Abstract
The retinoblastoma tumor suppressor protein pRB is conventionally regarded as an inhibitor of the E2F family of transcription factors. Conversely, pRB is also recognized as an activator of tissue-specific gene expression along various lineages including osteoblastogenesis. During osteoblast differentiation, pRB directly targets Alpl and Bglap, which encode the major markers of osteogenesis, alkaline phosphatase and osteocalcin. Surprisingly, p130 and repressor E2Fs were recently found to co-occupy and repress Alpl and Bglap in proliferating osteoblast precursors prior to differentiation. This raises the further question of whether these genes convert to E2F activation targets when differentiation begins, which would constitute a remarkable situation wherein pRB and E2F would be co-targeting genes for activation. Chromatin immunoprecipitation (ChIP) analysis in an osteoblast differentiation model shows Alpl and Bglap are indeed targeted by an activator E2F, i.e. E2F1. Promoter occupation of Alpl and Bglap by E2F1 occurs specifically during activation, and depletion of E2F1 severely impairs their induction. Mechanistically, promoter occupation by E2F1 and pRB is mutually dependent, and without this cooperative effect, activation steps previously shown to be dependent on pRB, including recruitment of RNA polymerase II, are impaired. Myocyte and adipocyte specific genes are also co-targeted by E2F1 and pRB during differentiation along their respective lineages. The finding that pRB and E2F1 cooperate to activate expression of tissue-specific genes is a paradigm distinct from the classical concept of pRB as an inhibitor of E2F1, but is consistent with the observed roles of these proteins in physiological models.
Keywords: retinoblastoma protein, pRB, E2F, osteoblasts, myogenesis, adipogenesis, alkaline phosphatase, osteocalcin, myosin heavy chain, C/EBPα, osteosarcoma
Introduction
The retinoblastoma tumor suppressor protein pRB is conventionally regarded as an inhibitor of the cell cycle promoting activity of the E2F family of transcription factors. The E2F family includes members that are predominantly activating, in particular E2F1, E2F2, and E2F3, or predominantly repressing, particularly E2F4 and E2F5 (reviewed in 1-2). E2F family members interact directly with pRB and its close relatives p107 and p130. In these pairings, p130 cooperates with repressor E2Fs to occupy and repress the promoters of cell cycle genes in G0, while pRB interacts preferentially with activator E2Fs (3). However, the consequences of the pRB interaction are not entirely clear, with evidence supporting alternative models of pRB-mediated repression enacted either directly at the promoters of E2F targets, or remotely (discussed in 2,4-5). Conversely, pRB is recognized as an activator of tissue-specific gene expression in various differentiation programs including myogenesis, adipogenesis, osteogenesis and neurogenesis (reviewed in 6-11). During osteoblast differentiation pRB is readily detected on the activated promoters of the Alpl and Bglap genes, which encode the major markers of osteoblast differentiation, i.e. alkaline phosphatase and osteocalcin, and pRB is required for efficient induction of these genes (12-17). In osteoblasts particularly, appreciation is growing that the pivotal physiological role of pRB may lie in its requirement as an activator of tissue-specific genes (17-20). This function of pRB has not previously been linked directly with E2F-mediated activation however.
In growth-arrested cells, p130 and repressor E2Fs occupy cell cycle regulated promoters as part of a multiprotein repressor complex termed DREAM (21). Recent findings revealed that p130 and repressor E2Fs also co-target Alpl and Bglap in proliferating osteoblast precursors prior to differentiation while these tissue-specific genes still await expression (22). This surprising identification of Alpl and Bglap as E2F repression targets raises the question of whether they convert to E2F activation targets when differentiation begins. If so, that would constitute a remarkable situation where pRB and E2F would be co-targeting genes for activation. This possibility has at least one precedent, seen in proliferating cells undergoing DNA damage, where pRB and E2F1 cooperate to activate expression of apoptotic genes (23).
Occupation of Alpl and Bglap by activator E2Fs was evaluated here by chromatin immunoprecipitation (ChIP) analysis in an osteoblast differentiation model. The results show these genes are indeed targeted by an activator E2F, in particular E2F1, and that promoter occupation by E2F1 occurs specifically during activation. Moreover, E2F1 plays an essential role in activation, as induction of Alpl and Bglap is severely impaired in E2F1-depleted cells even though other transcriptional activators also target these promoters. Mechanistically, this is explained by the finding that promoter occupation by E2F1 and pRB is mutually dependent. Without this cooperative effect, a series of activation events known to be dependent on pRB, including ultimately the recruitment of RNA polymerase II, is impaired. The finding that pRB and E2F1 cooperate to activate expression of tissue-specific genes in terminally differentiating cells is a new paradigm distinct from the classical concept of pRB as an inhibitor of E2F1, but is consistent with the observed roles of these proteins in physiological models.
Materials and Methods
Cells and protein expression
C3H/10T1/2 (CCL-226) and 3T3-L1 (CL-173) cells were obtained directly from the ATCC, and induced with standard protocols (24,25). MC3T3-E1 cell passaging, authentication, differentiation by exposure to ascorbic acid and β-glycerol phosphate, immunoblotting, and in situ staining for alkaline phosphatase activity were described previously (12,17,26-27), as were the phenotypes of the pRB, p130, BRM and BRG1 knockdown lines and the line expressing Adenovirus E1A 12S.YH47/928, which sequesters the p300 family (17;22;28). The E2F1- depleted line (E2F1_KD.AA12) was generated using the Origene HuSH system and puromycin selection with the knockdown sequence 5′-CCAAGAATCATATCCAGTGGCTAGGCAGC-3′.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays and primer sequences for Alpl (alias Akp2), Bglap, CDC2 and cyclin B have been described (28,29). Other promoter primers used are MHC (Forward: 5′ATGCCAGTGAGGAAGGAATG-3′; Reverse: 5′-GCCTTGCATCACTCTCGAAT-3′), and C/EBPα (Forward: 5′-CTGGAAGTGGGTGACTTAGAGG-3′; Reverse: 5′-GAGTGGGGAGCATAGTGCTAG-3′). Antibodies were obtained from Santa Cruz: pRB (sc-50x), p107 (sc-318), E2F1 (sc-193), E2F2 (sc-9967x), E2F3 (sc-228222), SP1 (sc-59), RNA polymerase-II (sc-9001), p300 (sc-584), E2F4 (sc-1082), E2F5 (sc-968), Lin9 (sc-130571), RBBP4 (sc-33170), BRM (sc-6450), BRG1 (sc-10768) and RUNX2 (sc12488); from BD Transduction: p130 (610261); from AbCam: KDM5A (ab65796); and from Cell Signaling: HDAC1 (2062); normal mouse IgG is a component of the EZ ChIP system (Upstate Cell Signaling Solutions). Differentiation was monitored with complementing biological assays.
