Abstract
Somatic mutations in the KEAP1 ubiquitin ligase or its substrate NRF2 (NFE2L2) commonly occur in human cancer, resulting in constitutive NRF2-mediated transcription of cytoprotective genes. However, many tumors display high NRF2 activity in the absence of mutation, supporting the hypothesis that alternative mechanisms of pathway activation exist. Previously, we and others discovered that via a competitive binding mechanism, the proteins WTX (AMER1), PALB2 and SQSTM1 bind KEAP1 to activate NRF2. Proteomic analysis of the KEAP1 protein interaction network revealed a significant enrichment of associated proteins containing an ETGE amino acid motif, which matches the KEAP1 interaction motif found in NRF2. Like WTX, PALB2, and SQSTM1, we found that the dipeptidyl peptidase 3 (DPP3) protein binds KEAP1 via an ‘ETGE’ motif to displace NRF2, thus inhibiting NRF2 ubiquitination and driving NRF2-dependent transcription. Comparing the spectrum of KEAP1 interacting proteins with the genomic profile of 178 squamous cell lung carcinomas characterized by The Cancer Genome Atlas revealed amplification and mRNA over-expression of the DPP3 gene in tumors with high NRF2 activity but lacking NRF2 stabilizing mutations. We further show that tumor-derived mutations in KEAP1 are hypomorphic with respect to NRF2 inhibition and that DPP3 over-expression in the presence of these mutants further promotes NRF2 activation. Collectively, our findings further support the competition model of NRF2 activation and suggest that ‘ETGE’-containing proteins like DPP3 contribute to NRF2 activity in cancer.
INTRODUCTION
Constitutive activation of the NF-E2-related factor 2 (NRF2) cap-n-collar transcription factor is emerging as a prominent molecular feature of many tumors. When active, NRF2 controls the expression of ~200 genes that collectively function to maintain a healthy intracellular reduction-oxidation (redox) balance, clear electrophilic xenobiotics, and degrade damaged and misfolded proteins (1, 2). The leading hypothesis posits that whereas short-term NRF2 activation antagonizes oncogenesis by curtailing oxidative damage, constitutive activation promotes the survival of metabolically stressed cancer cells, as well as cancer cells under chemotherapeutic insult. Indeed, depletion of NRF2 from cancer-derived cell lines results in apoptosis and increased sensitivity to chemotherapeutic agents (3). In human non-small cell lung cancer, tumors showing high levels of NRF2 protein are associated with a poor outcome and increased resistance to therapy (4–6).
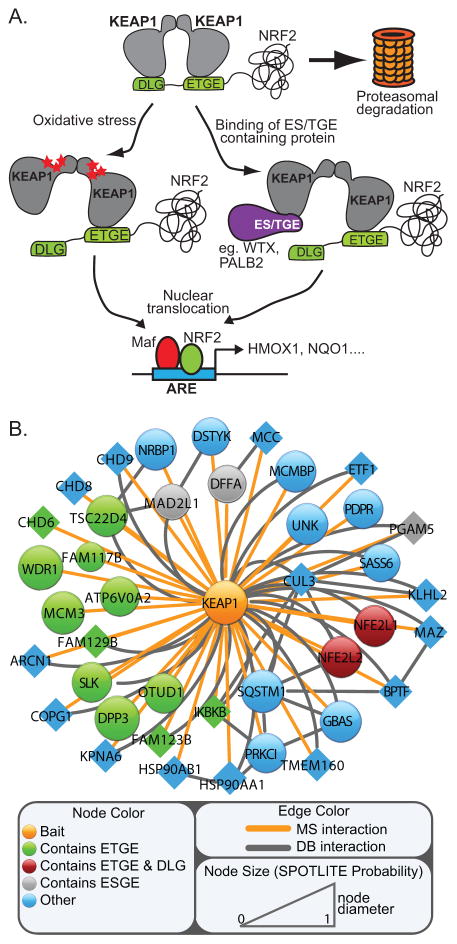
At basal state, NRF2 protein level and activity is maintained at low levels through ubiquitin-dependent proteosomal degradation (7–9). The mechanics of this ubiquitination, which is conceptualized in the ‘hinge-and-latch’ model, involves a homodimeric E3 ubiquitin ligase complex comprising the KEAP1 substrate recognition module and a cullin-3 scaffold (10, 11) (Figure 1A). An amino-terminal DLG and ETGE motif within NRF2 independently binds two KEAP1 monomers within the complex, yielding a 2:1 stoichiometry of KEAP1:NRF2. The intermolecular protein dynamics governing ubiquitination of NRF2 relies on the differential affinities between the ETGE and DLG motifs for KEAP1; the ETGE motif binds KEAP1 with approximately 100-fold greater affinity than the DLG (10). In response to oxidative stress, modification of reactive cysteines within KEAP1 induces a conformational change within the homodimer. This architectural re-structuring releases the low affinity DLG motif from KEAP1, thus re-positioning NRF2 in a conformation unfavorable for ubiquitination (10–13).
Figure 1. The KEAP1 protein interaction network is enriched for ETGE-containing proteins.
(A) Cartoon schematic of NRF2 ubiquitination by KEAP1. KEAP1 inactivation is shown through cysteine modification and the competitive association of ETGE-containing proteins. (B) Schematic representation of the KEAP1 protein interaction network as defined by affinity purification and mass spectrometry. Nodes and edges were sized and colored according to probabilistic scoring approach, sequence and source of data. Circular nodes were sized according to SPOTLITE probability (10% FDR). Triangular nodes represent borderline SPOTLITE scored interactions that were observed across multiple APMS runs. See also Table S1.
Recent cancer genomic studies reported somatic mutation of NRF2 or KEAP1 in 34% of squamous cell lung carcinoma and 12% of lung adenocarcinoma (5, 14). Consistent with the direct inhibition of NRF2 by KEAP1, mutations striking both KEAP1 and NRF2 within the same tumor are typically not observed (15). Moreover, whereas activating mutations within NRF2 almost invariably target the DLG or ETGE motifs, mutations within KEAP1 span the entire length of the protein (15, 16). Genomic alterations in KEAP1 or NRF2 have also been reported in a variety of other cancers, including gastric carcinoma, colorectal carcinoma, hepatocellular carcinoma, and ovarian cancer (17–21). In addition to mutation, hypermethylation of the KEAP1 promoter and NRF2 copy number amplifications promote NRF2 activity in lung, colon and prostate cancer (21–23).
Although we have an understanding of how oxidative stress and genetic mutation activate NRF2 signaling, the identity and function of proteins that physically interact with KEAP1 and NRF2 has been comparatively understudied. A growing body of evidence suggests that cancer-associated increases in NRF2 transcript and protein can occur in the absence of genomic alteration (15, 17, 24), underscoring the importance of identifying the full complement of regulatory mechanisms governing NRF2 activity. We recently reported that the WTX tumor suppressor protein physically binds KEAP1 to competitively inhibit NRF2 ubiquitination (25). Similarly, p62/SQSTM1, PALB2, and p21 bind KEAP1 or NRF2 to sterically inhibit NRF2 ubiquitination (26–28). For WTX and PALB2, the association with KEAP1 is achieved through an ETGE motif, which mimics the NRF2 binding interface. As expected, these proteins activate NRF2-mediated transcription in the absence of oxidative stress, through ETGE-dependent competition with NRF2 for KEAP1 binding. Here, we sought to comprehensively define all ETGE or ESGE containing proteins within the KEAP1 protein interaction network, determine whether they functionally control NRF2 and evaluate their expression within human tumors, particularly in relation to NRF2 activity.
