Abstract
The inhibitory protein SOCS3 plays a key role in the immune and hematopoietic systems by regulating signaling induced by specific cytokines. SOCS3 functions by inhibiting the catalytic activity of Janus Kinases (JAKs) that initiate signaling within the cell. We determined the crystal structure of a ternary complex between murine SOCS3, JAK2 (kinase domain) and a fragment of the IL-6 receptor β-chain. The structure shows that SOCS3 binds JAK2 and receptor simultaneously, using two opposing surfaces. Whilst the phosphotyrosine-binding groove on the SOCS3 SH2 domain is occupied by receptor, JAK2 binds in a phospho-independent manner to a non-canonical surface. The kinase inhibitory region of SOCS3 occludes the substrate-binding groove on JAK2 and biochemical studies show it blocks substrate association. These studies reveal that SOCS3 targets specific JAK-cytokine receptor pairs and explains the mechanism and specificity of SOCS action.
Introduction
Cytokines and growth factors utilize specific receptor-associated tyrosine kinases to initiate an intracellular signaling cascade. Whilst growth factors such as EGF interact with cell-surface receptors possessing intrinsic tyrosine kinase domains, the majority of cytokines utilize receptors that lack this but instead associate with a family of exogenous kinases called JAKs (Janus Kinases)1,2. Cytokine binding to these receptors allows JAK dimers to self-activate, in trans, from an inactive state and this initiates the signaling cascade3,4. In order to prevent aberrant or prolonged signaling that could lead to pathological proliferation and carcinogenesis there is a need for these receptor-associated kinases to be regulated tightly.
The principal regulators of JAK/STAT signaling are the SOCS (Suppressor of Cytokine Signaling) family of proteins5–8. The human genome encodes eight SOCS proteins (SOCS1-7 and CIS) and all share a similar architecture which includes a central SH2 domain followed by a SOCS box domain at their C-terminus. The SH2 domain recruits tyrosine-phosphorylated substrates whilst the SOCS box binds elongins B and C and Cullin5 which leads to the ubiquitination of these substrates9–13. Thus SOCS proteins can be considered the substrate recruitment modules of E3 ubiquitin ligases that act to shut down cytokine signaling by inducing the proteolytic degradation of signaling molecules.
The two most potent members of the family, SOCS1 and SOCS3, act via an additional mechanism. They contain a short motif termed the kinase inhibitory region (KIR) which allows them to suppress signaling by direct inhibition of JAK catalytic activity14,15. This is the primary mode-of-action of SOCS1 and SOCS3 as deletion of their SOCS box domain alone (and hence elimination of ubiquitination activity) results in a much milder phenotype12,16 than the full knockout.
There are four mammalian JAKs (JAK1-3 and TYK2); recently it has been shown that SOCS3 directly inhibits JAK1, JAK2 and TYK2 but does not inhibit JAK317. Despite the ability of SOCS3 to inhibit these JAKs, deletion of SOCS3 in mice has revealed specificity for particular cytokines, including LIF18 and IL-619 (which both signal through the gp130 shared co-receptor) as well as G-CSF20 and Leptin21. Specificity arises from the ability of SOCS3 to inhibit only JAKs associated with certain cytokine receptors. The gp130, LIF and Leptin receptors all contain phosphotyrosine motifs that act as SOCS3 binding sites22–23–24. Whether these motifs act to bring SOCS3 into close proximity with JAK before it shuttles off the receptor to bind JAK directly or whether SOCS3 can bind both JAK and receptor simultaneously has been unclear.
To determine the molecular mechanism of SOCS3 action we solved the crystal structure of a SOCS3/JAK2/gp130 complex which showed that SOCS3 is bound to both the kinase domain of JAK2 and a fragment of the IL-6 receptor (gp130) at the same time. The gp130 fragment resides in the canonical phosphotyrosine-binding groove of SOCS3 whilst a surface on the other face of the SH2 domain is used to anchor JAK2. Given that in vivo JAK is also bound to gp130, the structure indicated that the true (high affinity) target of SOCS3 is a JAK/gp130 complex rather than JAK or gp130 alone. This explains why SOCS3 is highly specific for IL-6 family cytokines and others, such as G-CSF, whose receptors also contain SOCS3 binding motifs. Structural and biochemical analysis also revealed that the KIR of SOCS3 occupies the substrate-binding groove on JAK2 and occludes the P+1 pocket. The arginine immediately upstream of the KIR acts as the pseudosubstrate residue, indicating that SOCS3 inhibits signaling by blocking the substrate-binding site of the kinase that initiates the intracellular signaling cascade.
Results
SOCS3 binds JAK and cytokine receptor simultaneously
In order to determine the molecular details of a SOCS3/JAK2/receptor interaction we solved the crystal structure of a SOCS3/JAK2/gp130 ternary complex (SOCS322-185/JAK2JH1/gp130750-764, see Fig. 1a). SOCS322-185, was used as previous work had defined it as the minimal fully active fragment14 and full-length SOCS3 is poorly soluble. The gp130 shared co-receptor contains a single SOCS3 binding site, centered on pTyr75724. As the full intracellular domain of the receptor is over 250 residues in length and unstructured25 we prepared a phosphorylated fragment of this receptor (gp130750-764: STASTVEpYSTVVHSG). The crystal structure of this peptide in complex with SOCS322-185 has been solved previously26. Finally, the tyrosine kinase (JH1) domain of JAK2 (JAK2JH1) was used as it contains the SOCS3 interaction site17. An ATP mimetic was necessary to successfully crystallize JAK2JH1 previously27 and therefore a stoichiometric equivalent of CMP-6 was added to several of our crystallization trials.
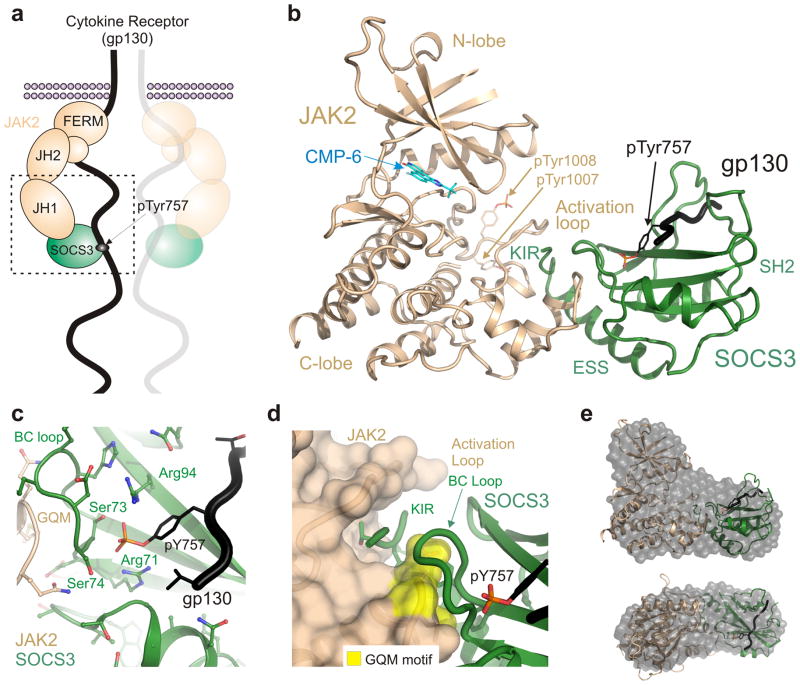
Figure 1. The structure of a JAK/SOCS/gp130 complex and the two interfaces.
(a) Schematic of SOCS3 inhibiting JAK2 whilst bound to gp130 (shared receptor for IL-6, LIF etc). JAK2 consists of four distinct domains and the C-terminal kinase domain (JH1) interacts with SOCS3. The boxed region indicates the ternary complex structure solved in this work (b) Ribbon diagram of the JAK2 (beige)/SOCS3 (green)/gp130 (black) complex. The gp130 peptide is located in the canonical pTyr-binding groove on the SH2 domain of SOCS3 whilst the opposing face of the SH2 domain contacts JAK2. (c) SOCS3 binds gp130 via the canonical pTyr-binding groove on the SH2 domain with the BC loop (S73-F79) of SOCS3 contacting both JAK2 and gp130 via opposing faces. The co-ordination of the phosphate moiety from gp130 pY757 is identical to that seen in the absence of JAK2. (d) The short KIR motif of SOCS3 sits in a groove bordered by the activation loop and GQM motif (yellow) of JAK2. (e) The molecular envelope of the JAK2/SOCS3/gp130 complex in solution calculated from SAXS data by performing 10 independent DAMMIF ab initio bead reconstructions (grey spheres) superimposed with the complex crystal structure (as shown in panel a). Upper panel shown in the same orientation as panel b; lower panel, top view. Data collection statistics and validation are shown as Supplementary materials.