Gene expression
Quantitative reverse transcription PCR (qRT-PCR) was performed as described previously with the Alpl, Bglap and Gapdh expression primers (22,30). Other primers used were: E2F1 (Forward: 5′-GGATCTGGAGACTGACCATCAG-3′; Reverse: 5′-GGTTTCATAGCGTGACTTCTCCC-3′), Rb1 (Forward 5′-CCTTGAACCTGCTTGTCCTCTC-3′; Reverse 5′-CTGAGGCTGCTTGTGTCTCTGT-3′), MHC (Forward: 5′-GCGACTTGAAGTTAGCCCAGGA-3′; Reverse: 5′-CTCGTCCTCAATCTTGCTCTGC-3′) C/EBPα (Forward: 5′-GTGCTGGAGTTGACCAGTGA-3′; Reverse 5′-AAACCATCCTCTGGGTCTCC-3′) and Beta-actin (Forward: 5′-CATTGCTGACAGGATGCAGAAGG-3′; Reverse: 5′-TGCTGGAAGGTGGACAGTGAGG-3′). Gapdh was used as an internal control. Data are presented as means ± the standard error of the mean.
Results
Alpl and Bglap are co-targeted by E2F1 and pRB during activation
Osteoblast-specific gene expression can be studied in MC3T3-E1 cells, which are cell culture adapted, non-transformed precursor cells committed to the osteoblast lineage but not yet differentiated. When stimulated by bone anabolic agents, they undergo terminal differentiation in a tightly synchronized program lasting more than two weeks (26). Induction of alkaline phosphatase (Alpl) is an early stage event, coordinated closely with differentiation-associated cell cycle arrest, while induction of osteocalcin (Bglap) peaks later, marking the onset of the mineralization stage (Fig. 1A). The Bglap promoter is tightly regulated and well characterized (reviewed in 31). In contrast, while induction of alkaline phosphatase is the major clinical marker of osteoblast differentiation, the Alpl promoter has not been well characterized in terms of direct occupation by transcriptional activators. Notably though, efficient induction of alkaline phosphatase, like osteocalcin, is dependent on pRB, which targets both promoters directly (13-14,17). Alpl is targeted by RUNX2 (30), and the presence of canonical Sp1 binding sites near the transcriptional start of Alpl (32) suggests Sp1 targets Alpl as well. Both factors have been reported on the osteocalcin promoter (13,15,28,33). We were particularly interested in the possibility that activator E2Fs might target these promoters during differentiation, due to recent findings that they are targeted by p130 and repressor E2Fs prior to differentiation (22).
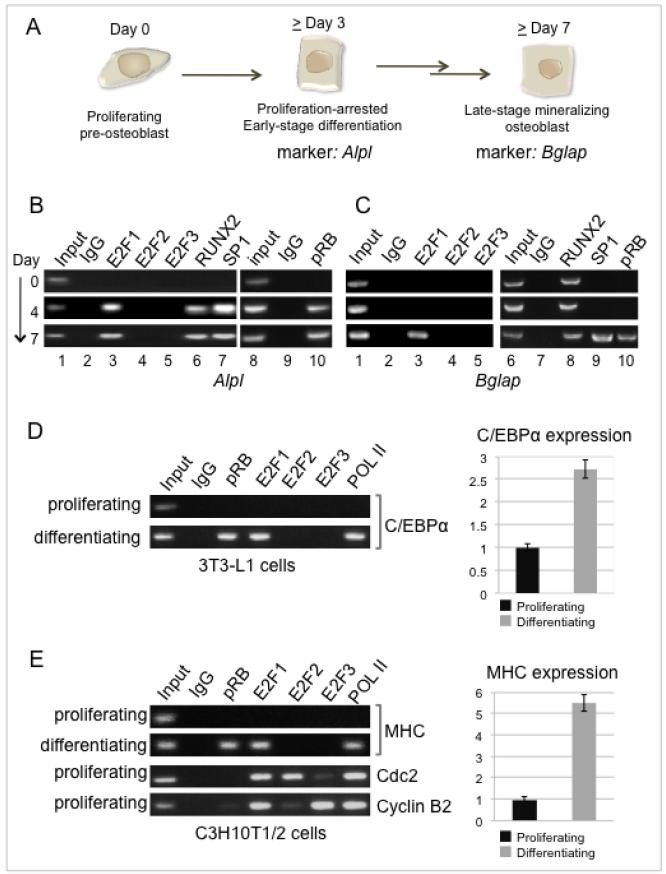
Figure 1. Analysis of transactivator binding to Alpl and Bglap.
A. The tightly synchronized process of osteoblast differentiation takes place over more than two weeks. Cell cycle arrest is apparent by day 4, and correlates with activation of the tissue-specific marker alkaline phosphatase encoded by Alpl. Differentiation culminates in the onset of a mineralizing phenotype marked by activation of the osteocalcin-encoding gene, Bglap.
B. ChIP analysis performed on the Alpl promoter from normal parental pre-osteoblasts prior to and during induction of differentiation (4 and 7 days post-induction) shows recruitment of the transactivators E2F1, RUNX2, and Sp1 coincident with promoter occupation by pRB. Typical expression patterns of Alpl and Bglap (osteocalcin) are shown in Figure 3C.
C. ChIP analysis of the Bglap promoter shows constitutive occupation by RUNX2 and recruitment of E2F1 and Sp1 in conjunction with pRB. This occurs later on Bglap than on Alpl, consistent with the delayed time of peak expression of Bglap.
D. ChIP analysis of the C/EBPα promoter in proliferating precursor 3T3-L1 cells compared with the same cells induced to differentiate along the adipocyte lineage. Expression of C/EBPα was monitored in tandem by quantitative qRT-PCR.
E. ChIP analysis of the myosin heavy chain (MHC) promoter in proliferating precursor C3H/10T1/2 cells compared with the same cells induced to differentiate along the myoblast lineage. Expression of MHC was monitored in tandem by quantitative qRT-PCR. The cdc2 and cyclin B promoters were probed in the proliferating cell lysates as positive controls for E2F2 and E2F3.