METHODS
Tissue Culture, Transfections, and Small Interfering RNAs
HEK293T and H2228 cells were obtained from the American Tissue and Culture Collection, which authenticates cells line using short tandem repeat analysis. Cell lines were not passaged for more than 6 months after resuscitation. HEK293T cells were grown in DMEM supplemented with 10% FBS and 1% GlutaMAX (Life Technologies) in a 37°C humidified incubator with 5% CO2. H2228 cells were grown in RPMI supplemented with 10% FBS. KEAP1 −/− mouse embryo fibroblasts (MEFs) were cultured in IMDM supplemented with 10% FBS. The KEAP1 −/− MEFs were kindly provided by Thomas Kensler and Nobunao Wakabayshi. Expression constructs were transfected in HEK293T cells with Lipofectamine 2000 (Life Technologies) and KEAP1 −/− MEFs as with Fugene HD (Roche). Transfection of siRNA was performed with Lipofectamine RNAiMAX (Life Technologies). siRNA sequences are provided in Supplemental Methods.
Affinity Pulldowns and Western Blotting
For Streptavidin and FLAG affinity purification, cells were lysed in 0.1% NP-40 lysis buffer. Cell lysates were cleared by centrifugation and incubated with streptavidin resin (GE Healthcare) or FLAG resin (Sigma) before washing with lysis buffer and eluting with NuPAGE loading buffer (Life Technologies). For siRNA, HEK293T cells were transiently transfected and lysed in RIPA buffer 60 h post-transfection. All antibodies and buffers used for Western analysis are listed in Supplemental Methods.
Plasmids, Expression Vectors, and Site-directed Mutagenesis
Expression constructs in the SBPHA backbone were generated with standard PCR techniques. Constructs for DPP3 and DPP3Y318F were a generous gift from Maja Abramić. The reporter gene fusion construct for human hNQO1-ARE-luciferase was a kind gift from Jeffrey Johnson. The SLK-HA construct was a generous gift from Dr. Andrey Cybulsky. Expression constructs for ETGE-containing proteins were obtained from Open Biosystems and cloned into a custom lentiviral vector (pHAGE-CMV-FLAG-DEST). ETGE deletion mutants were generated by PCR-based mutagenesis and sequence verified prior to use (GENEWIZ).
ARE-luciferase Quantification
For DNA, cells were transfected with expression constructs, FLAG-KEAP1, FLAG-NRF2, hNQO1-ARE luciferase, and a control plasmid containing Renilla luciferase driven by a constitutive cytomegalovirus (CMV) promoter. Approximately 24 h post-transfection, NRF2-mediated transcription was measured as the ratio of Firefly to Renilla luciferase activity (Promega Dual-Luciferase Reporter Assay System). For siRNA, HEK293T cells stably expressing the ARE-luciferase and Renilla control reporters were transfected with siRNA. Approximately 60 h post-transfection, activation was measured. For the assay depicted in Figure 6E, treatment with 50 μM tBHQ was performed 48 h post-transfection, and activation was measured 60 h post-transfection.
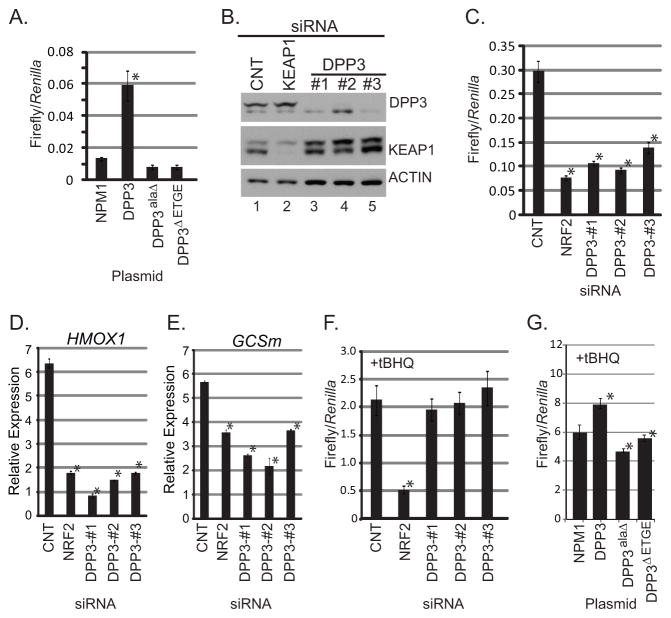
Figure 6. DPP3 is an activator of NRF2-mediated transcription.
(A) HEK293T cells were transfected with the indicated plasmid along with constitutively expressed Renilla luciferase and the NQO-1 promoter driving Firefly luciferase (NQO1-ARE). Cells were lysed and luciferase values were normalized to Renilla. Error bars represent standard deviation from the mean over 3 biological replicates, * p<0.05; Students T-test as compared to NPM1. (B) HEK293T cells were transfected with 10nM of the indicated siRNA. Protein lysate was analyzed by Western blot for the indicated endogenous protein. (C) HEK293T cells stably expressing the ARE reporter and Renilla luciferase were transfected with the indicated siRNA. Error bars represent standard deviation from the mean from 3 biological replicates. * p<0.05; Students T-test as compared to CNT. (D and E) HEK293T cells were transfected with siRNAs against the indicated mRNAs before mRNA isolation and qPCR for the indicated endogenous target genes. Relative expression was calculated based on expression of endogenous target gene transcript normalized to GAPDH. Error bars represent standard deviation from the mean from 3 biological replicates. * p<0.05; Students T-test as compared to CNT. (F and G) HEK293T cells were transfected with the indicated siRNAs or plasmids. Cells were treated with 50μM tBHQ 18 h priors to lysis. Error bars represent standard deviation from the mean from 3 biological replicates. * p<0.05; Students T-test.
Cell-based NRF2 ubiquitination experiments
HEK293T stably expressing SBPHA-KEAP1 cells were transfected with VSV-UB1, FLAG-NRF2, and SBPHA-DPP3. Venus-NPM1 was used such that each condition received the same mass of DNA. Cells were lysed in 0.1% NP-40 lysis buffer.
RNA Isolation, Reverse Transcription, and Semi-quantitative Real Time-PCR
Total RNA from cells was harvested in TRIzol (Life Technologies) reagent according to the manufacturer’s instructions. RNA was quantified by UV spectrophotometry, and cDNA was created using the RevertAid First Strand cDNA synthesis Kit (Fermentas). PCR was performed in triplicate with 30 cycles of amplification with 1 s denaturation at 95 °C and 5 s annealing at 60 °C, on an ABI 7900HT Fast Realtime PCR machine. Quantitative light cycler PCR primers are listed in the Supplemental Methods.
Crystallographic Modeling
The coordinates for the KEAP1-NRF2 peptide complex and DPP3 were downloaded from the RCSB Protein Data Bank (PDB IDs 1X2R and 3FVY, respectively). The superposition of the ETGE motifs of NRF2 and DPP3 was done in PyMOL (The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC). PyMOL was used to prepare the images used in Figure 4D–G.