Small crystals of a 1:1:1 (SOCS3/JAK2/gp130) complex were obtained, the best of which diffracted to 3.9Å. Phasing was achieved by molecular replacement using the higher resolution JAK2 (PDB: 3FUP) and SOCS3 structures (PDB: 2HMH). The structure was refined to Rwork and Rfree values of 0.2491 and 0.2808 respectively (Table 1). Despite this relatively low resolution, the fundamental details of the JAK2–SOCS3–gp130 interaction are clear. Electron density is shown in Supplemental Data.
Table 1.
Data collection and refinement statistics (molecular replacement)
| SOCS3/JAK2-JH1/gp130/CMP-6 | |
|---|---|
| Data collection | |
| Space group | P65 |
| Cell dimensions | |
| a, b, c (Å) | 139.29, 139.29, 316.81 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) ≠ | 95.9-3.9 (3.99-3.90) |
| Rmeas (%)≠ | 47.6 (665.8) |
| I/σI | 5.29 (0.52) |
| Completeness (%) | 99.98 (99.99) |
| Redundancy | 11.6 (11.6) |
| Refinement | |
| Resolution (Å) ≠ | 45.59-3.90 |
| No. reflections | 31682 |
| Rwork/Rfree | 24.91/28.08 |
| No. atoms | |
| Protein | 13585 |
| Ligand/ion | 92 |
| Water | 0 |
| B-factors | |
| Protein | 198.0 |
| Ligand | 198.3 |
| Water | N/A |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.594 |
Values in parentheses are for highest-resolution shell.
Criteria for data cut-off from 42. CC½ = 99.3 (15.3)
It was unclear whether SOCS3 would remain bound to the gp130 fragment in the presence of JAK2. After initial rounds of refinement, clear difference density in Fo-Fc maps for the gp130 fragment could be observed in the canonical phosphotyrosine (pTyr) binding groove on the SH2 domain of SOCS3 (Fig. 1b Supplementary Fig. 1). The gp130 fragment lies across the central three-stranded beta-sheet of the SH2 domain with the phosphotyrosine co-ordinated by the conserved R71 in βB, the serines in the BC loop and R94 in βD (Fig. 1c), just as seen in the absence of JAK226.
The SOCS3 BC loop that helps co-ordinate the pTyr also contacts JAK2 (Fig 1d). In fact gp130pY757 is located within 7Å of JAK2 at its closest point. To investigate whether binding of JAK2 influences the binding of gp130 or vice-versa we attempted to determine the structure of a SOCS3/JAK2 complex in the absence of gp130. However crystals obtained only diffracted to 7Å. Whilst this resolution is too low for structure determination, these SOCS3–JAK2 crystals grew in the same conditions as SOCS3–JAK2–gp130 and had essentially identical cell dimensions, suggesting that gp130 does not induce any large conformational changes.
The SOCS3 binding site on JAK2 is centered on the GQM motif
We observed four SOCS3–JAK2–gp130 trimers in the asymmetric unit and two potential SOCS3–JAK2 interfaces. The interface with the higher buried surface area (980 cf. 660 Å2) mapped to the region of SOCS3 identified by NMR to bind JAK2 and was consistent with mutagenesis data17. Further support for this assembly being representative of the biologically functional complex in solution was obtained using small angle X-ray scattering (SAXS). The SOCS3–JAK2–gp130 complex crystal structure is consistent with an ab initio bead model calculated from experimental scattering data (Fig. 1e). In addition, the theoretical scattering curve calculated for the crystal structure is in agreement with the experimental scattering curve (see Supplementary Fig. 2; χ=0.658 for fit). SAXS data collection statistics are presented in Supplementary Table 1.
The SOCS3/JAK2 interface is predominantly hydrophobic and centered upon the GQM motif17 in JAK2. This short motif (residues Gly1071, Gln1072, Met1073 of JAK2) is responsible for the ability of SOCS3 to selectively bind JAK1, JAK2 and TYK2 but not JAK3 and it sits at the junction of the “JAK insertion loop”27 and the αG helix28 (Met1073 is located on the first turn of this helix). SOCS3 docks onto this motif using segments of the SH2 domain, ESS helix and KIR.
Within the GQM motif, Gln1072 and Met1073 are buried deeply at the interface with SOCS3 (Fig. 2a,b). Gln1072 is stacked against the conserved SOCS3 residue Phe79, whilst Met1073 sits in a hydrophobic pocket formed by the SOCS3 ESS helix and two adjacent phenylalanines on the BC loop (Phe79–Phe80). Gly1071 allows the BC loop of SOCS3 to stack against the peptide backbone of JAK2 as well as providing the torsional flexibility for a tight turn immediately preceding the αG helix. Mutation of either Gly1071 or Met1073 renders JAK2 resistant to inhibition by SOCS317. The interface extends out from the GQM motif into the αG helix of JAK2 where Met1073 and Phe1076 form a non-polar surface that packs against a hydrophobic surface on SOCS3. It appears that the adjacent D1080 on the third turn of this helix in JAK2 forms a hydrogen bond with Y31 on SOCS3, however the electron density for that sidechain is not resolved well enough to state this unequivocally. Only minor conformational changes in the JAK2 GQM motif can be seen upon binding SOCS3. In contrast, this region adopts a very different orientation in JAK3, which lacks a GQM motif (Supplementary Fig. 3).
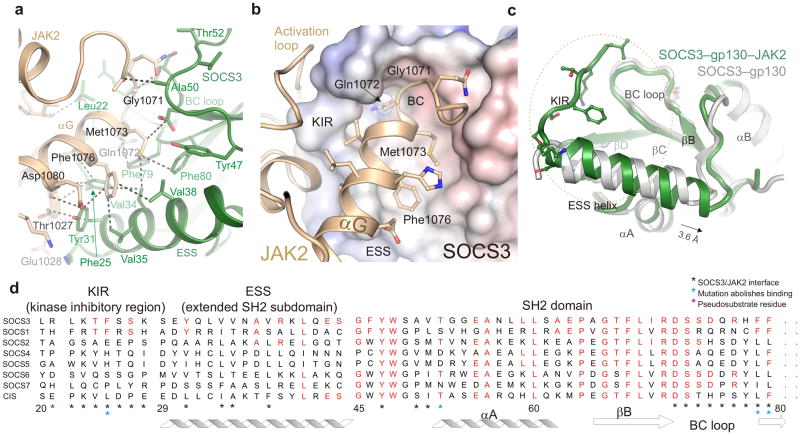
Figure 2. The SOCS3/JAK2 interaction.
(a) The hydrophobic SOCS3/JAK2 interface. Important residues are labeled and a selection of van der Waals contacts shown as dotted lines. Color scheme is the same as Figure 1. (b) Residues from the GQM motif and αG28 helix of JAK2 bind a concave hydrophobic surface on SOCS3 formed by the KIR, BC loop and extended SH2 subdomain (ESS). JAK2 is shown in ribbon representation and SOCS3 as an electrostatic surface (+/− 250mV). The GQM motif and Phe1076 from αG of JAK2 are highlighted. (c) Comparison of the SOCS3 structure in isolation (PDB:2HMH26, white) and in complex with JAK2 (this work, green). The ESS helix can be seen to undergo a translation of half a helical turn upon binding JAK2 whilst the KIR (which is unstructured in the absence of JAK2) adopts an extended structure. The orientation of SOCS3 is the same as in (b). (d) Sequence conservation of the JAK2-binding interaction surface between SOCS1 and SOCS3. Conserved residues are shown in red.
The JAK2 binding site on SOCS3
The SOCS3–JAK2–gp130 structure revealed that the majority of the JAK2 binding surface on SOCS3 is a concave hydrophobic region formed by the extended SH2 subdomain (ESS) and the BC loop. This loop is responsible for coordinating pTyr757 from gp13026 and its opposite face contacts JAK2. In particular, Asp72, Ser73, Phe79 and Phe80 from this loop all contact JAK2 directly. The SOCS3 ESS is an amphipathic α-helix and the hydrophobic face of this helix contacts residues from the similarly hydrophobic face of JAK2αG. JAK2 binding induces an extra helical turn at the beginning of the ESS helix and the whole region undergoes a translation of half a helical turn (Fig. 2c). This reconfiguration leads to a slightly larger hydrophobic face than in the absence of JAK2.