ChIP assays here show Alpl is targeted by Sp1 along with RUNX2 coincident with the onset of differentiation (Figure 1B, lanes 6 and 7). Parallel probes for activating E2Fs identify Alpl as an E2F1 target as well (lane 3), and show E2F1 occupying the promoter with similar dynamics as RUNX2 and Sp1. Neither E2F2 nor E2F3 was detected. Association of the activators is concordant with promoter occupation by pRB (lane 10). An analogous probe of Bglap (Fig. 1C) re-affirms the constitutive presence of RUNX2 (lane 8) described previously in these cells (28), and confirms the recruitment of Sp1 (lane 9) reported in related systems (33). Most significantly, the ChIP assay identifies Bglap as another target of E2F1 (lane 3), which again occupies the promoter specifically during activation, and with dynamics similar to Sp1 and pRB. Occupation of the Bglap promoter by Sp1, E2F1, and pRB occurs later than on Alpl, consistent with the relative peak activation times of the respective genes. The difficulty in detecting pRB on repressed E2F targets in cell cycle studies is sometimes attributed to the nature of available antibodies (discussed in 5), but pRB occupation signals are robust on the E2F1 activation targets.
pRB and E2F1 co-target myoblast and adipocyte differentiation markers during activation
Co-targeting of induced tissue-specific genes by pRB and E2F1 may be a more general phenomenon, given that genome-wide approaches show E2F1 targets a much wider range of genes than those involved in cell cycle progression (34-36). If E2F1 is a general activator, there is potential for co-targeting in any cell system where pRB is a co-activator, such as myoblasts and adipocytes. The genes most closely linked with pRB-mediated activation in these systems are C/EBPα in adipocytes and myosin heavy chain (MHC) in myoblasts (37;38). Probing C/EBPα in differentiating 3T3-L1 cells shows co-occupation by pRB and E2F1 specifically on the active promoter, defined by the presence of RNA polymerase II and by up-regulated expression (Fig. 1D). The MHC promoter was probed in C3H/10T1/2 cells induced to differentiate along the myoblast lineage. ChIP assays again show co-occupation by pRB and E2F1, correlating with occupation by RNA polymerase II and activated expression monitored by qRT-PCR (Fig. 1E). Neither E2F2 nor E2F3 was detected, consistent with the pattern seen on the osteogenic markers. Analysis of the cdc2 and cyclin B2 promoters in proliferating C3H/10T1/2 cells provides a positive control for the E2F2 and E2F3 immune complexes. The E2F proteins have a range of targets in these systems (e.g. 39-40), while pRB targets a specific subset of key differentiation markers. The result is co-targeting of key tissue-specific genes by pRB and E2F1 during activation, which is readily detectable in at least the osteoblast, adipocyte, and myoblast differentiation programs.
Stable E2F1 occupation of osteogenic genes is pRB-dependent
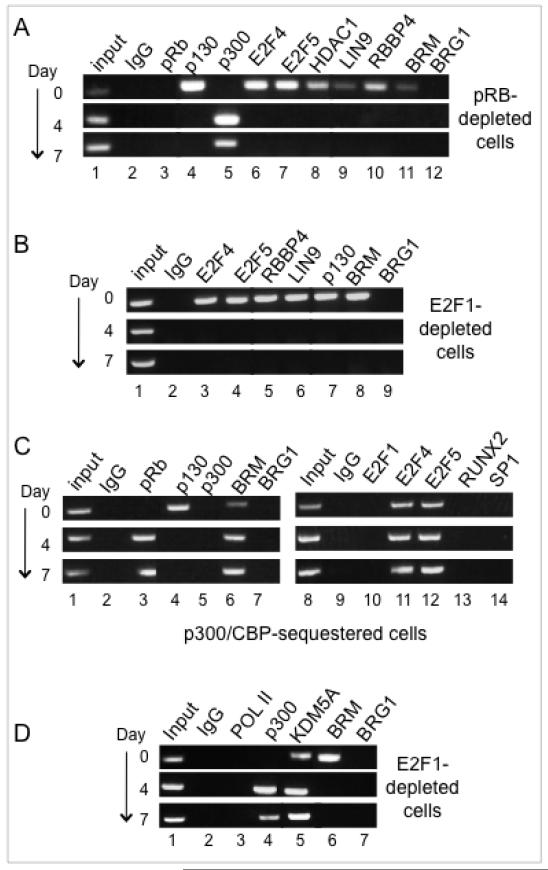
Osteoblast models have particular physiological relevance for pRB function because of the strong association between human germ-line mutation of Rb1 and osteosarcoma (41). pRB is recognized for its role in the enhanced recruitment of certain transcription factors to osteogenic genes, but a role regarding E2F1 recruitment during activation of tissue specific genes would be a novel paradigm. This activity was probed in an MC3T3-E1 derived cell line stably expressing an shRNA sequence targeting pRB. These cells were described previously and show severely impaired activation of Alpl and Bglap (17). ChIP analysis here shows E2F1, RUNX2 and Sp1 all fail to bind the Alpl promoter detectably in the pRB-depleted cells (Fig. 2A, lanes 3 through 5). The occupation pattern on Bglap likewise shows E2F1 and Sp1 romoter occupation dependent on pRB (Fig. 2B, lanes 3 and 5) (constitutive binding of RUNX2 on Bglap was discussed above). Expression of pRB in these cells is compared with other expression profiles in Figure 3, below. The conclusion that pRB facilitates binding of specific transcription factors is consistent with earlier reports regarding RUNX2 (13,16) and Sp1 (42). However, the finding that pRB plays a similar role in enhancing promoter occupation of tissue-specific genes by E2F1 is novel.
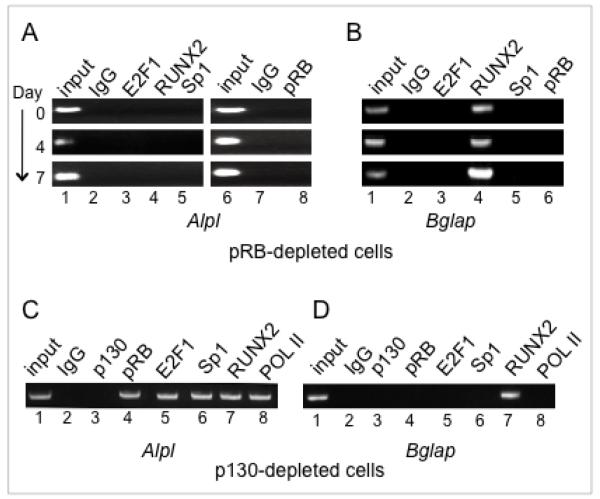
Figure 2. Promoter occupation by E2F1 in pRB-depleted or p130-depleted cells.
A. ChIP analysis of the Alpl promoter in pRB-depleted cells shows impaired recruitment of E2F1, RUNX2, and Sp1 (compare with Figure 1B). The Alpl expression pattern is shown in Figure 3C.
B. ChIP analysis of Bglap in the pRB-depleted background shows impaired recruitment of E2F1 and Sp1 (compare with Figure 1C). Bglap expression is shown in Figure 3C.
C. ChIP analysis of Alpl in p130-depleted cells shows constitutive occupation by E2F1 along with pRB, as well as by other transcriptional activators. Alpl is constitutively up-regulated in p130-depleted cells, as discussed in the text.