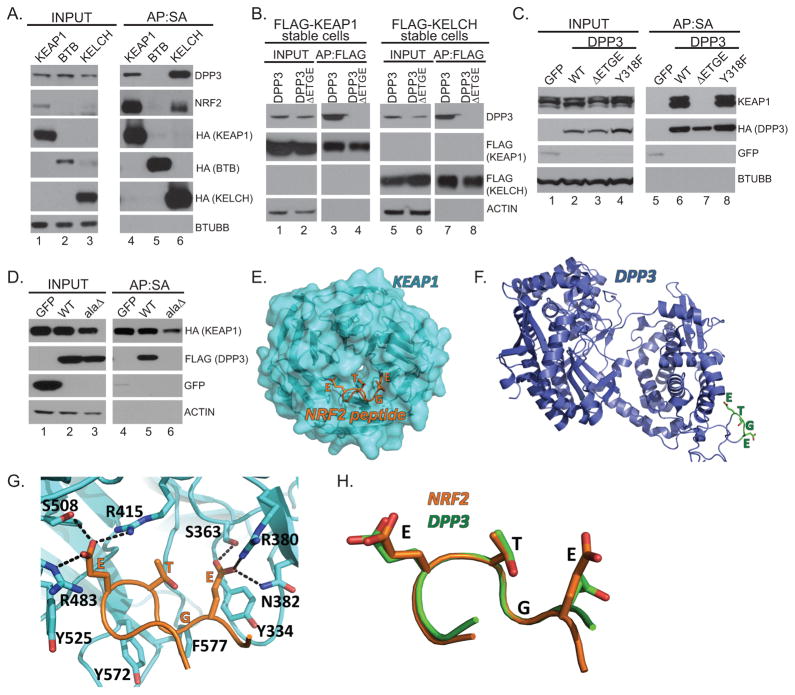
Figure 4. DPP3 interacts with the KELCH domain of KEAP1 via its ETGE motif.
(A) Protein complexes from HEK293T cells stably expressing SBPHA-KEAP1, SBPHA-BTB, and SPBHA-KELCH were Streptavidin affinity purified and analyzed by Western blot. (B) Cells stably expressing FLAG-KEAP1 or the FLAG-KELCH domains of KEAP1 were transfected with the indicated SBPHA-DPP3 construct before affinity purification and Western blot. (C) Protein complexes from HEK293T cells stably expressing the indicated fusion protein were Streptavidin affinity purified and analyzed by Western blot. (D) HEK293T cells were transiently co-transfected with FLAG-GFP, FLAG-DPP3-WT, or FLAG-DPP3-AAGE (alaΔ) before FLAG-affinity purification and Western blot. (E) The KELCH domain of Keap1 (PDB ID 1X2R) adopts a six-bladed β-propeller structure (cyan). The ETGE motif of NRF2 (orange) binds near the central pore of the β-propeller. (F) The structure of human DPP3 (PDB ID 3FVY, blue) reveals an ETGE motif (residues 480–483, green) in an unstructured surface loop. (G) KEAP1 binding to the ETGE peptide (orange sticks) is stabilized by both hydrogen bonds (to serine and asparagine residues, cyan sticks) and electrostatic interactions (to arginine residues, cyan sticks) with KEAP1. (H) Superposition of the NRF2 ETGE motif bound to KEAP1 with the ETGE motif of DPP3 reveals similar conformations.
Affinity Purification and Mass Spectrometry
For Streptavidin and FLAG affinity purification, cells were lysed in 0.1% NP-40 lysis. Cell lysates were incubated with Streptavidin or FLAG resin and washed 5X with lysis buffer. The precipitated proteins were trypsinized directly off beads using the FASP Protein Digestion Kit (Protein Discovery). For tandem purification of the FLAG-KEAP1 and SBPHA-DPP3 complex (Figure 3B), protein complexes were eluted after the first affinity purification with either 150 μg/μl FLAG peptide or 50 mM biotin.
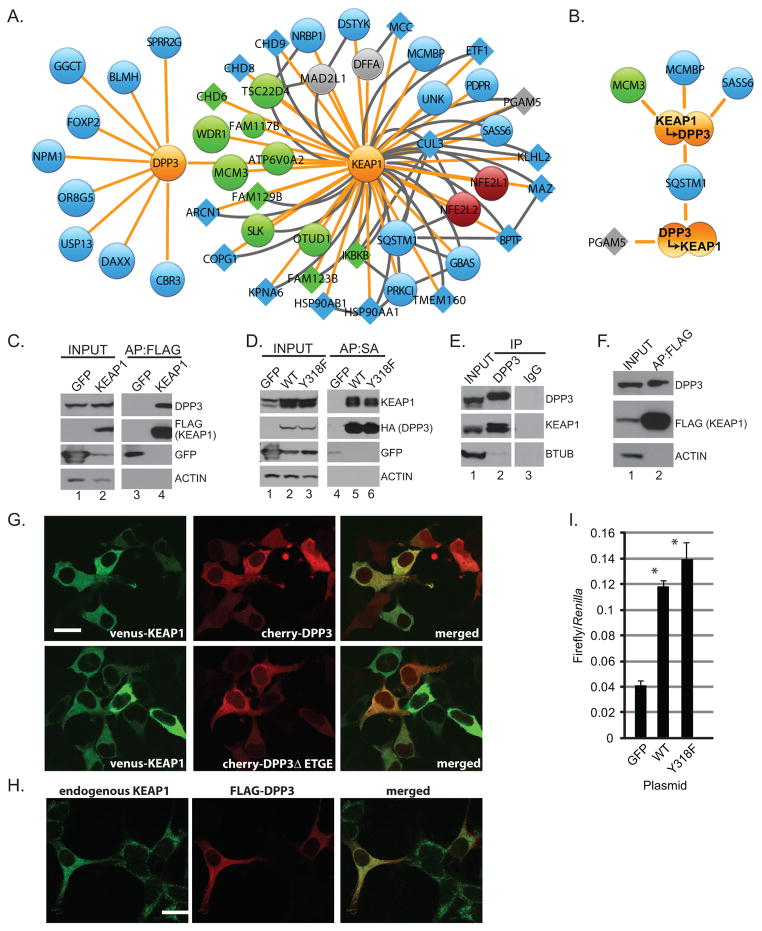
Figure 3. DPP3 is a KEAP1 interacting protein.
(A) Schematic protein interaction network for DPP3 and KEAP1. Node and edge coloring and sizing are consistent with Figure 1. (B) HEK293T cells stably expressing KEAP1 and DPP3 were lysed and subjected to two sequential rounds of affinity purification before mass spectrometry. Data shown represent biological duplicate experiments, wherein the order of affinity purifications was reversed. (C) Protein complexes from HEK293T cells stably expressing FLAG-KEAP1 were affinity purified and analyzed by Western blot. (D) Protein complexes from HEK293T cells stably expressing SBPHA-DPP3 or SBPHA-DPP3-Y318F were streptavidin affinity purified and analyzed by Western blot. (E) Endogenous DPP3 from HEK293T cells was immunopurified and analyzed by Western blot for the indicated endogenous proteins. (F) Protein complexes were FLAG affinity purified from the lung adenocarcinoma cell line H2228 stably expressing FLAG-KEAP1 and analyzed by Western blot. (G) HEK293 cells were transfected with venus-KEAP1 and the indicated mCherry-fused DPP3 expression construct. (H) HEK293T cells transfected with FLAG-DPP3 and stained for FLAG and endogenous KEAP1. Scale =20 μm. (I) HEK293T cells were transfected with NQO1-ARE-luciferase, constitutively active Renilla luciferase and the indicated expression plasmid before lysis and luciferase quantification (* P<0.05 across three biological replicate experiments).
Protein Identification, Filtering and Bioinformatics
Detailed methods for the mass spectrometry and peptide identification are provided in Supplemental Methods. Filtering of false interactions from non-tandem, wildtype experiments was accomplished using SPOTLITE, with an internal lab dataset of 158 Streptavidin experiments on 60 different baits, and using a 10% FDR for the entire dataset. FLAG-based APMS data were not scored with SPOTLITE because our FLAG-specific reference dataset is prohibitively small. Proteins identified in tandem or mutant experiments were accepted if they passed the SPOTLITE filtering on the non-tandem, wildtype experiments. Unfiltered data and associated SPOTLITE results are provided as Table S1.