The key feature of the JAK2 binding epitope involves the SOCS3 KIR. The eight residue KIR lies immediately upstream of the ESS (see Fig. 2d) and is unstructured in isolation26,29. However in our complex structure it was sharply folded back underneath the BC loop with its three N-terminal residues (Leu22–Thr24) occupying a deep groove on the JAK2 surface (Fig. 3a, Supplementary Fig. 4). Whilst these contribute few inter-molecular hydrogen bonds, there are many van der Waals contacts which make up over 20% of the total buried surface area within the complex. Within the KIR, Phe25 is particularly important, as it is placed in a deep hydrophobic pocket at the interface of the two proteins that is formed by residues from both SOCS3 and JAK2 and this residue is known to be required for SOCS activity14. Collectively, the KIR and residues from the ESS and the BC loop of the SH2 domain form the JAK binding epitope.
Figure 3. The JAK2 binding epitope is composed of the KIR, ESS and SH2 domain and is conserved between SOCS3 and SOCS1.
(a) Cut-away view of the JAK2-binding epitope on SOCS3. SOCS3 is shown as a ribbon diagram and JAK2 as an electrostatic surface (+/− 250mV). The SOCS3 KIR (Leu22–Ser29), which is unstructured in isolation, folds back underneath the BC loop and sits in a groove formed by the JAK2 activation loop and GQM motif (labeled). SOCS3 Phe25 sits in a deep hydrophobic pocket formed by residues from both SOCS3 and JAK2. The interface is mostly hydrophobic. (b) IC50 plots of SOCS3 point mutants. These assays highlight SOCS3 Phe25, Phe79 and Phe80 as being required for inhibition. The curves shown are an average of duplicate experiments. (c) An isolated KIR peptide from SOCS1 inhibits JAK2 with a 0.1mM IC50. Error bars are SD from triplicate experiments. (d) SOCS3/JAK2 interface highlights the conservation of residues in the JAK2-binding epitope. Conserved residues between SOCS3 and SOCS1 shown in yellow. (e) Close-up of the interface shown in (d). Color scheme as in (d).
To fully characterize this epitope, an alanine scan was performed on SOCS3 residues that contact JAK2 and the ability of these mutants to inhibit JAK2 was tested. As shown in Table 2 and Figure 3b three residues were found to be essential: Phe25 from the KIR and Phe79, Phe80 from the BC loop. These are absolutely conserved in SOCS3 and SOCS1 in all vertebrates. Of the remaining residues, mutation of Glu30 resulted in a 20-fold increase in the IC50, possibly because it helps to position the SOCS3 KIR at 90° to the ESS helix by hydrogen bonding Ser26. Mutation of Tyr47 impaired activity, however it hydrogen bonds Asp72 and this pair of residues is conserved across all SOCS proteins, even those that do not bind JAK2, and likely has a structural role within the SH2 domain.
Table 2.
IC50 values for SOCS3 mutants.
| Mutation | Region | IC50mutant/IC50WT* |
|---|---|---|
| WT | KIR | 1.0 +/− 0.3 |
| SOCS1-3 | 0.1 +/− 0.05 | |
| L22F | 0.1 +/− 0.03 | |
| L22A | 1.2 +/− 0.2 | |
| K23A | 1.4 +/− 0.1 | |
| T24A | 3.3 +/− 0.5 | |
| F25A | no inhibition | |
| S26A | 1.0 +/− 0.04 | |
| S27A | 2.5 +/− 0.2 | |
| K28A | 2.8 +/− 0.7 | |
| S29A | ESS | 2.0 +/− 0.3 |
| E30A | 23.0 +/− 2.9 | |
| Y31A | 6.0 +/− 0.1 | |
| Y47F | βA | 80 +/− 20 |
| A50E | 1.1 +/− 0.2 | |
| A50K | 6.0 +/− 2.0 | |
| T52A | 10 +/− 2.0 | |
| R77A | BC loop | 1.4 +/− 0.2 |
| F79A | no inhibition | |
| F80A | no inhibition |
Errors are SD from three independent experiments.
SOCS1-3 refers to a SOCS1/SOCS3 chimera which binds to JAK2 with 5–10 fold higher affinity30
In order to further characterize the KIR we investigated whether, on its own, it was capable of inhibiting JAK2. The SOCS3 KIR as an isolated peptide could not inhibit the kinase activity of JAK2. However the KIR of SOCS1 (the only other SOCS protein known to directly inhibit JAK215) inhibited JAK2, albeit with low (100 μM) affinity (Fig. 3c). As shown in Figure 3d,e, although the sequence identity between SOCS1 and SOCS3 is only 33%, the SOCS/JAK interface site is almost completely conserved. This suggests that SOCS1 will share the same mode-of-interaction with JAK2 as does SOCS3.
The Kinase Inhibitory Region is required for JAK binding
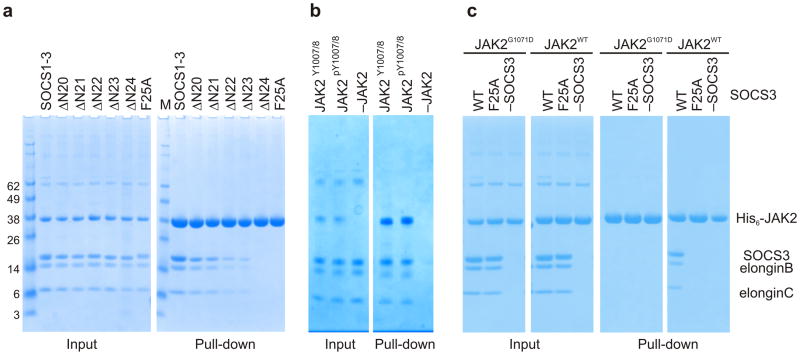
The failure of the F25A KIR mutant to inhibit JAK2 indicates that the KIR is required for inhibition but does not necessarily indicate that it is required for binding to JAK2. In order to investigate this, a series of mutants with truncated KIRs was constructed and co-precipitation experiments were employed. The concentration of JAK2 used in each pull down was 5μM with a 2-fold molar excess of SOCS3–elonginBC. The elonginBC complex is the physiogical ligand for the SOCS box of SOCS proteins and increases their solubility. The Kd of the SOCS3-JAK2 interaction is approximately 1μM30 and these concentrations were chosen to ensure that a near-stoichiometric (85%) pull-down of SOCS3 would occur for the wild-type construct whilst any reduction in affinity >5-fold for the mutant constructs should lead to a visible reduction in the pull-down efficiency. As shown in Figure 4a, there was a gradual loss of JAK2 binding as residues were removed, with SOCS3ΔN24, which begins at Phe25, showing no detectable interaction with JAK2. The importance of Phe25 is demonstrated by the fact that the interaction between JAK2 and SOCS3 is abolished by mutation of this residue to alanine. To date, there has been an assumption that SOCS3 would bind directly to JAK2 pY1007 or 1008 via its SH2 domain as part of its inhibitory mechanism, even if it was not the sole site of binding. However, our crystal structure showed no contact between SOCS3 and pY1007,8 and SOCS3 bound to dephosphorylated JAK2 (JAK2Y1007/Y1008) with similar affinity to phosphorylated JAK2 (JAK2pY1007/pY1008) (Fig. 4b, Supplementary Fig. 5). Moreover, as shown in Figure 4c, there was no binding to JAK2 when the JAK2 GQM motif was mutated (G1071D) even though the activation loop was phosphorylated as determined by western blot with a pY1007 specific antibody (data not shown).
Figure 4. The KIR is required for JAK binding.
(a) Co-precipitation experiments show that SOCS3 binds JAKJH1 provided the kinase inhibitory region is intact. Lanes 2–6 show there is a gradual loss of binding as the N-terminal residues of the SOCS3 KIR are removed, as well as when Phe25 is mutated to Alanine. The SOCS1-SOCS3 chimera is shown as a positive control (b) Co-precipitation experiments show that SOCS3binds activated (pY1007/8) and dephosphorylated (Y1007/8) JAK2JH1 with similar affinity. Dephosphorylated JAK2JH1 was prepared by co-expressing it with the phosphatase PTP1B and then used in co-precipitation experiments with SOCS3 as described in panel a. A western blot of this experiment probed with a pY1007/8 specific antibody shows that JAK2 co-expressed with PTB1B was >95% dephosphorylated (Supplemental Figure 5) (c) Co-precipitation experiments show that SOCS3 does not bind JAK2JH1 if the JAK2 GQM motif is mutated (G1071D). SOCS3F25A is shown as a negative control.