D. ChIP analysis of Bglap shows p130 depletion is not sufficient for constitutive occupation by pRB and/or E2F1 of this late-stage marker.
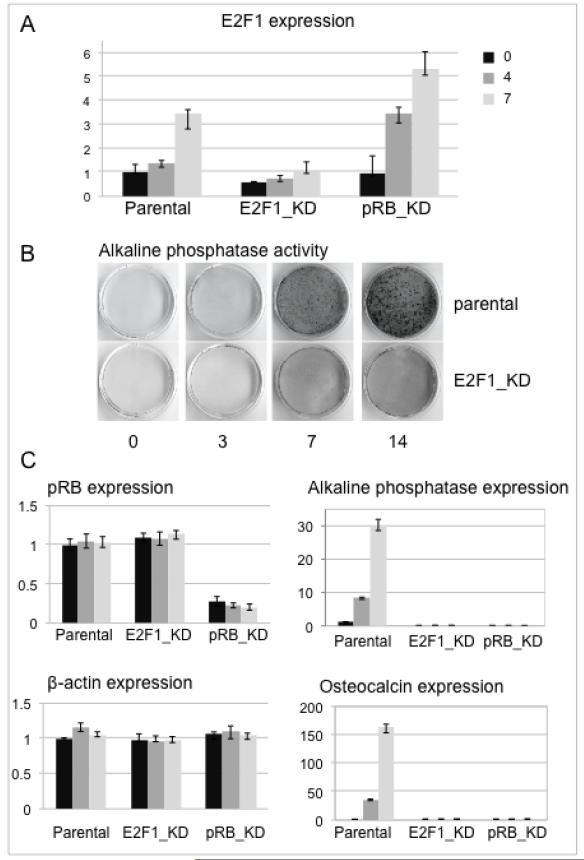
Figure 3. E2F1-depleted osteoblast precursors show impaired induction of osteogenic genes.
A. E2F1 expression determined by qRT-PCR at days 0, 4, and 7 post-induction in cells stably expressing an shRNA sequence targeting E2F1 (E2F1_KD), or pRB (pRB_KD), is shown relative to normal parental cells.
B. Alkaline phosphatase activity, monitored colorimetrically in situ shows sharply increased activity by day 7 post-induction in parental cells, while induction is impaired in E2F1 depleted cells.
C. Quantification of expression levels at days 0, 4, and 7 post-induction, as indicated in panel A, shows E2F1 depletion does not affect Rb1 expression, but has a similar effect as pRB depletion on expression of Alpl (alkaline phosphatase) and Bglap (osteocalcin). Expression values were normalized to Gapdh; β-actin is included as an additional control.
Prior to differentiation, p130 and repressor E2Fs occupy the Alpl and Bglap promoters in osteoblast precursors; depletion of p130 results in dissociation of the E2F4/5 repressor E2Fs from both promoters, and is sufficient to permit pRB occupancy and constitutively up-regulated expression of the early marker Alpl, but not the late-stage marker Bglap (22). ChIP assays here show occupation by E2F1 parallels pRB in the p130-deficient cells in the absence of a differentiation signal (Fig. 2C and 2D). E2F1 constitutively occupies the promoter of the early marker Alpl along with pRB, Sp1, RUNX2 and RNA polymerase II, but is not detected on the promoter of the late-stage marker Bglap in the non-induced cells. Promoter occupation by pRB and E2F1 is thus closely related to, but not necessarily an automatic consequence of dissociation of the p130 and E2F4/5 repressor complexes.
E2F1 is required for efficient activation of osteogenic promoters
The promoter occupation dynamics imply that E2F1 plays an activating role in Alpl and Bglap expression, but do not show this directly. To test the functional contribution of E2F1, a stable cell line expressing an E2F1-targeting shRNA was derived from the osteoblast precursors. Expression of E2F1 increases a few-fold post-differentiation in the parental cells as they undergo differentiation-associated cell cycle arrest (Fig. 3A, day 7 vs day 0), consistent with E2F1 continuing to play a significant role even when the cells are no longer cycling. E2F1 mRNA levels are not completely ablated in the shRNA expressing line (E2F1_KD), but are reduced below 50% of normal. E2F1 expression is at or above normal levels in pRB-deficient cells analyzed in parallel, so the lack of E2F1 on target promoters in Fig. 2 is not due to impaired expression. The biological effect of E2F1 depletion was assessed by monitoring alkaline phosphatase activity in an in situ colorimetric assay (Fig. 3B). Activity increases detectably in parental cells by day three of differentiation, and peaks by day seven to ten, while the E2F1-depleted cells show markedly less enzyme activity. Directly comparing the effect of depleting E2F1 vs pRB (Fig. 3C) shows E2F1 depletion does not affect pRB expression, but impedes expression of the tissue specific genes to a similar extent as pRB depletion. Induction of Alpl and Bglap (osteocalcin) is profoundly impaired in both genetic backgrounds. Taken together, the results show E2F1 is recruited to the Alpl and Bglap promoters dependent on pRB, and plays a significant role in activation of their expression. The requirement for E2F1 is relatively specific; the related E2F family members E2F2 and E2F3 were not detected on the promoter, and seem unable to compensate for E2F1 deficiency in the activation of these specific target genes. Specificity within the E2F 1-3 subgroup in regard to transcriptional activation targets has been observed in other systems as well, although mouse models suggest that more functional overlap occurs in long-term development models (36,40,43).