Motif Analysis
Identification of enriched 4-mer amino acid sequences was performed using a 1-tail Fisher’s exact test (Table S2). We individually tested each of the 13265 4-mer sequences present among the KEAP1 interactors, taking into account the number of interacting proteins, interacting proteins having the motif, total proteins in the UniProtKB/SwissProt database, and the total number of proteins having the motif within UniProtKB/SwissProt. Bonferroni correction was applied due to multiple hypothesis testing.
Immunostaining
For subcellular localization of exogenously expressed proteins, cells were co-transfected with the indicated plasmids and plated on 10ug/ml fibronectin-coated coverslips. Cells were fixed in 4% paraformaldehyde in cytoskeletal buffer for 15 minutes, and coverslips were mounted to slides using the Prolong Gold antifade reagent (Molecular Probes). Images were acquired using a Zeiss LSM5 Pascal Confocal Laser Scanning Microscope equipped with a 63X/1.42 Oil PlanApo objective lenses. Localization of endogenous KEAP1 was determined by immunostaining cells as described above, except: 1) cells were fixed in 4% PFA in cytoskeletal buffer for 15 minutes and permeabilized with 0.1% Triton in PBS for 5 minutes, 2) after blocking in 1% BSA/PBS for 1h, cells were double stained for KEAP1 (Proteintech) and flag (Sigma) at 4°C, overnight, followed by incubation with FITC-conjugated-donkey anti-rabbit IgG and RRX-conjugated-donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories) at room temperature for 2h and 3), images were acquired using a Zeiss LSM710 Spectral Confocal Laser Scanning Microscope.
RESULTS
Proteomic analysis of the KEAP1 protein interaction network
We defined the KEAP1 protein interaction network by affinity purification and shotgun mass spectrometry (APMS) (Figure 1B). In total, the KEAP1 complex was analyzed 13 times, where variations in affinity purification, detergent solubilization and cell treatment helped to maximize comprehensive network mapping (Table S1). True interactions were identified from false positives using SPOTLITE, a novel probabilistic scoring algorithm that couples direct and indirect data to identify false positive interactions within APMS data (Figure 1B and Table S1) (manuscript under review). Of 42 high confidence KEAP1-interacting proteins identified, 17 contain an ETGE, ESGE or both. To determine if this motif is enriched within the KEAP1 protein interaction network (PIN) beyond chance observation, we performed a Fisher’s exact test. The ETGE motif was identified as the only significant 4 amino acid sequence within the KEAP1 PIN (Table S2). Together, these data support and expand the ‘ETGE’ competition model of KEAP1 regulation.
The ETGE motif is required for binding to KEAP1
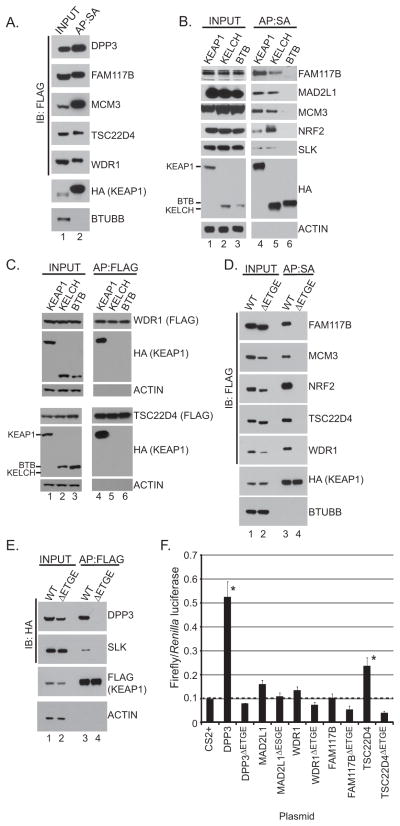
We selected eight ETGE-containing proteins and validated their association with KEAP1 (Figure 2A–B). Western blot analysis of affinity purified KEAP1 protein complexes revealed the presence of endogenously expressed DPP3, FAM117B, MCM3, SLK and MAD2L1 (Figure 2B). Expression of exogenous TSC22D4 and WDR1 also showed interaction with KEAP1 (Figure 2C). To map the domain within KEAP1 responsible for binding the ETGE proteins, full-length KEAP1, the KEAP1 KELCH domain, or the KEAP1 BTB domain were purified and endogenous associated proteins were detected by Western blot. With the exception of WDR1 and TSC22D4 which bound only full length KEAP1, DPP3, FAM117B, MCM3, SLK and MAD2L1 bound the KELCH domain of KEAP1 (Figure 2B–C). To directly evaluate a role for the ETGE motif in binding KEAP1, we generated ETGE-deletion mutants for FAM117B, MCM3, TSC22D4, WDR1, DPP3 and SLK. Like WTX and PALB2, deletion of the ETGE (ΔETGE) motif within these proteins abrogated KEAP1 binding (Figure 2D and E). Finally, functional impact of the ETGE-containing proteins on NRF2-mediated transcription was evaluated. Of the proteins tested, DPP3 and TSC22D4 strongly activated NRF2-mediated transcription in an ETGE-dependent manner, the former of which was previously identified as an activator of NRF2-dependent transcription in a gain-of-function screen (29) (Figure 2F). Over-expression of SLK also activated NRF2-mediated transcription, although this activation was independent of the ETGE motif (Figure S1).
Figure 2. The ETGE motif is required for KEAP1 association.
(A) Streptavidin affinity purified KEAP1 protein complexes from HEK293T cells were analyzed by Western blot for the indicated FLAG-tagged proteins (SBP, streptavidin binding peptide; HA, hemagglutinin). (B) Streptavidin affinity purified protein complexes from HEK293T cells stably expressing the indicated SBPHA-tagged proteins were analysed by Western blot for the indicated proteins. (C) HEK293T cells stably expressing FLAG-tagged WDR1 or TSC22D4 were transfected with SBPHA-tagged KEAP1, KEAP1 BTB, or KEAP1 KELCH before FLAG affinity purification and Western blot (D) FLAG-tagged wild-type or ET/SGE-deletion constructs were expressed in HEK293T cells stably expressing SBPHA-KEAP1 before streptavidin affinity purification and Western blot. (E) HEK293T cells stably expressing FLAG-tagged KEAP1 were transfected with the indicated expression constructs; associated proteins were revealed by FLAG immunoprecipitation and Western blot. (F) HEK293T cells were transfected with the indicated ET/SGE-containing protein, constitutively expressed Renilla luciferase and the NRF2-driven Firefly luciferase (NQO1-ARE). Error bars represent standard deviation from the mean from 3 biological replicates (* p<0.05; Students T-test).
DPP3 is a KEAP1 interacting protein
The protein dipeptidyl peptidase III (DPP3) had the greatest impact on NRF2-dependent transcription and was the most abundant protein within the KEAP1 PIN (Figure 2F and Table S1, respectively)(30). To further explore DPP3, we defined and compared the DPP3 PIN to the KEAP1 PIN (Figure 3A). With the exception of the observed interaction between KEAP1 and DPP3, the integrated PIN revealed no common interacting proteins. We also defined the PIN for DPP3ΔETGE; as expected KEAP1 was not observed (Table S1). To more rigorously characterize the KEAP1-DPP3 protein complex, we performed sequential affinity purifications for FLAG-KEAP1 and SBPHA-DPP3. Using HEK293T cells stably expressing both proteins, we purified FLAG-KEAP1 complexes and then from the resulting eluate, purified DPP3 with Streptavidin (Figure 3B, top). The reciprocal sequential purification was done and analyzed by APMS (Figure 3B, bottom). Despite observing over 1500 spectral counts representing each bait protein, the only protein identified in both APMS experiments was SQSTM1, represented by 15 and 1 total spectra, respectively (Figure 3B and Table S1). Together these data argue that the KEAP1-DPP3 complex is largely exclusive from other interacting proteins.