SOCS3 inhibits JAK2 by blocking substrate binding
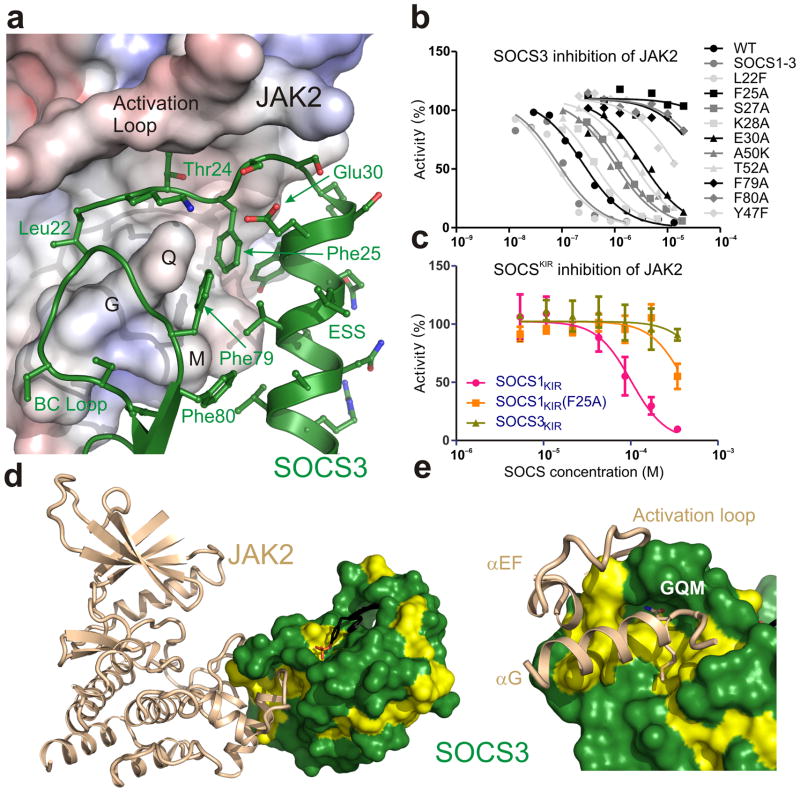
The substrate binding site of JAK2 can be modeled using the IRK/IRS-1 complex (PDB: 1IR3)31. This indicated that the KIR of SOCS3 partially occupies the substrate binding groove (Fig. 5a). In particular, the first residue of the KIR, Leu22, sits in the predicted P+1 binding site, one residue downstream from the substrate tyrosine. Therefore, we hypothesized that SOCS3 inhibits JAK2 by blocking substrate binding. However, given that our previous work had shown that SOCS3 displayed apparently non-competitive inhibition kinetics as regards substrate17, this hypothesis required further validation.
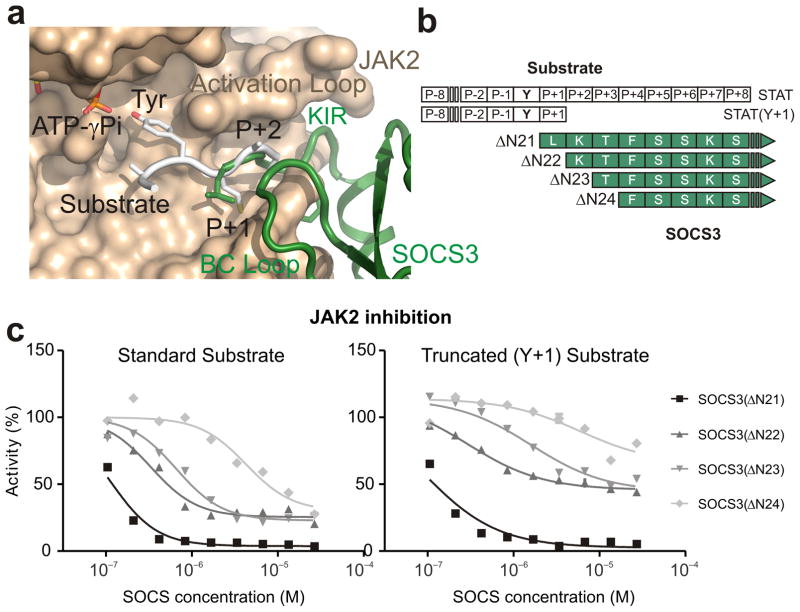
Figure 5. SOCS3 inhibits JAK2 by blocking substrate binding.
(a) Model of a substrate peptide (white) bound to JAK2 based on the IRK–substrate–ATP structure (PDB 1IR3) shows that the KIR of SOCS3 would block substrate binding. L22 is located where the P+1 residue would reside (both are shown in stick representation). (b) Schematic of the predicted overlap with substrate binding for the constructs and substrates used in these experiments. (c) Kinase inhibition assays performed with constructs of SOCS3 truncated at the N-terminal end of the KIR show that constructs lacking 1–3 residues only partially inhibit JAK2 activity. Left, kinase inhibition experiments using the standard STAT5b peptide (Y+8) as a substrate. Right, as for left but using a C-terminally truncated (Y+1) substrate. Inhibition of STAT5bY+1 by SOCS3ΔN22,23 is only 50% complete under saturating conditions.
We reasoned that if the SOCS3 KIR functions by blocking substrate binding then truncating one or more residues from its N-terminal end (see Fig. 5b) would reduce the ability of SOCS3 to inhibit JAK2. As shown in Figure 5c and Supplementary Figure 6, SOCS3 mutants that lacked the first 1–3 residues of the KIR showed quantitative and qualitative differences compared to wild-type (WT) SOCS3. Deleting the first one or two residues led to a 10-fold increase in the IC50, whilst deleting the third residue increased this by a further 10-fold. Owing to the high concentrations of SOCS3 proteins used in these assays, inhibition can be observed even for SOCS3ΔN24. Changes in IC50 indicate changes in the affinity of the interaction. Of greater interest was the fact that these shorter constructs could not achieve 100% inhibition of JAK, even at saturating concentrations. For example, when JAK2 is fully bound by SOCS3ΔN22 and SOCS3ΔN23 it retained 25% of its activity. These data are consistent with a model in which these truncated forms of SOCS3 cannot completely block substrate binding because of the reduced overlap between the N-terminus of the KIR and the substrate. For example, a reduction in the affinity of substrate for a SOCS3ΔN22–JAK2 complex (compared to its affinity for JAK2 alone) would lead to the observed residual activity. In contrast, the affinity of substrate for a SOCS3ΔN21/JAK2 complex is zero as the binding site is completely blocked. In support of this, we found that when this overlap was reduced even further by using a C-terminally truncated form of the substrate, which only contained a single residue downstream of the tyrosine, inhibition was even less complete (50%), see Figure 5c.
Residues upstream of the KIR can act as a pseudosubstrate
One characteristic of substrate blocking inhibitors is that they act as pseudosubstrates. The SOCS3–JAK2–gp130 structure showed that the first residue of the SOCS3 KIR, Leu22, sits in the P+1 binding site, not the tyrosine binding site itself. This suggested that a residue upstream of the KIR, rather than any residue within it, would be the true pseudosubstrate residue.