Mechanisms of promoter occupation
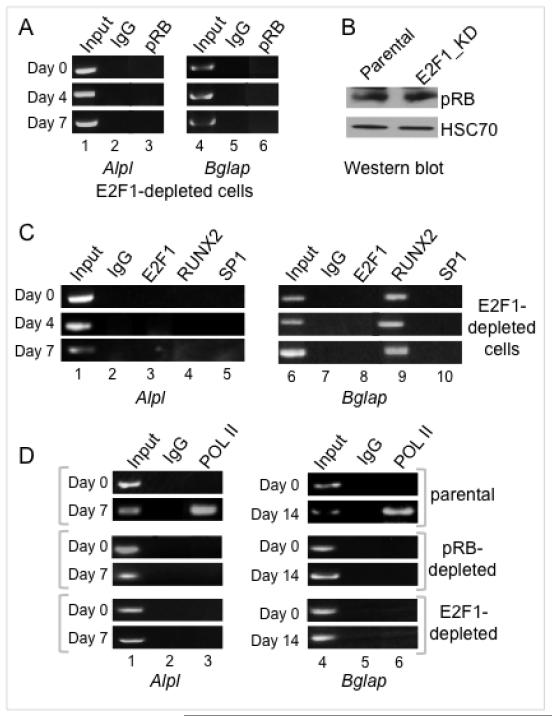
Figure 2 showed pRB deficiency impairs the ability of E2F1 to occupy the Alpl and Bglap promoters. A converse requirement can be seen in the E2F1-depleted cells where ChIP assays show pRB association with both promoters is sharply impaired (Fig. 4A compared with Figs. 1B and 1C) although pRB levels remain constant (Fig. 4B). To determine whether this mutual dependence is key to pRB transcriptional activation functions, events known to require pRB were probed in further ChIP assays. These reveal that Alpl occupation by RUNX2 and Sp1 also depends on E2F1 (Fig. 4C lanes 4 and 5), as does Bglap occupation by Sp1 (Fig. 4C lane 10). Thus, deficiency of E2F1 has the extended effect of impairing occupation by other, unrelated, transcriptional activators. Direct binding between E2F1 and Sp1 has been reported for cooperative activation of a cell cycle related gene (44). The concept of a facilitating network of promoter-occupying factors is broadened here, with pRB and E2F1 mutually required for promoter access by various other transactivators. pRB is also required for recruitment of chromatin-remodeling complexes that facilitate RNA polymerase II (RNA pol II) association with Alpl (17). Extending this analysis to Bglap (Fig. 4D) shows a similar requirement for pRB in RNA Pol II occupation (lane 6). RNA Pol II recruitment at both promoters is likewise dependent on E2F1 (lanes 3 and 6). The requirement for E2F1 to cooperate with pRB in activation events that include recruitment of other transcriptional activators and the basal transcription machinery helps explain how expression of Alpl and Bglap is so profoundly impaired in E2F1-depleted cells even though E2F1 is not the only transcription factor that targets these promoters.
Figure 4. E2F1 is required for occupation by other specific transcriptional activators and RNA polymerase II.
A. ChIP analysis performed on Alpl and Bglap in E2F1-depleted cells shows impaired recruitment of pRB (compare with Figures 1B/1C).
B. Western blotting shows pRB levels in E2F1-depleted cells similar to normal parental levels.
C. ChIP analysis of Alpl and Bglap in E2F1-depleted cells shows impaired recruitment of RUNX2 and Sp1 on Alpl, and of Sp1 on Bglap (compare with Figures 1B/1C).
D. ChIP analysis of the Alpl and Bglap promoters shows sharply impaired recruitment of RNA polymerase II in either a pRB-depleted or E2F1-depleted cell background.
The promoter dynamics indicate a specific transition from occupation by p130 and repressor E2Fs to occupation by pRB and E2F1. For the early marker Alpl, depletion of p130 is sufficient to propel the transition without the late-stage signals required to initiate activation of Bglap (Fig. 2C), thus providing an accessible system for further mechanistic analysis. Dissociation of the BRM-containing SWI/SNF repressor complex from Alpl is independent of pRB (17 and Fig. 5A, lane 11). A mechanism of pRB-dependent displacement of p130 is conceivable, but is excluded here by the ChIP dynamics showing successful dissociation of p130 along with E2F4/E2F5 in pRB-depleted cells (Fig. 5A, lanes 4,6,7). Tissue-specific promoter repression by p130 and E2F4/5 can involve a larger complex termed DREAM, similar to the configuration seen on cell cycle specific genes, in which p130 and E2F4/5 are stably associated with a module of the development-regulating LIN complex (21,22). Monitoring of the key LIN complex components, LIN9 and RBBP4, shows their expected dissociation along with p130 (Fig. 5A, lanes 9,10). Monitoring of HDAC1 further affirms the general dissociation of repressor complexes (Fig. 5A, lane 8). Similar dynamics are seen in the E2F1-depleted cells, where dissociation of p130, E2F4/5, LIN9, RBBP4, and BRM also proceeds normally (Fig. 5B). Thus the mechanism of p130 and E2F4/5 dissociation does not require direct competition from either pRB or E2F1.
Figure 5. p130 and E2F4/5 are replaced on the Alpl promoter in a two-step mechanism by which pRB occupies the promoter with E2F4/5 prior to recruitment of E2F1, RUNX2 or Sp1.
A. ChIP analysis of Alpl shows pRB is not required for dissociation of p130 and repressor E2Fs, or associated LIN complex proteins.
B. ChIP analysis of Alpl shows E2F1 likewise is not required for dissociation of p130 and repressor E2Fs or associated LIN complex proteins.
C. Compromising p300 family function reveals a transition state in which pRB occupies the promoter simultaneously with E2F4/5, and without E2F1, RUNX2, or Sp1. These cells show sharply impaired expression of Alpl, as discussed in the text.
D. ChIP analysis of Alpl in E2F1-depleted cells shows E2F1 is required for dissociation of KDM5A, and for association of BRG1.
Further insight into the transition mechanism comes from considering the role of the co-activating histone acetyltransferase p300. p300 (but not the closely related protein CBP) moves onto the Alpl promoter during activation (seen in Fig. 5A, lane 5) and is implicated in dissociation of the repressors. Compromising p300 family function by expression of a specific Adenovirus E1A variant sharply impairs expression of Alpl, and impedes dissociation of BRM-SWI/SNF while permitting p130 to dissociate successfully, and permitting pRB to occupy the promoter simultaneously with E2F4/5 (17,22; also seen in Fig. 5C, lanes 3,4,6,11,12,15). Further analysis here reveals that activation stalls at this step in p300-compromised cells without recruitment of E2F1, Sp1 or RUNX2 (lanes 10,13,14). This intermediate step apparently transitions rapidly in a wild-type genetic background, but is distinguished by the requirement for a discrete function necessary to dissociate BRM-SWI/SNF and E2F4/5. The latter step fails when the p300 family is compromised, so by implication may involve acetylation of one or more targets. Given that stable association of pRB is not seen in E2F1-depleted cells, we can infer that pRB dissociates along with E2F4/5, then returns in concert with E2F1 to enable subsequent activation steps. pRB is not necessary for dissociation of E2F4/5 and BRM-SWI/SNF, so presumably enacts some other function(s) at this point, which likely includes dissociation of the KDM5A demethylase. KDM5A (aliases: JARID1A or RBP2) specifically removes a methyl group from di- or tri-methylated H3K4 to maintain a repression signal at targeted promoters. KDM5A is removed from osteogenic promoters in a pRB-dependent manner (14), presumably prior to recruitment of p300 because KDM5A dissociation from Alpl is not impaired in p300-compromised cells (17). Exclusion of KDM5A is dependent on E2F1, however, (Fig 5D, lane 5), likely because a continued presence of pRB at the promoter is required to maintain dissociation of KDM5A.