We next tested whether endogenously expressed DPP3 and KEAP1 associate. First, endogenous DPP3 was detected within FLAG-KEAP1 affinity purified protein complexes from HEK293T cells (Figure 3C). Second, endogenous KEAP1 affinity purified with SBPHA-DPP3 (Figure 3D). Finally, we detected KEAP1 within immunopurified endogenous DPP3 protein complexes (Figure 3E). In addition to these studies in HEK293T cells, DPP3 was also detected within KEAP1 protein complexes isolated from H2228 lung cancer cells (Figure 3F). As protein complex purification is subject to post-lysis interactions, we determined if DPP3 and KEAP1 co-localized within cells. Although discrete subcellular localizations were not observed, exogenously expressed DPP3 co-localized with both exogenous and endogenous KEAP1 protein; DPP3ΔETGE also co-localized with KEAP1 (Figure 3G and 3H). Comparing transfected versus untransfected cells, the expression of DPP3 did not affect KEAP1 subcellular localization (Figure 3G). Finally, we tested whether the catalytic activity of DPP3 affected its association with KEAP1. Stably expressed wild-type (WT) and the catalytically inactive (Y318F) mutant (31) of DPP3 bound endogenous KEAP1 (Figure 3D), indicating that the catalytic activity of DPP3 is not required for KEAP1 binding. Consistent with this, wildtype DPP3 or DPP3Y318F similarly activated NRF2-dependent transcription when over-expressed (Figure 3I).
The ETGE motif is required for DPP3 binding to KEAP1
Like WTX, PALB2, NRF2 and most of the ETGE-containing proteins evaluated, endogenous DPP3 associated with the KELCH domain of KEAP1 (Figure 4A). To validate that the ETGE motif is required for this binding, constructs encoding SBPHA-DPP3 or SBPHA-DPP3ΔETGE were transiently transfected into cells stably expressing FLAG-tagged full length KEAP1 or the KELCH domain. Wild-type DPP3 bound full length KEAP1 and the KELCH domain; however, DPP3ΔETGE was unable to bind KEAP1 or the KEAP1 KELCH domain (Figure 4B). Similarly, whereas endogenous KEAP1 failed to immunopurify with DPP3ΔETGE, it did co-purify with DPP3-WT and the catalytic mutant DPP3Y318F (Figure 4C). These data were confirmed by APMS of DPP3ΔETGE and DPP3Y318F (Table S1). Deletion of the ETGE motif within DPP3 may render the protein unstable and/or misfolded, which could account for lack of binding to KEAP1. To address this possibility, we tested the ability of a DPP3 alanine mutant to bind KEAP1. Like DPP3ΔETGE, alanine point mutations within the domain (ETGE AAGE) abolished KEAP1 binding (Figure 4D).
Crystallographic modeling revealed that NRF2 binds KEAP1 near the central pore of the KELCHβ-propeller (Figure 4E) (10, 13, 32). Using the crystal structure of DPP3, we asked if the ETGE motif within DPP3 and NRF2 adopt similar tertiary conformations. The ETGE motif in DPP3 lies on an unstructured loop on the surface of the protein (Figure 4F) (33). It is therefore in a sterically favorable position to bind to KEAP1, as opposed to being buried within the globular domains. Similar to the NRF2 peptide, three tyrosine residues and one phenylalanine residue defines the binding surface that accommodates the specific conformation of the ETGE peptide of DPP3 (Figure 4G). Strikingly, the ETGE motif of DPP3 and NRF2 adopt identical conformations when superimposed, suggesting that the ETGE motif of both proteins may interact with KEAP1 in a similar manner (Figure 4H) (root mean square deviation ≈ 0.05 Å between Cα atoms).
DPP3 competes with endogenous NRF2 for binding to KEAP1
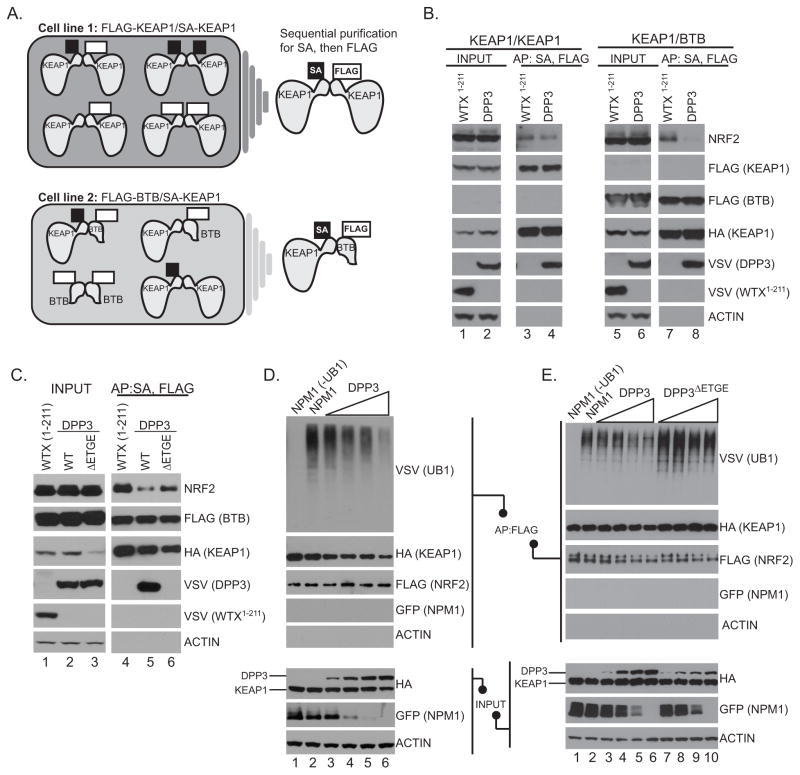
We tested whether DPP3 association with KEAP1 displaces NRF2. As homodimeric KEAP1 binds a single NRF2 molecule via two amino acid motifs (Figure 1A), competition experiments required the isolation of monomeric KEAP1. We created two double stable cell lines: the first expressed both SBPHA-KEAP1 and FLAG-KEAP1, and the second expressed SBPHA-KEAP1 and the FLAG-tagged BTB domain of KEAP1. Sequential affinity purification with streptavidin and FLAG resins purified KEAP1 homodimer or a KEAP1-BTB “pseudo-monomer”, allowing us to test whether DPP3 competes with NRF2 for KEAP1 binding (Figure 5A). Compared to a truncated form of WTX that does not interact with KEAP1 (25), DPP3 over-expression resulted in reduced NRF2 binding to the pseudo-monomer KEAP1 but not to the KEAP1 homodimer (Figure 5B; compare lanes 3 and 4 to lanes 7 and 8). In contrast, when DPP3ΔETGE was introduced into each double-stable cell line, NRF2 binding to both the KEAP1-KEAP1 homodimer and KEAP1-BTB heterodimer was maintained (Figure 5C; compare lane 5 to 6). These findings suggest that DPP3 competes with NRF2 for binding to KEAP1 in an ETGE-dependent manner.