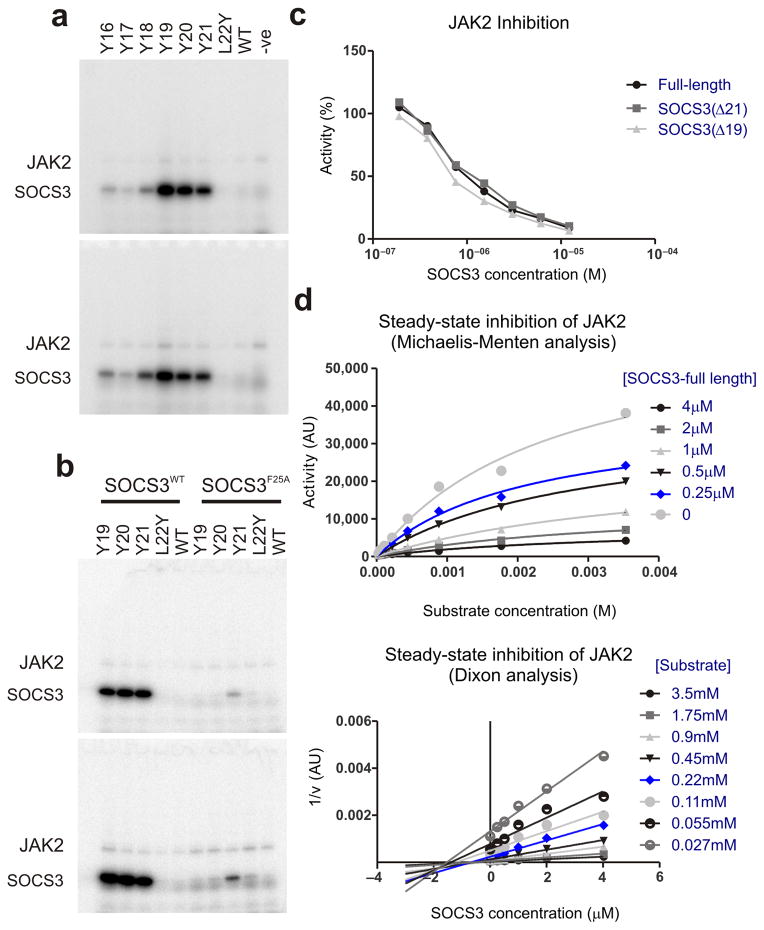
In order to determine whether this is the case we constructed various mutant forms of SOCS3, with a tyrosine residue 1–6 residues upstream of L22. Glycine was used the spacer residue(s) between the tyrosine and L22. A mutant containing tyrosine at position 22 (L22Y SOCS3) was present as a negative control, since based on our structure it should not be phosphorylated. As shown in Figure 6a and Supplementary Figure 6b, tyrosines at positions 21, 20 and 19 of SOCS3 were very efficiently phosphorylated by JAK2. This efficiency is due to the fact that they are bound in a specific orientation on JAK2 which localizes them to the active site, as F25A versions of these mutants were not phosphorylated to the same extent (Fig. 6b).
Figure 6. Residues upstream of the SOCS3 KIR act as a pseudosubstrate.
(a) Radioactive kinase assays show that constructs of SOCS3 with a tyrosine 1–3 residues upstream of the KIR are good substrates for JAK2. Reactions were performed for 1 minute (upper panels) and 2 minutes (lower panels) and then analyzed via SDS-PAGE followed by autoradiography. Experiments performed in the absence of SOCS3 (−ve) are shown as a control. (b) As for (a) but SOCS3 F25A mutants were used as controls to show that tyrosines upstream of the KIR are only good substrates for JAK2 when forced into close proximity of the active site by the remainder of SOCS3. (c) Full-length SOCS3 inhibits JAK2 with an identical IC50 to SOCS3 lacking the first 19 or 21 residues. (d) Steady-state inhibition of JAK2 by SOCS3 gives non-competitive inhibition as analyzed by Michaelis-Menten (upper) and Dixon (lower) plots. The average of duplicate experiments are shown in each case.
For solubility reasons, all our biochemical and structural studies to date have used constructs of SOCS3 beginning at residue 22, the N-terminus of the KIR, rather than residue one. Given that residue 21 is the true pseudosubstrate residue we were concerned that the SOCS3 KIR may have been mis-defined as only consisting of residues 22 onwards and that full-length SOCS3 may be a more potent inhibitor and perhaps display competitive kinetics. Therefore, we purified full-length SOCS3 and performed a full steady-state kinetic analysis. SOCS31-225 inhibited JAK2 with an identical IC50 to SOCS322-225 and was also apparently non-competitive as regards substrate (Fig. 6c,d). When combined with previous cellular data14,32, to our knowledge there are no experiments that can distinguish between full-length SOCS3, and SOCS3 lacking the first 21 residues.
The fact that JAK2 is active when bound by SOCS3 further supports the hypothesis that SOCS3 functions by blocking substrate binding and not by preventing catalysis per se. These data, in addition to the correlation between the degree of overlap between SOCS3 and the substrate and the degree of inhibition (Fig. 5), alongside the structure of the SOCS3–JAK2–gp130 complex leads us to conclude that SOCS3 inhibits JAK2 by blocking substrate binding.
Discussion
SOCS3 is a potent inhibitor of JAK14 and yet, in a biological context, shows remarkable specificity for inhibiting JAK signaling only when stimulated by particular cytokines. The SOCS3–JAK2–gp130 structure, along with supporting biochemical data, elucidates both the mechanism of SOCS3 inhibition as well as providing the molecular basis of its specificity. In short, SOCS3 inhibits JAK’s enzymatic activity by blocking substrate binding and gains specificity of action by only binding tightly to JAK when the kinase is attached to specific receptors.
Given that our previous data had shown that SOCS3 inhibits JAK2 with non-competitive kinetics towards substrate17, the above model required further validation. We reasoned that there were two testable aspects to the hypothesis: (A) if SOCS3 blocks substrate binding using the KIR then truncating this region should lead to impaired inhibition; and (B) if SOCS3 acts as a pseudosubstrate then it should be convertible to a substrate by mutating the appropriate residue to tyrosine. The outcome of both of these experiments support our model: Even under saturating SOCS3 concentrations, truncation of the KIR led to impaired inhibition and mutating residues 19–21 to tyrosine converted SOCS3 into a high-affinity JAK2 substrate.
That tyrosine placed in three consecutive positions were equally good substrates was surprising. There are two major interactions that exist between a tyrosine kinase and a substrate tyrosine: (1) The tyrosine ring stacks on a conserved proline (Pro1017 in JAK2) and (2) a hydrogen bond forms between the tyrosine hydroxyl and the catalytic aspartate (Asp976 in JAK2). It is unlikely that the Tyr-Pro interaction could occur for all three of our mutants but there may be enough flexibility in the main chain and tyrosine sidechain χ1 torsion angle to allow the Tyr-Asp hydrogen bond. If the Tyr-Pro interaction is not essential then other hydroxyl containing residues might act as substrates when forced near the JAK active site. Indeed, we observed that a threonine placed at position 19 of SOCS3 was also phosphorylated (Supplementary Fig. 7).
Whilst tyrosines at position 19, 20 and 21 were equally good substrates, our structure suggests residue 21 is the true pseudosubstrate residue. This residue in SOCS3 and SOCS1 is conserved throughout evolution as an arginine or histidine. Modeling these into our structure suggest both of these are long enough to hydrogen-bond Asp976 and contain a planar element on their sidechains that could stack onto Pro1017. However Arg21 does not seem to provide any extra affinity toward JAK2 and is not required for complete inhibition. Therefore any interactions it makes seem to be dispensable as regards SOCS3 function.
Blat describes several mechanisms via which active site binders can display apparently non-competitive kinetics33. The two that appear most plausible here are either that: (A) SOCS3 binding blocks ADP release; or (B) there is a slow step in SOCS3 binding to JAK2 that breaks the rapid equilibrium assumption. In the latter case, for example, SOCS3 may form a rapid encounter complex with JAK2 within milliseconds followed by a slower (>seconds) re-orientation of the SOCS3 KIR into the substrate binding groove. The initial encounter complex would not involve the KIR and hence would not compete with substrate binding. Either of these scenarios could explain the kinetic data.
The structure of the SOCS3–JAK2–gp130 complex demonstrates explicitly that SOCS3 binds JAK2 and cytokine receptor simultaneously. This has important functional consequences as it allows the formation of an unusual ternary complex (JAK–SOCS–Receptor) in which each moiety is bound directly to the other two. Such a complex will show an overall stability (affinity) that is much greater than the sum of its individual interactions. In effect, whilst SOCS3 binds JAK2 with micromolar affinity, it will bind a JAK2/receptor complex with much higher affinity provided the receptor has a SOCS3-interaction motif. Genetic deletion of SOCS3 has shown it to be a critical inhibitor of LIF, G-CSF, Leptin and IL-6 (which binds to gp130) and all of these cytokines act through receptors with a known SOCS3-interaction motif. Our data predict that if the concentration of SOCS3 in the cytoplasm is high enough (i.e. approaching 1 μM, the Kd of the SOCS3/JAK2 interaction) it will circumvent the need for receptor-mediated JAK binding and will bind and inhibit all of JAK1, JAK2 and TYK2. This is supported by the fact that SOCS3 is known to inhibit a multitude of different cytokines when artificially over-expressed (for review see 34). Notably, our model of SOCS3 action and specificity does not require it to be bound to the same receptor chain as JAK2, as the opposing chain(s) in a receptor dimer or oligomer would also induce the same high affinity ternary complex.