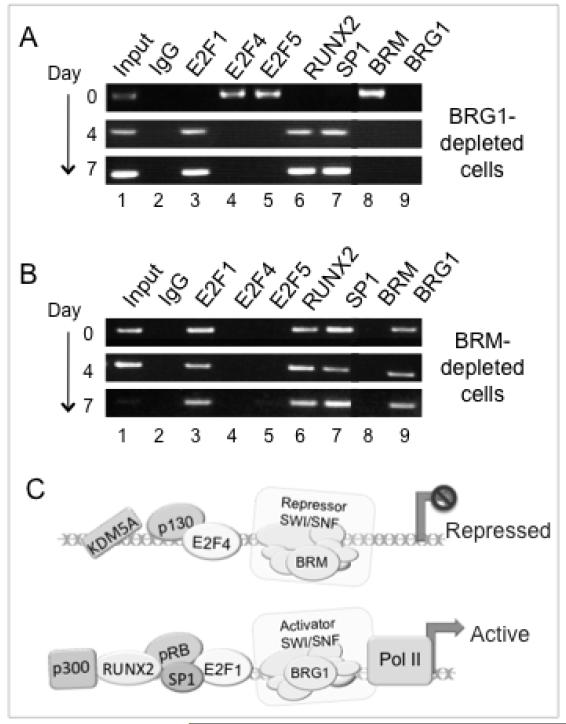
Ultimately, recruitment of RNA polymerase II is dependent on BRG1-containing SWI/SNF, whose recruitment in turn depends on pRB (17). If recruitment of BRG1-SWI/SNF requires the direct presence of pRB at the promoter, it would presumably depend on E2F1 as well. This is confirmed here in the E2F1-depleted cells (Fig. 6A, lane 7). Promoter occupation by pRB is unimpaired in BRG1-depleted cells, so mechanistically pRB occupation precedes recruitment of BRG1-SWI/SNF (17). Probes here show E2F1, RUNX2 and Sp1, likewise, all occupy the promoter successfully in a BRG1-deficient background (Fig. 6A, lanes 3,6,7), indicating that BRG1-dependent chromatin remodeling is not a prerequisite for DNA access by these factors. The role of SWI/SNF with respect to these factors on Alpl is essentially a negative one in which they are actively excluded by BRM-SWI/SNF acting coordinately with p130-associated complexes. Depletion of either BRM or p130 results in constitutive activation of Alpl expression (22), and binding of E2F1, Sp1 and RUNX2 occurs constitutively on Alpl in p130-depleted cells in a p300-competent background (Fig. 2C). Similarly E2F1, Sp1 and RUNX2 occupy the promoter constitutively (day 0) in BRM-depleted cells (Fig. 6B, lanes 3,6,7), confirming the required role of BRM-SWI/SNF in excluding these factors prior to induction.
Figure 6. Promoter occupation by E2F1 is excluded by BRM-SWI/SNF, and not dependent on BRG1-SWI/SNF.
A. ChIP analysis of Alpl cells shows BRG1-SWI/SNF is not required for association of transcription factors E2F1, RUNX2, or Sp1, although BRG1 is required for Alpl activation, as discussed in the text.
B. ChIP analysis shows BRM-depletion is sufficient for Alpl occupation by activators E2F1, RUNX2, and Sp1. Constitutive expression of Alpl in BRM-depleted cells is discussed in the text.
C. Schematic illustration of Alpl occupation patterns. For illustration, proteins are shown associated with recognized binding partners, but various other combinations are known, and whether these specific interactions occur, or are maintained, directly on the promoter cannot be inferred from promoter occupation analysis.
A schematic of the transition from Alpl repression to activation is shown in Fig. 6C. Alpl in the precursor cells is actively repressed by the presence of BRM-SWI/SNF and p130 complexes linked with repressor E2Fs. Confluence and anabolic signals induce dissociation of p130 from Alpl, after which pRB transiently occupies the promoter in the presence of E2F4/5. Notably, dissociation of p130 and its E2F4/5 binding partners are separate mechanistic steps. Dissociation of KDM5A likely occurs at this stage, as it is independent of p300 (17). A p300-linked event, possibly involving acetylation of one or more targets, then facilitates dissociation of BRM-SWI/SNF along with the repressor E2Fs and pRB. Removal of the repressors permits E2F1 and pRB to establish stable occupation of the promoter along with RUNX2 and Sp1, and cooperatively facilitate recruitment of an activating BRG1-containing SWI/SNF, which in turn facilitates recruitment of RNA polymerase II.
Discussion
This study identifies key tissue-specific markers in the myoblast, adipocyte, and osteoblast lineages that are targeted jointly by E2F1 and pRB during activation. Analysis in the osteoblast program shows E2F1 and pRB are mutually required for recruitment of RNA polymerase II and efficient activation of their common targets.
pRB is thought to access tissue-specific promoters primarily through its ability to bind to certain transcriptional activators including RUNX2 and Sp1, but on key promoters these factors cannot provide efficient promoter access for pRB in an E2F1-depleted background. Far from repressing E2F1-mediated transactivation of co-targeted genes during differentiation, pRB facilitates E2F1 access to key promoters where pRB plays an essential co-activator role. While this paradigm contrasts with the conventional concept of pRB as an inhibitor of E2F1, it is concordant with two converging lines of evidence: the findings in multiple labs that pRB acts directly to up-regulate tissue-specific gene expression in various cell types (8-15), and the recognition that the transcriptional activation functions of E2F1 extend in some cases to tissue specific genes during terminal differentiation (34-36).
Co-operation between pRB and E2F1 for transcriptional activation is not without precedent. Complexes containing E2F1 and pRB form in proliferating cells following DNA damage and participate in the activation of pro-apoptotic genes (23). It now appears cooperation between pRB and E2F1 for activation may be a fairly common phenomenon that occurs in normally differentiating cells, at least in the mesenchymal stem cell derived lineages examined here, even as they undergo differentiation-associated cell cycle arrest and repression of E2F1-mediated cell cycle functions.
The combination of ChIP assays and genetics applied here gives a powerful view of protein occupation patterns on endogenous promoters, and of their relative timing and order of dependence. These approaches do not address whether the various factors interact directly and/or occupy specific sites on DNA, but these concepts have become more fluid as the understanding of promoter occupation has grown to encompass chromatin remodeling and the assembly of large protein complexes connected by networks of interactions among themselves, histones, and DNA. ChIP-seq approaches have revealed that cognate recognition sites identified with reporter plasmids and DNA pull-downs in vitro are not necessarily an accurate representation of binding sites in the physiological context of the genome. This is strikingly true for E2F1 where the classic E2F consensus (TTTG/CG/CCGC) was found at only about 12% of E2F1 binding sites identified in vivo (35). Alpl and Bglap have similar patterns of occupation by p130, pRB and the E2Fs, but no clear similarities in their promoter structure with respect to potential E2F binding sites. Alpl contains a highly GC-rich region surrounding the transcriptional start and extending approximately 1.5 kilobases upstream that contains at least fifteen motifs matching within 15% stringency to the in vitro consensus TTTG/CG/CCGC, along with several overlapping Sp1 binding sequences CCGCCC (Terao et al). Clustering of potential sites near the transcriptional start is a frequent characteristic of confirmed E2F1 target genes, but not all consensus motifs are bound in vivo (35,45). The osteocalcin (Bglap) promoter contains only scattered sequences similar to the in vitro E2F consensus, but this remains consistent with findings that only a small percentage of E2F binding sites identified in vivo contain a consensus E2F motif (35,45).