Figure 5. DPP3 competes with NRF2 for KEAP1 binding.
(A) Schematic representation of the sequential affinity purification approach employed to purify a KEAP1-KEAP1 homodimer or KEAP1-BTB domain pseudo-monomer. (B) HEK293T cells stably expressing FLAG-KEAP1 and SBPHA-KEAP1 or FLAG-BTB and SBPHA- KEAP1 were transfected with VSV-DPP3-WT or VSV-WTX1–211 truncation mutant before sequential streptavidin and FLAG affinity purification and Western blot. (C) HEK293T cells stably expressing FLAG-BTB and SBPHA-KEAP1 were co-transfected with VSV-WTX(1–211), VSV-DPP3 or VSV-DPP3ΔETGE. Protein complexes were affinity purified with Streptavidin then FLAG and analyzed by Western blot. (D and E) HEK293T cells stably expressing SBPHA-KEAP1 were co-transfected with VSV-UB1, FLAG-NRF2, and either SBPHA-DPP3-WT, SBPHA-DPP3ΔETGE or negative control Venus-NPM1. NRF2 was FLAG affinity purified and analyzed by Western blot.
The ‘hinge-and-latch’ model predicts that loss of binding of the NRF2 DLG motif to KEAP1 results in a reduction of NRF2 ubiquitination and subsequent degradation. Given that DPP3 competes for binding to KEAP1 via the ETGE motif (Figure 5A–C), we tested if DPP3 over-expression reduced NRF2 ubiquitination in an ETGE-dependent manner. Affinity purification of exogenous NRF2 followed by Western blot analyses revealed relative levels of NRF2 ubiquitination. As the amount of wild-type DPP3 increased, ubiquitination of NRF2 decreased, as compared to control (Figure 5D; compare lanes 2 and 6). Consistent with its inability to bind KEAP1, DPP3ΔETGE did not reduce NRF2 ubiquitination (Figure 5E). These data suggest that over-expression of DPP3 alters the architecture of NRF2 bound to dimeric KEAP1, and ultimately decreases NRF2 ubiquitination.
DPP3 activates NRF2 signaling in an ETGE-dependent fashion
To establish functional significance of DPP3 as a regulator of NRF2 activity, we determined whether DPP3 gain-of-function and loss-of-function impacted NRF2 transcriptional activity. Over-expression of DPP3, but not DPP3ΔETGE or DPP3alaΔ significantly induced NRF2-dependent expression of an antioxidant responsive firefly luciferase reporter (Figure 6A). For loss-of-function, we designed and tested the silencing efficacy of three non-overlapping siRNAs targeting DPP3 (Figure 6B). siRNA-mediated silencing of DPP3 suppressed NRF2-mediated transcription of the ARE reporter, similar to that of NRF2 silencing (Figure 6C). To validate this phenotype using endogenous NRF2 readouts, qPCR was employed to monitor the expression of two well-established NRF2 target genes: heme oxgenase-1 (HMOX1) and glutamate-cysteine ligase modifier (GCSm). In agreement with the reporter data, DPP3 siRNAs reduced HMOX1 and GCSm transcript levels similar to that of NRF2 silencing (Figure 6D). Finally, the model predicts that DPP3 gain-of-function or loss-of-function would not affect NRF2 activity after treatment with a pathway agonist, as NRF2 would already be in a sterically unfavorable conformation for KEAP1-mediated ubiquitination and degradation (Figure 1A). Consistent with this hypothesis, neither DPP3 siRNAs (Figure 6F) nor over-expression of DPP3 (Figure 6G) were able to suppress NRF2-mediated transcription after treatment with the small molecule, tert-butylhydroquinone (tBHQ).
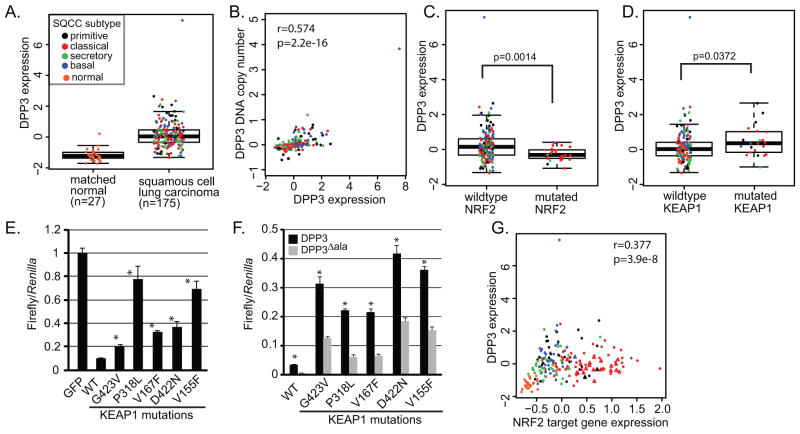
DPP3 expression and DNA copy number positively correlates with NRF2 activity in squamous cell lung cancer
To establish physiological significance for DPP3 in controlling KEAP1-NRF2 signaling, we evaluated DPP3 mRNA abundance and gene alterations in squamous cell lung carcinoma, using data from the TCGA consortium (15). First, we found that DPP3 mRNA expression is increased in lung SQCC as compared to matched normal tissue (Figure 7A). Of the four established lung SQCC subtypes, DPP3 expression is highest in primitive-type tumors (Figure 7A and S2)(34). Second, DPP3 genomic copy number and mRNA expression positively correlated, suggesting that DPP3 gene amplification may drive DPP3 over-expression in lung SQCC (Figure 7B). Third, when segregated by genotype, DPP3 mRNA levels were higher in NRF2 wildtype lung SQCC as compared to tumors with mutated NRF2, which is consistent the proposed DPP3-competition model (Figure 7C). Surprisingly, DPP3 mRNA abundance was found to be increased in KEAP1 mutant tumors as compared to KEAP1 wildtype tumors (Figure 7D). This co-occurrence might be explained if the mutations in KEAP1 are hypomorphic, resulting in a partially compromised ability to suppress NRF2. If so, the presence of DPP3 may further drive NRF2 activity. To test this hypothesis, we cloned and expressed five distinct KEAP1 mutations from the TCGA lung SQCC dataset and evaluated their impact on NRF2 function. In both HEK293T cells and KEAP1−/− mouse embryo fibroblasts, all five KEAP1 mutants displayed reduced but not absent activity in suppressing NRF2, thus supporting the notion that these somatic mutations are hypomorphic (Figure 7E and Figure S3). Impressively, over-expression of DPP3 further activated NRF2 in the presence of all five KEAP1 hypomorphs (Figure 7F). Importantly, the KEAP1 somatic mutants analyzed maintained association with both DPP3 and NRF2 (Figure S3). These data support a model wherein somatic mutation of KEAP1 partially impairs its ability to suppress NRF2, and the presence of ‘ETGE’-containing proteins like DPP3 may further drive pathway activity in KEAP1 mutant tumors.
Figure 7. DPP3 expression positively associates with NRF2 activity in squamous cell lung cancer.