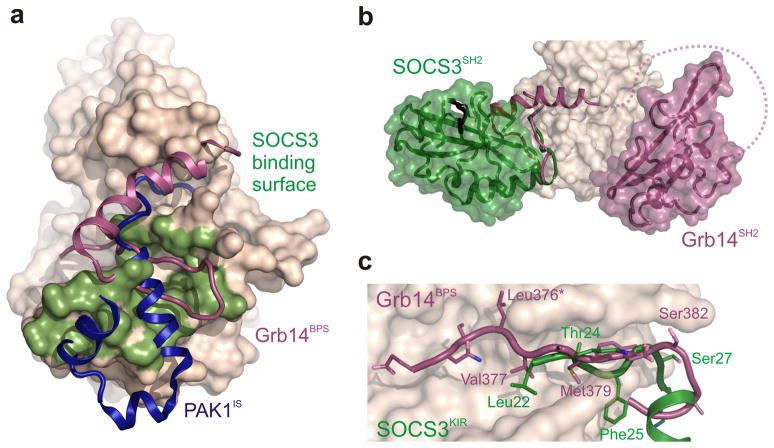
The SOCS3–JAK2–gp130 structure reveals many parallels between SOCS3 inhibition of JAK signaling and Grb14 inhibition of the insulin RTK (Fig. 7): Grb14 is anchored to the insulin receptor kinase domain (IRK) via its SH2 domain and it blocks substrate binding via a KIR-like region N-terminal to this. The KIR-like motif in Grb14 is unstructured in the absence of IRK in the same way that the SOCS3 KIR is unstructured in the absence of JAK35; and finally Grb14 also acts as a pseudosubstrate without displaying competitive kinetics36,37. The major difference between Grb14 and SOCS3 is that the former anchors itself to IRK by binding its phosphorylated activation loop using the canonical phosphotyrosine binding groove whereas SOCS3 binds JAK2 via a non-canonical surface which frees the pTyr binding groove to bind receptor. Interestingly, the SOCS3/JAK2 interaction does not involve phosphotyrosines on the activation loop of JAK2 as previously supposed15 but instead is mediated by a hydrophobic surface on JAK2. Overall this surface is very similar to that used by the inhibitory switch (IS) region of PAK138. However autoinhibition of PAK1 disrupts the catalytic site whereas SOCS3 and Grb14 act by blocking substrate. Whilst there is a relatively long (23 residue) flexible linker between the SH2 domain of Grb14 and its KIR-like region, in SOCS3 the two are attached via a short, rigid, helical linker (the ESS) which is likely necessary to ensure that the KIR remains tightly bound to the substrate binding groove (given that the KIR on its own has a relatively low affinity for JAK2). Therefore, in SOCS3, the SH2 domain both tethers and positions the KIR for binding whereas in Grb14 it merely tethers.
Figure 7. Comparison of three kinase inhibitors: SOCS3, PAK1IS and Grb14.
(a) SOCS3 binds to a surface on the JAK2 kinase domain (shaded green) similar to that used by Grb14 on the insulin receptor kinase (magenta) and by the autoregulatory (IS) region of PAK1 (blue). (b) SOCS3 and Grb14 are tethered to their target kinase via a different surface. The SH2 domain of Grb14 binds the activation loop of IRK using the canonical pTyr binding groove. In contrast, SOCS3 binds JAK2 using the opposite face of the SH2 domain which leaves the canonical pTyr binding groove available for binding receptor (black). (c) SOCS3, like Grb14, inhibits its target kinase by blocking substrate binding. The BPS region of Grb14 and the KIR of SOCS3 occupy the substrate-binding groove of their target kinases. Leu376 of Grb14 acts as the pseudosubstrate residue (asterisk).
Activating mutations in JAK2, particularly V617F, are associated with the majority of cases of myeloproliferative neoplasms such as polycythemia vera39, and are also found in a number of acute leukemias40. As such, JAK2 has long been recognized as an important drug target for the treatment of a number of hematological malignancies and currently there at least six different JAK inhibitors in clinical trials for myeloid disease41. All of these trials are using compounds that bind to the ATP binding site of JAK2 and are therefore ATP competitive. Such compounds are outcompeted by high intracellular ATP concentrations and are prone to off-target effects as their site of interaction is structurally similar throughout the kinome. SOCS3, by virtue of being non-competitive towards ATP, is unaffected by the high concentration of ATP within the cytoplasm and targets only JAK1, JAK2 and TYK2. The structural details presented here provide useful information toward the development of a small-molecule mimetic of the SOCS3 KIR which would offer distinct advantages over all currently available JAK targeted therapeutics.
Although the structure presented here is of a specific SOCS (SOCS3) bound to a specific JAK (JAK2) and receptor (gp130), it has wider significance in terms of SOCS–JAK–Receptor biology. It seems clear that SOCS3 will bind to the same surface on JAK1 and TYK2 as it does on JAK217 and given the sequence similarity between SOCS1 and SOCS3, especially in the JAK-binding surface, we believe SOCS1 will interact in the same way with these three JAKs. As all cytokines that signal via the JAK/STAT pathway use at least one of these three kinases then the structure presented here provides the molecular mechanism for all SOCS1 and SOCS3 based signaling inhibition. The specificity within the SOCS1,3/JAK/Receptor system is provided by the receptor and therefore it is important that the full repertoire of SOCS1 and SOCS3 binding sites on these receptors be identified. Our current efforts lie in this direction.
ONLINE METHODS
Expression and purification of a JAK2JH1:SOCS3:gp130750-764 complex
The JH1 of JAK2, residues 836–1132 (Genbank: Protein, AAH54807; cDNA, BC054807), was cloned into pFastBac HTb (Life Technologies), and the resulting bacmid used to transfect Sf21 cells. High titer baculovirus was used to infect 1–5 liters Sf21 cells grown to a density of 2 × 106 mL−1 in the presence of 0.4 μM of the JAK inhibitor 2-(1,1-Dimethylethyl)-9-fluoro-3,6-dihydro-7H-benz[h]-imidaz[4,5-f]isoquinolin-7-one (EMD biosciences, also called CMP-6). Cells were collected 48 h after infection and snap frozen. Cells were lysed by sonication and His6-JAK purified by IMAC using standard protocols. All SOCS3 constructs were engineered such that the PEST motif was replaced by a Gly-Ser x4 linker to aid solubility and stability and were expressed in inclusion bodies in E. coli and refolded as described previously43. At this stage a 2x molar excess of murine gp130 phosphopeptide (gp130750-764:STASTVEpYSTVVHSG) was added. The His6 tag from SOCS3 and JAK2 was then removed by treatment with TEV protease and the proteins mixed in a 1:2:2 ratio (JAK:SOCS3:gp130) and the complex purified by gel filtration in Tris-buffered saline (TBS) containing 1mM DTT on a Superdex 200 26/60 column. A final purification step using anion exchange on a Mono-Q column with a 0–500mM NaCl elution in 10mM Tris pH 8.5, 1mM DTT was performed and the complex concentrated to 10mg/mL.
Crystallization and structure determination
Crystallization was accomplished by hanging-drop vapor diffusion at 20°C, using protein at 5–10 mg/ml, and a drop ratio of 2:1 protein:precipitant. Hexagonal prism-shaped crystals of various sizes (100–400μm in length) were obtained from 0.3–0.5M K2HPO4, 1.7–1.5M NaH2PO4 (constant 2M phosphate) and 0.1M phosphate-citrate pH 4.2, but smaller crystals consistently gave better diffraction. Crystals were flash-frozen in liquid nitrogen, using mother liquor as cryoprotectant. Diffraction data were collected on beamline MX2 at the Australian Synchrotron using a wavelength of 0.95371 Å. Data were integrated using XDS44 and scaled using XSCALE. The cutoff for data used in refinement was determined using the Pearson correlation coefficient, as represented in the XSCALE output42. Data in resolution shell 3.99 to 3.90 Å had CC½ of 15.3%. A molecular replacement solution was obtained using PHASER45, using JAK2 (3FUP, chain A) and SOCS3 (2HMH, chain A) as search models. The phases obtained using four copies of JAK2 revealed clear density for the three helices in each copy of SOCS3. Two SOCS3 molecules were subsequently placed using PHASER whilst the other two copies were positioned manually. The gp130 peptide was initially absent from the search model and was inserted during refinement once electron density could be clearly discerned. Refinement was performed using PHENIX46 and model building performed in COOT47. Refinement converged with Rwork of 0.249 and Rfree of 0.281 for data to 3.9 Å resolution. 96% of residues in the final structure are in the favored area of Ramachandran space and 0.12% are outliers. Buried surface area was calculated using PISA48. Further details are provided in supplemental data.
