. doi: 10.1016/j.febslet.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaguchi Y, Nariya H, Park JH, Inouye M. Inhibition of specific gene expressions by protein-mediated mRNA interference. Nature communications. 2012;3:607. doi: 10.1038/ncomms1621. [DOI] [PubMed] [Google Scholar]
- 12.Kamada K, Hanaoka F, Burley SK. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol Cell. 2003;11(4):875–884. doi: 10.1016/s1097-2765(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 13.Hazan R, Sat B, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. Journal of bacteriology. 2004;186(11):3663–3669. doi: 10.1128/JB.186.11.3663-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li GY, Zhang Y, Chan MC, Mal TK, Hoeflich KP, Inouye M, Ikura M. Characterization of dual substrate binding sites in the homodimeric structure of Escherichia coli mRNA interferase MazF. Journal of molecular biology. 2006;357(1):139–150. doi: 10.1016/j.jmb.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, Braberg H, Yang Z, Meng EC, Pettersen EF, Huang CC, Datta RS, Sampathkumar P, Madhusudhan MS, Sjolander K, Ferrin TE, Burley SK, Sali A. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic acids research. 2011;39(Database issue):D465–474. doi: 10.1093/nar/gkq1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. Journal of the American Chemical Society. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 17.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 18.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wurch T, Lestienne F, Pauwels PJ. A modified overlap extension PCR method to create chimeric genes in the absence of restriction enzymes. Biotechnology Techniques. 1998;12(9):653–657. [Google Scholar]
- 21.Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J Biol Chem. 2005;280(5):3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- 22.van Dijk M, Bonvin AMJJ. Pushing the limits of what is achievable in protein-DNA docking: benchmarking HADDOCK’s performance. Nucleic acids research. 2010;38(17):5634–5647. doi: 10.1093/nar/gkq222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamphuis MB, Bonvin AM, Monti MC, Lemonnier M, Munoz-Gomez A, van den Heuvel RH, Diaz-Orejas R, Boelens R. Model for RNA binding and the catalytic site of the RNase Kid of the bacterial parD toxin-antitoxin system. Journal of molecular biology. 2006;357(1):115–126. doi: 10.1016/j.jmb.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Molecular cell. 2003;12(4):913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 25.Zorzini V, Haesaerts S, Cheung A, Loris R, van Nuland NAJ. (1)H, (13)C, and (15)N backbone and side-chain chemical shift assignment of the staphylococcal MazF mRNA interferase. Biomolecular NMR assignments. 2011;5(2):157–160. doi: 10.1007/s12104-010-9290-1. [DOI] [PubMed] [Google Scholar]
- 26.Gogos A, Mu H, Bahna F, Gomez CA, Shapiro L. Crystal structure of YdcE protein from Bacillus subtilis. Proteins. 2003;53(2):320–322. doi: 10.1002/prot.10457. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Wang K, Gao X, Zhang X, Li L, Su X, Zhang J. Biochemical and Structural Analysis of E. coli MazF Toxin. New York Structural GenomiX Research Consortium (NYSGXRC) 2011 [Google Scholar]
- 28.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta crystallographica Section D, Biological crystallography. 2004;60(Pt 12 Pt 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 29.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li GY, Zhang YL, Chan MCY, Mal TK, Hoeflich KP, Inouye M, Ikura M. Characterization of dual substrate binding sites in the homodimeric structure of Escherichia coli mRNA interferase MazF. Journal of molecular biology. 2006;357(1):139–150. doi: 10.1016/j.jmb.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi Y, Inouye M. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nature reviews Microbiology. 2011;9(11):779–790. doi: 10.1038/nrmicro2651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence alignments based on overlaid tertiary structures between MazF-ec and Kid
Sequence alignments were carried out based on the overlaid tertiary structures between MazF-ec (PDB ID: 1UB4) and Kid from Escherichia coli (PDB ID: 1M1F). White and black boxes indicate β-sheets (S) and α-helices (H), respectively, on the basis of the secondary structures of MazF-ec (PDB ID: 1UB4).
Figure S2. The helix/strand regions of MazF-ec and MazF-sa based on NMR assignments
Sequence alignments were carried out using ClustalW2. The helix/strand regions of MazF-ec and MazF-sa were analyzed by NMR assignments. White and black boxes indicate β-sheets (S) and α-helices (H), respectively.
Figure S3. Characterization of protein solubility of MazF-ec/loop1+2-mt3.
mazF-ec/loop1+2-mt3 was cloned into peT28a with a C-terminal His-tag and was introduced into E. coli BL21(DE3). Protein expression was induced by the addition of 1M isopropyl-β-□-1-thiogalactoside (IPTG) for 5 h (lane 3 to 7) at 37°C in M9 media including 0.4% glucose. The cells were disrupted by sonication in 500μl buffer (20 mM Tris-HCl pH 8.0). The supernatants and pellet were separated by centrifugation at 18,000 × g for 10 min. Lane1, whole-cell lysate before induction; lane 2, whole-cell lysate after incubation with IPTG for 3 h at 37°C; lane 3, whole-cell lysate after incubation with IPTG for 5 h at 37°C; lane 4, supernatants (sup) without NaCl added; lane 5, pellet (pel) without NaCl added; lane 6, sup with 0.5M NaCl added to extract protein; lane 7, pel with 0.5M NaCl added to extract protein; lane 8, molecular mass markers. Arrow indicates the band size of MazF-ec/loop1+2-mt3.