(A) DPP3 mRNA abundance in 175 squamous cell lung carcinomas and 27 matched normal tissues (p=4.601e-14; Kruskal-Wallis test). Normal or lung SQCC subtype is indicated by color. With respect to tumor subtype, DPP3 is over-expression is enriched within the primitive subtype (p=0.03334; Kruskal-Wallis test; see also Figure S2). (B) DPP3 mRNA expression positively correlates with DPP3 genomic copy number (Spearman rank correlation). (C) Correlation of DPP3 mRNA expression with NRF2 mutational status (p=0.00141; Kruskal-Wallis test). (D) Correlation of DPP3 mRNA expression with KEAP1 mutational status (p=0.03718; Kruskal-Wallis test). (E and F) HEK293T cells were transiently transfected with NQO1-ARE-luciferase reporter, constitutively active Renilla reporter and the indicated expression plasmids (* P<0.05 across at least three biological triplicate experiments) (G) DPP3 mRNA expression positively associates with NRF2 target gene expression (Spearman rank correlation test). The NRF2 gene signature consists of 15 genes (3). Triangles represent tumors with NRF2 mutations. (See also Figure S2 and S3).
Finally, we tested whether DPP3 expression associated with NRF2 transcriptional activity across the lung SQCC cohort, as defined by the expression of a gene set signature consisting of 15 NRF2 target genes (3). DPP3 expression and the NRF2 signature score strongly associated (Figure 7H). Together, these data suggest that through competitive binding to KEAP1, DPP3 genomic amplification and over-expression may promote NRF2 activity in squamous cell carcinoma of the lung.
DISCUSSION
Aberrant KEAP1/NRF2 signaling has emerged as a critical regulatory pathway in a multitude of disease pathologies, most notably cancer. Although substantial progress has been made to define how reactive cysteines within KEAP1 govern its ability to ubiquitinate NRF2, the role of proteins peripheral to the KEAP1/NRF2/CUL3 core complex has only just begun to be explored. Recent studies have revealed four proteins that bind KEAP1 or NRF2 and ultimately inhibit NRF2 ubiquitination. Of these, WTX and PALB2 employ an ETGE motif to directly bind KEAP1, thus displacing and stabilizing NRF2. Given these discoveries and the likelihood that KEAP1-associated proteins contribute to NRF2 perturbation in human disease, particularly when genomic alterations within KEAP1 and NRF2 are lacking, we sought to establish the ETGE motif as a defining characteristic in KEAP1 associated proteins that functionally control NRF2 stability.
We found that the ETGE motif defines the most frequently observed four amino acid sequence within the KEAP1 protein interaction network (Table S2; Fisher’s exact test, 1-tail, p=5.8e-13). Of 13 ETGE containing proteins identified, we tested 7 and found that all required the ETGE motif to bind KEAP1 (Figure 2E). Aside from this ETGE-dependent KEAP1 binding however, we noted very few similarities. For example, with the exception of WTX, DPP3, and PALB2, none of the proteins have been previously reported to contribute or respond to oxidative stress. Additionally, functional annotations for identified proteins are surprisingly diversified: DNA replication and licensing (MCM3, MCMBP), cytoskeletal dynamics (SLK, WDR1), transcription (TSC22D4), and apoptosis (SLK) (35–45). Within the context of our data, these observations suggest that each ETGE protein may function to control KEAP1 activity or be controlled by KEAP1 in a context-dependent fashion.
Because it robustly activates NRF2-mediated transcription (Figure 6A), binds KEAP1 with near exclusivity (Figure 3A and B) and has established catalytic activity, we chose to focus our mechanistic studies on DPP3. Although DPP3 possesses exopeptidase activity in vitro (33, 46), we were unable to reveal a role for its catalytic activity in either contributing to the KEAP1 interaction (Figure 3D and 4C) or regulating NRF2-mediated transcription (Figure 3I). That said, our studies did not examine the temporal effects of DPP3 expression on NRF2 activity, but rather assessed pathway activity at steady-state. Focused studies are needed to determine whether DPP3 catalytic activity functions to control KEAP1 and NRF2 dynamics, as well as pathway activity in vivo. Given the pressing need of identifying new drug targets within the KEAP1-NRF2 pathway, the possibility of targeting DPP3 catalytic function to control KEAP1-NRF2 remains an important opportunity.
The ETGE motif of DPP3 resides in a flexible loop on the surface of the protein and adopts a similar conformation to the NRF2 ETGE peptide when bound to KEAP1 (Figure 4F and G). Loss of this motif, either through deletion or point mutation, abrogates the KEAP1-DPP3 protein interaction (Figure 4), perturbs the interaction between NRF2 and KEAP1 (Figure 5), as well as alters NRF2 ubiquitination (Figure 5). We interpret these data to support a model wherein DPP3 competes the low-affinity DLG motif of NRF2 off of the KEAP1 KELCH domain, resulting in a complex of KEAP1, DPP3, and NRF2. Additional experiments are needed to determine protein stoichiometry within complexes containing KEAP1, NRF2, and DPP3. These experiments will reveal whether competitors like DPP3 specifically compete off the DLG of NRF2, as opposed to the ETGE motif; based on the relative affinities of the DLG and ETGE motifs, competition with the DLG motif of NRF2 is most probable (10, 11).
In cancer, over-expression of an ETGE-containing protein may promote NRF2 activity in the absence of inactivating KEAP1 mutations or activating NRF2 mutations. Indeed, high NRF2 activity in tumors lacking KEAP1 or NRF2 mutation has been reported in ovarian cancer, sarcoma and squamous cell lung cancer (5, 17, 47). After defining the ETGE-containing proteins within the KEAP1 protein interaction network (Figure 1), we surveyed their expression and DNA copy number across tumor samples taken from the TCGA SQCC lung cohort. DPP3 demonstrated copy number gains and mRNA over-expression, and importantly both of which positively correlated with NRF2 activity (Figure 7). Whether genomic amplification of DPP3 constitutes a ‘driver’ event in cancer remains an important question for future research. Interestingly, two studies have demonstrated DPP3 over-expression in ovarian cancer. Given our data in lung SQCC, DPP3 expression may similarly be driven by genomic amplification in ovarian carcinoma, possibly functioning as a NRF2 agonist (48, 49).
Our cancer genomic analyses and functional annotation of cancer-derived mutations in KEAP1 suggest that some KEAP1 somatic mutations are hypomorphic, resulting in partial NRF2 activation. This contrasts NRF2 mutation, which we believe yields a maximally activated pathway, one insensitive to KEAP1 modifiers such as DPP3. This hypothesis is supported by the following: 1) DPP3 genomic amplification and mRNA over-expression was largely restricted to NRF2 wild type tumors, 2) DPP3 over-expression positively associated with KEAP1 mutant tumors, 3) DPP3 over-expression in the presence of mutant KEAP1 further activated NRF2 signaling, and 4) as a tumor suppressor frequently targeted in cancer, KEAP1 is somewhat unique in that it is rarely deleted through homozygotic loss. Therefore, from a therapeutic and prognostic perspective, KEAP1 mutant tumors are not equivalent to NRF2 mutants. Future studies are needed to challenge this model. For example, does forced DPP3 expression drive NRF2 activity in mouse models of lung cancer, and would synergy be seen between DPP3 expression and KEAP1 mutation in vivo? Given that multiple different cancer types have recently been found to exhibit constitutive NRF2 activation—some of which in the absence of NRF2 or KEAP1 mutations—our data collectively support a model where the expression of ETGE-containing proteins drive NRF2-mediated signaling via a competitive binding mechanism.
Supplementary Material
Acknowledgments
We thank members of the Major lab for critical review of the manuscript. M.B.M. is supported by the NIH through the NIH Director’s New Innovator Award, 1-DP2-OD007149-01 and a Scholar Award from the Sidney Kimmel Cancer Foundation. D.N.H is supported by grants from The Cancer Genome Atlas (NIH U24 CA143848 and NIH U24 CA143848-02S1). D.N.H and M.B.M are additionally supported by a grant from the Greensboro Golfers Against Cancer.