Mutagenesis
Mutagenesis of SOCS3 was performed using either the Quikchange site-directed mutagenesis kit (Stratagene) for internal mutations or by incorporating the mutation in the 5′ primer and using standard PCR. All mutant SOCS3 proteins used in kinase assays and pseudosubstrate assays were co-expressed with elonginB/C as described previously9. ElonginB/C is the physiological ligand for the SOCS box of SOCS3 and aids both solubility and stability. JAK2 mutants were generated as reported previously17.
Pseudosubstrate assays
10 μM mutant SOCS3/elonginBC complexes (with tyrosine residues inserted 0–3 residues upstream of L22 in SOCS3) were incubated with 1 μM JAK in TBS containing 2mM MgCl2, 1mM ATP, 1mM DTT and 1 μCi γ-[32P]ATP for 10–120s. The reaction was stopped by the addition of boiling SDS-PAGE buffer and analyzed by SDS-PAGE followed by Coomassie staining and autoradiography.
JAK inhibition assays
JAK2JH1 used for enzymatic assays was expressed and purified as described above except that CMP-6 was omitted from the growth media. Inhibition assays were essentially as described previously17. Briefly, 10 nM JAK2JH1 was incubated with either 0–2 mM substrate peptide for 10–20 min at 25°C. The substrate used was STAT5b693–708 (RRAKAADGYVKPQIKQVV) The reaction buffer was TBS containing 2mM MgCl2, 1mM ATP, 1mM DTT and 1 μCi γ-[32P]ATP. After incubation, the reactions were spotted onto P81 phosphocellulose paper and washed extensively (4 × 200 ml, 15 min) with 5% H3PO4 then exposed to a phosphorimager plate (Fuji).
Co-precipitation assays
5 μM JAK was incubated with 10 μM of each SOCS3/elonginBC complex in buffer A (TBS containing 5mM imidazole and 1mM DTT) for 5 minutes in 100 μl total volume. 20 μl of a 50% slurry of Ni-NTA resin (Qiagen) was added and the incubation continued for a further 5 minutes before the tube was centrifuged for 1 minute at 1000 ×g in a 0.22 μm spin-filter to remove the supernatant. The beads were washed twice using 100 μl of buffer A containing 20 mM imidazole and then the proteins eluted from the beads by the addition of 25 μl of buffer A containing 250mM imidazole. Results were analyzed via SDS-PAGE and Coomassie blue staining.
Expression of dephosphorylated Jak2 kinase domain
Mouse JAK2 JH1 domain (residues 836–1132) was co-expressed with the phosphatase, PTP1B (residues 2–321), in Sf21 insect cells to obtain activation loop-dephosphorylated JAK2 JH1. A composite expression construct encoding N-terminally His-tagged mJAK2 JH1 domain and N-terminally FLAG-tagged human PTP1B (residues 2–321) was generated using the MultiBac Turbo system (ATG Biosynthetics). This expression construct was prepared by (i) cloning a cDNA encoding His-mJAK2 (836-1132) into the vector, pAceBac1 (ATG Biosynthetics), which encodes a polh promoter; (ii) cloning a cDNA encoding N-terminally FLAG-tagged human PTP1B (2-321) PCR amplified from IMAGE clone 4844022 into the vector pAceBac2 (ATG Biosynthetics), which includes a p10 promoter; (iii) ligation of a I-CeuI to BstXI fragment of the clone described in (ii) into a BstXI digested preparation of the construct described in (i). The resulting expression construct was transformed into chemically competent E. coli (DH10MultiBac strain; sourced from ATG Biosynthetics) to generate bacmids for baculovirus production. Bacmid DNA and P1 virus were generated using standard protocols and P2 virus was generated by infecting Sf21 cells (density 1.5 × 106 cells/mL) in shaking culture with 1% v/v P1 virus and growing for 4 days. P2 viral supernatant was used to infect 0.5L cultures of Sf21 cells (at 3–4×106 cells/mL density) and grown in shaking culture for 48 hours. Cell pellets were harvested by centrifugation and purification of dephosphorylated His-mJAK2 JH1 performed using Ni-NTA and gel filtration chromatography. Dephosphorylation was assessed by quantitative infrared western blot (Odyssey IR scanner) using a pY1007/8 specific antibody (Cell Signalling Technologies C80C3 used at 1/2000 dilution).
Small angle X-ray scattering (SAXS) measurements and data analysis
SAXS data collection was performed at the Australian Synchrotron SAXS/WAXS beamline using an inline gel filtration chromatography setup, essentially as described previously49. Summary statistics for data collection are presented in Supplementary Table 1. Protein samples (initial concentration of 5mg/mL) were injected on to an inline Superdex 200 5/150 column (GE Healthcare) pre-equilibrated with 150 mM NaCl, 20 mM HEPES pH 7.5 and eluted via a 1.5 mm glass capillary positioned in the X-ray beam. Scattering data were collected in 2sec quanta over the course of the elution and 2D intensity plots from the peak of the SEC run were radially averaged, normalized to sample transmission, with scattering profiles from earlier in the size exclusion chromatography elution averaged and used to perform background subtraction of 1D profiles. All data analyses were performed using the ATSAS suite50: Guinier analysis using PRIMUS51; indirect Fourier transform using GNOM52 to obtain the distance distribution function, P(r), and the maximum dimension of the scattering particle, Dmax. Theoretical scattering curves were calculated from atomic coordinates and compared with experimental scattering curves using CRYSOL53. Low resolution shape envelopes were generated using the ab initio bead-modeling program, DAMMIF54, by performing 10 independent reconstructions, aligning all with the most probable model using DAMSEL and DAMSUP, averaging models using DAMAVER and adjustment to correspond with the experimentally determine excluded volume using DAMFILT55. The final bead model was superimposed upon the X-ray crystal structure coordinates using SUPCOMB1356.
Supplementary Material
Acknowledgments
We thank P. Colman, P. Czabotar and M. Lawrence for advice and I. Segel for helpful discussions. This work was supported in part by the National Health and Medical Research Council of Australia program grants 461219 and 1016647 (N.A.N) and project grant 1011804 (J.J.B), the U.S. National Institutes of Health CA22556 (N.A.N), the Victorian State Government Operational Infrastructure Support Grant, and the NHMRC Independent Research Institutes Infrastructure Support Scheme (361646). N.A.N. acknowledges fellowship support from the National Health and Medical Research Council, J.M.M and J.J.B. from the Australian Research Council and L.N.V. from the Leukaemia Foundation of Australia and the Australian Stem Cell Centre. N.A.N. is a founder and member of the scientific advisory board of MuriGen Pty Ltd. We thank the scientists at the MX and SAXS/WAXS beamlines at the Australian Synchrotron. Crystallization trials were performed at CSIRO collaborative crystallization centre (C3).
Footnotes
Accession codes. The atomic coordinates of the SOCS3/JAK2/gp130/CMP6 complex can be found at the Protein Data Bank, entry 4GL9
Author Contributions
N.J.K. carried out crystallographic data collection, structure determination and refinement. J.J.B performed inhibition and co-precipitation assays. J.M.M. and L.N.V carried out SAXS data collection and analysis. J.M.M, A.L, I.S.L and E.L.W performed protein expression and purification experiments. All authors commented on the manuscript. J.J.B, N.J.K and N.A.N designed experiments, analyzed data, supervised the project and wrote the paper.