All of the factors considered here have been identified in pull-down assays with multiple others, and such interactions may play a role during recruitment whether or not they are maintained once the promoter is accessed. The key mechanistic information comes from the ChIP assays whose exceptional advantage lies in revealing the dynamics of assembly at a specific promoter, and providing a functional readout for genetic approaches. While pRB and E2F1 have the ability to interact directly, the high-affinity interaction seen in pull-down assays is unlikely to be required for the co-activation function seen here. Naturally occurring pRB mutants identified in low-penetrance retinoblastoma retain the ability to promote differentiation even though defective for E2F binding in electromobility-shift assays (46). The reduced tumor susceptibility associated with these pRB variants supports the idea that promoting differentiation is a fundamental part of the pRB tumor suppressor role, while their existence indicates that stable interaction with E2F1 is not essential for pRB-dependent activation of tissue-specific genes. E2F1 mutants are consistent with this conclusion. Mutation analysis of E2F1 indicates that no individual protein interaction domain is a key determinant for E2F1 binding to most of its target sites in the genome (47). Cooperation between E2F1 and pRB may require only a transient or indirect interaction, possibly bridged by mutual binding partners such as Sp1 (42, 44), and SWI/SNF, which co-precipitates with E2F1, E2F4 and E2F5 (48), as well as pRB (10), or assisted by other pRB binding partners that occupy the promoter with similar dynamics. Whether or not direct interaction is strictly essential, the mutual dependence of E2F1 and pRB for promoter occupation and association of transactivators including RNA polymerase-II shows they have a cooperative activation function, and that their co-occupation of target promoters is not simply fortuitous.
During differentiation, pRB is subject to acetylation and/or methylation at sites near the C-terminus, and the integrity of the modification sites is important for the differentiation function of pRB (9). The modifications impede interaction with E2F1 in some contexts (3,9,49), consistent with the earlier evidence that E2F1 pull down activity is not essential for pRB-dependent activation of tissue-specific genes (46). Whether the modifications favor other activities of pRB in the differentiation process is an intriguing question for future studies.
Deficiency of E2F1 is linked with tumor susceptibility (50), a finding at odds with the classical role of E2F1 as a driver of cell cycle specific gene expression, but possibly explained by the wider roles of E2F1 in regulating apoptosis and/or the DNA damage response. The studies presented here suggest it may also derive from a fundamental role for E2F1 in cooperating with pRB to promote terminal differentiation.
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Science and the National Cancer Institute of the National Institutes of Health under award numbers R01GM073257 (EM) and T32-CA134268 (SF). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest Statement: The authors certify they have no relationship they believe could be construed as resulting in an actual, potential, or perceived conflict of interest with regard to this manuscript.
References
- 1.van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol. 2008;9:713–24. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- 2.Biswas A, Johnson DG. Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 2012;72:13–17. doi: 10.1158/0008-5472.CAN-11-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munro S, Carr SM, La Thangue NB. Diversity within the pRb pathway: is there a code of conduct? Oncogene. 2012;31:4343–52. doi: 10.1038/onc.2011.603. [DOI] [PubMed] [Google Scholar]
- 4.Burkhart D, Sage J. Cellular mechanisms of tumor suppression by the retinoblastoma gene. Nature Reviews Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rizzolio F, Esposito L, Muresu D, Fratamico R, Jaraha R, Caprioli GV, et al. RB gene family: genome-wide ChIP approaches could open undiscovered roads. J Cell Biochem. 2010;109:839–43. doi: 10.1002/jcb.22448. [DOI] [PubMed] [Google Scholar]
- 6.Thomas DM, Yang HS, Alexander K, Hinds PW. Role of the retinoblastoma protein in differentiation and senescence. Cancer Biol Ther. 2003;2:124–30. [PubMed] [Google Scholar]
- 7.Liu H, Dibling B, Spike B, Dirlam A, Macleod K. New roles for the RB tumor suppressor protein. Curr Opin Genet Dev. 2004;14:55–64. doi: 10.1016/j.gde.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Korenjak M, Brehm A. E2F-Rb complexes regulating transcription of genes important for differentiation and development. Curr Opin Genet Dev. 2005;15:520–7. doi: 10.1016/j.gde.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen DX, McCance DJ. Role of the retinoblastoma tumor suppressor protein in cellular differentiation. J Cell Biochem. 2005;94:870–9. doi: 10.1002/jcb.20375. [DOI] [PubMed] [Google Scholar]
- 10.Zhu L. Tumour suppressor retinoblastoma protein Rb: a transcriptional regulator. Eur J Cancer. 2005;41:2415–27. doi: 10.1016/j.ejca.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Poznic M. Retinoblastoma protein: a central processing unit. J Biosci. 2009;34:305–12. doi: 10.1007/s12038-009-0034-2. [DOI] [PubMed] [Google Scholar]
- 12.Beck GR, Jr, Sullivan EC, Moran E, Zerler B. Relationship between Alkaline phosphatase levels, osteopontin expression and mineralization in differentiating MC3T3-E1 osteoblasts. J Cell Biochem. 1998;68:269–80. doi: 10.1002/(sici)1097-4644(19980201)68:2<269::aid-jcb13>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 13.Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, et al. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8:303–16. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- 14.Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG., Jr. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell. 2005;18:623–35. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Lee JS, Thomas DM, Gutierrez G, Carty SA, Yanagawa S, Hinds PW. HES1 cooperates with pRb to activate RUNX2-dependent transcription. J Bone Miner Res. 2006;21:921–33. doi: 10.1359/jbmr.060303. [DOI] [PubMed] [Google Scholar]
- 16.Luan Y, Yu XP, Xu K, Ding B, Yu J, Huang Y, et al. The retinoblastoma protein is an essential mediator of osteogenesis that links the p204 protein to the Cbfa1 transcription factor thereby increasing its activity. J Biol Chem. 2007;282:16860–70. doi: 10.1074/jbc.M610943200. [DOI] [PubMed] [Google Scholar]