References
- 1.Wilks AF. Two putative protein-tyrosine kinases identified by application of the polymerase chain reaction. Proc Natl Acad Sci U S A. 1989;86:1603–7. doi: 10.1073/pnas.86.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilks AF, Oates AC. The JAK/STAT pathway. Cancer Surv. 1996;27:139–63. [PubMed] [Google Scholar]
- 3.Shuai K, et al. Polypeptide signalling to the nucleus through tyrosine phosphorylation of Jak and Stat proteins. Nature. 1993;366:580–3. doi: 10.1038/366580a0. [DOI] [PubMed] [Google Scholar]
- 4.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–21. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 5.Endo TA, et al. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387:921–4. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 6.Hilton DJ, et al. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc Natl Acad Sci U S A. 1998;95:114–9. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naka T, et al. Structure and function of a new STAT-induced STAT inhibitor. Nature. 1997;387:924–9. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 8.Starr R, et al. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–21. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 9.Babon JJ, et al. The SOCS box domain of SOCS3: Structure and interaction with the elonginBC-cullin5 ubiquitin ligase. Journal of Molecular Biology. 2008;381:928–940. doi: 10.1016/j.jmb.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babon JJ, Sabo JK, Zhang JG, Nicola NA, Norton RS. The SOCS Box Encodes a Hierarchy of Affinities for Cullin5: Implications for Ubiquitin Ligase Formation and Cytokine Signalling Suppression. Journal of Molecular Biology. 2009;387:162–174. doi: 10.1016/j.jmb.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamizono S, et al. The SOCS box of SOCS-1 accelerates ubiquitin-dependent proteolysis of TEL-JAK2. J Biol Chem. 2001;276:12530–8. doi: 10.1074/jbc.M010074200. [DOI] [PubMed] [Google Scholar]
- 12.Zhang JG, et al. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc Natl Acad Sci U S A. 1999;96:2071–6. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang JG, et al. The SOCS box of suppressor of cytokine signaling-1 is important for inhibition of cytokine action in vivo. Proc Natl Acad Sci U S A. 2001;98:13261–5. doi: 10.1073/pnas.231486498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki A, et al. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes to Cells. 1999;4:339–351. doi: 10.1046/j.1365-2443.1999.00263.x. [DOI] [PubMed] [Google Scholar]
- 15.Yasukawa H, et al. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. Embo J. 1999;18:1309–20. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyle K, et al. Deletion of the SOCS box of suppressor of cytokine signaling 3 (SOCS3) in embryonic stem cells reveals SOCS box-dependent regulation of JAK but not STAT phosphorylation. Cellular Signalling. 2009;21:394–404. doi: 10.1016/j.cellsig.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babon JJ, et al. Suppression of Cytokine Signaling by SOCS3: Characterization of the Mode of Inhibition and the Basis of Its Specificity. Immunity. 2012;36:239–250. doi: 10.1016/j.immuni.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts AW, et al. Placental defects and embryonic lethality in mice lacking suppressor of cytokine signaling 3. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9324–9329. doi: 10.1073/pnas.161271798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yasukawa H, et al. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nature Immunology. 2003;4:551–556. doi: 10.1038/ni938. [DOI] [PubMed] [Google Scholar]
- 20.Croker BA, et al. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 2004;20:153–165. doi: 10.1016/s1074-7613(04)00022-6. [DOI] [PubMed] [Google Scholar]
- 21.Mori H, et al. Socs3 deficiency in the brain elevates leptin sensitivity and confers resistance to diet-induced obesity. Nature Medicine. 2004;10:739–743. doi: 10.1038/nm1071. [DOI] [PubMed] [Google Scholar]
- 22.Hortner M, et al. Suppressor of cytokine signaling-3 is recruited to the activated granulocyte-colony stimulating factor receptor and modulates its signal transduction. Journal of Immunology. 2002;169:1219–1227. doi: 10.4049/jimmunol.169.3.1219. [DOI] [PubMed] [Google Scholar]
- 23.De Souza D, et al. SH2 domains from suppressor of cytokine signaling-3 and protein tyrosine phosphatase SHP-2 have similar binding specificities. Biochemistry. 2002;41:9229–36. doi: 10.1021/bi0259507. [DOI] [PubMed] [Google Scholar]
- 24.Nicholson SE, et al. Suppressor of cytokine signaling-3 preferentially binds to the SHP-2-binding site on the shared cytokine receptor subunit gp130. Proc Natl Acad Sci U S A. 2000;97:6493–8. doi: 10.1073/pnas.100135197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skiniotis G, Lupardus PJ, Martick M, Walz T, Garcia KC. Structural organization of a full-length gp130/LIF-R cytokine receptor transmembrane complex. Mol Cell. 2008;31:737–48. doi: 10.1016/j.molcel.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergamin E, Wu J, Hubbard SR. Structural basis for phosphotyrosine recognition by suppressor of cytokine signaling-3. Structure. 2006;14:1285–92. doi: 10.1016/j.str.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Lucet IS, et al. The structural basis of Janus kinase 2 inhibition by a potent and specific pan-Janus kinase inhibitor. Blood. 2006;107:176–83. doi: 10.1182/blood-2005-06-2413. [DOI] [PubMed] [Google Scholar]
- 28.Knighton DR, et al. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–14. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 29.Babon JJ, et al. The structure of SOCS3 reveals the basis of the extended SH2 domain function and identifies an unstructured insertion that regulates stability. Molecular Cell. 2006;22:205–216. doi: 10.1016/j.molcel.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 30.Babon JJ, et al. Suppression of cytokine signaling by SOCS3: characterization of the mode of inhibition and the basis of its specificity. Immunity. 2012;36:239–50. doi: 10.1016/j.immuni.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. Embo J. 1997;16:5572–81. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholson SE, et al. Mutational analyses of the SOCS proteins suggest a dual domain requirement but distinct mechanisms for inhibition of LIF and IL-6 signal transduction. Embo J. 1999;18:375–85. doi: 10.1093/emboj/18.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blat Y. Non-competitive inhibition by active site binders. Chem Biol Drug Des. 2010;75:535–40. doi: 10.1111/j.1747-0285.2010.00972.x. [DOI] [PubMed] [Google Scholar]
- 34.Yoshimura A. The CIS family: negative regulators of JAK-STAT signaling. Cytokine Growth Factor Rev. 1998;9:197–204. doi: 10.1016/s1359-6101(98)00019-7. [DOI] [PubMed] [Google Scholar]
- 35.Moncoq K, et al. The PIR domain of Grb14 is an intrinsically unstructured protein: implication in insulin signaling. FEBS Lett. 2003;554:240–6. doi: 10.1016/s0014-5793(03)01095-0. [DOI] [PubMed] [Google Scholar]
- 36.Depetris RS, et al. Structural basis for inhibition of the insulin receptor by the adaptor protein Grb14. Mol Cell. 2005;20:325–33. doi: 10.1016/j.molcel.2005.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bereziat V, et al. Inhibition of insulin receptor catalytic activity by the molecular adapter Grb14. J Biol Chem. 2002;277:4845–52. doi: 10.1074/jbc.M106574200. [DOI] [PubMed] [Google Scholar]
- 38.Lei M, et al. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–97. doi: 10.1016/s0092-8674(00)00043-x. [DOI] [PubMed] [Google Scholar]
- 39.James C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–8. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 40.Chen E, Staudt LM, Green AR. Janus kinase deregulation in leukemia and lymphoma. Immunity. 2012;36:529–41. doi: 10.1016/j.immuni.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tefferi A, Pardanani A. JAK inhibitors in myeloproliferative neoplasms: rationale, current data and perspective. Blood Rev. 2011;25:229–37. doi: 10.1016/j.blre.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–3. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Babon JJ, et al. Secondary structure assignment of mouse SOCS3 by NMR defines the domain boundaries and identifies an unstructured insertion in the SH2 domain. Febs Journal. 2005;272:6120–6130. doi: 10.1111/j.1742-4658.2005.05010.x. [DOI] [PubMed] [Google Scholar]
- 44.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–44. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 49.Griffin MD, et al. Characterisation of the first enzymes committed to lysine biosynthesis in Arabidopsis thaliana. Plos One. 2012;7:e40318. doi: 10.1371/journal.pone.0040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1 -towards automated and web-supported small-angle scattering data analysis. Journal of Applied Crystallography. 2007;40:S223–S228. [Google Scholar]
- 51.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of Applied Crystallography. 2003;36:1277–1282. [Google Scholar]
- 52.Svergun DI. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. Journal of Applied Crystallography. 1992;25:495–503. [Google Scholar]
- 53.Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. Journal of Applied Crystallography. 1995;28:768–773. [Google Scholar]
- 54.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. Journal of Applied Crystallography. 2001;34:33–41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.