- 17.Flowers S, Beck GR, Jr, Moran E. Transcriptional activation by pRB, and its coordination with SWI/SNF recruitment. Cancer Res. 2010;70:8282–87. doi: 10.1158/0008-5472.CAN-10-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutierrez GM, Kong E, Sabbagh Y, Brown NE, Lee JS, Demay MB, et al. Impaired bone development and increased mesenchymal progenitor cells in calvaria of RB1−/− mice. Proc Natl Acad Sci USA. 2008;105:18402–7. doi: 10.1073/pnas.0805925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berman SD, Yuan TL, Miller ES, Lee EY, Caron A, Lees JA. The retinoblastoma protein tumor suppressor is important for appropriate osteoblast differentiation and bone development. Mol Cancer Res. 2008;6:1440–51. doi: 10.1158/1541-7786.MCR-08-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, Lees JA. Rb regulates fate choice and lineage commitment in vivo. Nature. 2010;466:1110–1114. doi: 10.1038/nature09264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, et al. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26:539–51. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 22.Flowers S, Beck GR, Jr, Moran E. Tissue-specific gene targeting by the multiprotein mammalian DREAM complex. J. Biol. Chem. 2011;286:27867–71. doi: 10.1074/jbc.C111.255091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ianari A, Natale T, Calo E, Ferretti E, Alesse E, Screpanti I, et al. Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell. 2009;15:184–94. doi: 10.1016/j.ccr.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madsen L, Petersen RK, Sørensen MB, Jørgensen C, Hallenborg P, Pridal L, et al. Adipocyte differentiation of 3T3-L1 preadipocytes is dependent on lipoxygenase activity during the initial stages of the differentiation process. Biochem J. 2003;375(Pt 3):539–49. doi: 10.1042/bj20030503. [DOI] [PubMed] [Google Scholar]
- 25.Sun Y, Ge Y, Drnevich J, Zhao Y, Band M, Chen J. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J Cell Biol. 2010;189(7):1157–69. doi: 10.1083/jcb.200912093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beck GR, Jr, Zerler B, Moran E. Gene array analysis of osteoblast differentiation. Cell Growth Differ. 2001;12:61–83. [PubMed] [Google Scholar]
- 27.Yaciuk P, Moran E. Analysis with specific polyclonal antiserum indicates that the E1A-associated 300 kilodalton product is a stable nuclear phosphoprotein that undergoes cell cycle phase-specific modification. Mol Cell Biol. 1991;11:5389–97. doi: 10.1128/mcb.11.11.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flowers S, Nagl NG, Jr, Beck GR, Jr, Moran E. Antagonistic roles for BRM and BRG1 SWI/SNF complexes in differentiation. J Biol Chem. 2009;284:10067–75. doi: 10.1074/jbc.M808782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagl NG, Jr, Zweitzig DR, Thimmapaya B, Beck GR, Jr, Moran E. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 2006;66:1289–1293. doi: 10.1158/0008-5472.CAN-05-3427. [DOI] [PubMed] [Google Scholar]
- 30.Xu F, Flowers S, Moran E. Essential role of ARID2 protein-containing SWI/SNF complex in tissue-specific gene expression. J Biol Chem. 2012;287:5033–41. doi: 10.1074/jbc.M111.279968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villafán-Bernal JR, Sánchez-Enríquez S, Muñoz-Valle JF. Molecular modulation of osteocalcin and its relevance in diabetes. Int J Mol Med. 2011;28:283–93. doi: 10.3892/ijmm.2011.706. [DOI] [PubMed] [Google Scholar]
- 32.Terao M, Studer M, Gianni M, Garattini E. Isolation and characterization of the mouse liver/bone/kidney-type alkaline phosphatase gene. Biochem. 1990;268:641–48. doi: 10.1042/bj2680641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stains JP, Lecanda F, Screen J, Towler DA, Civitelli R. Gap junctional communication modulates gene transcription by altering the recruitment of Sp1 and Sp3 to connexin-response elements in osteoblast promoters. J Biol Chem. 2003;278:24377–87. doi: 10.1074/jbc.M212554200. [DOI] [PubMed] [Google Scholar]
- 34.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29:409–17. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McClellan KA, Slack RS. Specific in vivo roles for E2Fs in differentiation and development. Cell Cycle. 2007;6:2917–27. doi: 10.4161/cc.6.23.4997. [DOI] [PubMed] [Google Scholar]
- 37.Classon M, Kennedy BK, Mulloy R, Harlow E. Opposing roles of pRB and p107 in adipocyte differentiation. Proc Natl Acad Sci USA. 2000;97:10826–31. doi: 10.1073/pnas.190343597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novitch BG, Mulligan GJ, Jacks T, Lassar AB. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fajas L, Landsberg RL, Huss-Garcia Y, Sardet C, Lees JA, Auwerx J. E2Fs Regulate Adipocyte Differentiation. Dev Cell. 2002;3:39–49. doi: 10.1016/s1534-5807(02)00190-9. [DOI] [PubMed] [Google Scholar]
- 40.Asp P, Acosta-Alvear D, Tsikitis M, van Oevelen C, Dynlacht BD. E2f3b plays an essential role in myogenic differentiation through isoform-specific gene regulation. Genes Dev. 2009;23:37–53. doi: 10.1101/gad.1727309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang N, Song WX, Luo J, Haydon RC, He TC. Osteosarcoma development and stem cell differentiation. Clin Orthop Relat Res. 2008;466:2114–30. doi: 10.1007/s11999-008-0335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noé V, Alemany C, Chasin LA, Ciudad CJ. Retinoblastoma protein associates with SP1 and activates the hamster dihydrofolate reductase promoter. Oncogene. 1998;16:1931–8. doi: 10.1038/sj.onc.1201718. [DOI] [PubMed] [Google Scholar]
- 43.Tsai SY, Opavsky R, Sharma N, Wu L, Naidu S, Nolan E, et al. Mouse development with a single E2F activator. Nature. 2008;454:1137–41. doi: 10.1038/nature07066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karlseder J, Rotheneder H, Wintersberger E. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, et al. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550–61. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sellers WR, Novitch BG, Miyake S, Heith A, Otterson GA, Kaye FJ, et al. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao AR, Rabinovich R, Xu M, Xu X, Jin VX, Farnham PJ. Genome-wide analysis of transcription factor E2F1 mutant proteins reveals that N- and C-terminal protein interaction domains do not participate in targeting E2F1 to the human genome. J Biol Chem. 2011;286:11985–96. doi: 10.1074/jbc.M110.217158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagl NG, Jr, Wang X, Patsialou A, Van Scoy M, Moran E. Distinct Mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell cycle control. EMBO J. 2007;26:752–63. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pickard A, Wong PP, McCance DJ. Acetylation of Rb by PCAF is required for nuclear localization and keratinocyte differentiation. J Cell Sci. 2010;123(Pt 21):3718–26. doi: 10.1242/jcs.068924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson NJ. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–48. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]