Footnotes
The authors declare no conflicts of interest.
References
- 1.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 2.Sykiotis GP, Bohmann D. Stress-activated cap’n’collar transcription factors in aging and human disease. Sci Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–84. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–9. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 5.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi T, Sonobe M, Menju T, Nakayama E, Mino N, Iwakiri S, et al. Mutations in Keap1 are a potential prognostic factor in resected non-small cell lung cancer. J Surg Oncol. 2010;101:500–6. doi: 10.1002/jso.21520. [DOI] [PubMed] [Google Scholar]
- 7.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–86. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–71. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–9. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogura T, Tong KI, Mio K, Maruyama Y, Kurokawa H, Sato C, et al. Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc Natl Acad Sci U S A. 2010;107:2842–7. doi: 10.1073/pnas.0914036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281:24756–68. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 12.Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biological chemistry. 2006;387:1311–20. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 13.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26:2887–900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–88. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Hammerman PS, Lawrence MS, Voet D, Jing R, Cibulskis K, Sivachenko A, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012 doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konstantinopoulos PA, Spentzos D, Fountzilas E, Francoeur N, Sanisetty S, Grammatikos AP, et al. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res. 2011;71:5081–9. doi: 10.1158/0008-5472.CAN-10-4668. [DOI] [PubMed] [Google Scholar]
- 18.Lister A, Nedjadi T, Kitteringham NR, Campbell F, Costello E, Lloyd B, et al. Nrf2 is overexpressed in pancreatic cancer: implications for cell proliferation and therapy. Mol Cancer. 2011;10:37. doi: 10.1186/1476-4598-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li QK, Singh A, Biswal S, Askin F, Gabrielson E. KEAP1 gene mutations and NRF2 activation are common in pulmonary papillary adenocarcinoma. J Hum Genet. 2011;56:230–4. doi: 10.1038/jhg.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–9. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanada N, Takahata T, Zhou Q, Ye X, Sun R, Itoh J, et al. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC cancer. 2012;12:66. doi: 10.1186/1471-2407-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frohlich DA, McCabe MT, Arnold RS, Day ML. The role of Nrf2 in increased reactive oxygen species and DNA damage in prostate tumorigenesis. Oncogene. 2008;27:4353–62. doi: 10.1038/onc.2008.79. [DOI] [PubMed] [Google Scholar]
- 23.Muscarella LA, Barbano R, D’Angelo V, Copetti M, Coco M, Balsamo T, et al. Regulation of KEAP1 expression by promoter methylation in malignant gliomas and association with patient’s outcome. Epigenetics : official journal of the DNA Methylation Society. 2011;6:317–25. doi: 10.4161/epi.6.3.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Je EM, An CH, Yoo NJ, Lee SH. Mutational and expressional analyses of NRF2 and KEAP1 in sarcomas. Tumori. 2012;98:510–5. doi: 10.1177/030089161209800417. [DOI] [PubMed] [Google Scholar]
- 25.Camp ND, James RG, Dawson DW, Yan F, Davison JM, Houck SA, et al. Wilms tumor gene on the X chromosome (WTX) inhibits the degradation of NRF2 through competitive binding to KEAP1. J Biol Chem. 2012 doi: 10.1074/jbc.M111.316471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W, Sun Z, Wang XJ, Jiang T, Huang Z, Fang D, et al. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34:663–73. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature cell biology. 2010;12:213–23. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 28.Ma J, Cai H, Wu T, Sobhian B, Huo Y, Alcivar A, et al. PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Mol Cell Biol. 2012;32:1506–17. doi: 10.1128/MCB.06271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Kern JT, Walker JR, Johnson JA, Schultz PG, Luesch H. A genomic screen for activators of the antioxidant response element. Proc Natl Acad Sci U S A. 2007;104:5205–10. doi: 10.1073/pnas.0700898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sardiu ME, Cai Y, Jin J, Swanson SK, Conaway RC, Conaway JW, et al. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105:1454–9. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salopek-Sondi B, Vukelic B, Spoljaric J, Simaga S, Vujaklija D, Makarevic J, et al. Functional tyrosine residue in the active center of human dipeptidyl peptidase III. Biological chemistry. 2008;389:163–7. doi: 10.1515/BC.2008.021. [DOI] [PubMed] [Google Scholar]
- 32.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. The EMBO journal. 2006;25:3605–17. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baral PK, Jajcanin-Jozic N, Deller S, Macheroux P, Abramic M, Gruber K. The first structure of dipeptidyl-peptidase III provides insight into the catalytic mechanism and mode of substrate binding. J Biol Chem. 2008;283:22316–24. doi: 10.1074/jbc.M803522200. [DOI] [PubMed] [Google Scholar]
- 34.Wilkerson MD, Yin X, Hoadley KA, Liu Y, Hayward MC, Cabanski CR, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res. 2010;16:4864–75. doi: 10.1158/1078-0432.CCR-10-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canterini S, Bosco A, Carletti V, Fuso A, Curci A, Mangia F, et al. Subcellular TSC22D4 localization in cerebellum granule neurons of the mouse depends on development and differentiation. Cerebellum. 2012;11:28–40. doi: 10.1007/s12311-010-0211-8. [DOI] [PubMed] [Google Scholar]
- 36.Gibson SI, Surosky RT, Tye BK. The phenotype of the minichromosome maintenance mutant mcm3 is characteristic of mutants defective in DNA replication. Mol Cell Biol. 1990;10:5707–20. doi: 10.1128/mcb.10.11.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madine MA, Khoo CY, Mills AD, Laskey RA. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature. 1995;375:421–4. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen T, Jagannathan M, Shire K, Frappier L. Interactions of the human MCM-BP protein with MCM complex components and Dbf4. PLoS One. 2012;7:e35931. doi: 10.1371/journal.pone.0035931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi N, Quimbaya M, Schubert V, Lammens T, Vandepoele K, Schubert I, et al. The MCM-binding protein ETG1 aids sister chromatid cohesion required for postreplicative homologous recombination repair. PLoS Genet. 2010;6:e1000817. doi: 10.1371/journal.pgen.1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakwe AM, Nguyen T, Athanasopoulos V, Shire K, Frappier L. Identification and characterization of a novel component of the human minichromosome maintenance complex. Mol Cell Biol. 2007;27:3044–55. doi: 10.1128/MCB.02384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cybulsky AV, Takano T, Guillemette J, Papillon J, Volpini RA, Di Battista JA. The Ste20-like kinase SLK promotes p53 transactivation and apoptosis. Am J Physiol Renal Physiol. 2009;297:F971–80. doi: 10.1152/ajprenal.00294.2009. [DOI] [PubMed] [Google Scholar]
- 42.Pombo CM, Bonventre JV, Molnar A, Kyriakis J, Force T. Activation of a human Ste20-like kinase by oxidant stress defines a novel stress response pathway. EMBO J. 1996;15:4537–46. [PMC free article] [PubMed] [Google Scholar]
- 43.Cvrckova F, De Virgilio C, Manser E, Pringle JR, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–30. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- 44.Kueh HY, Charras GT, Mitchison TJ, Brieher WM. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J Cell Biol. 2008;182:341–53. doi: 10.1083/jcb.200801027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujibuchi T, Abe Y, Takeuchi T, Imai Y, Kamei Y, Murase R, et al. AIP1/WDR1 supports mitotic cell rounding. Biochem Biophys Res Commun. 2005;327:268–75. doi: 10.1016/j.bbrc.2004.11.156. [DOI] [PubMed] [Google Scholar]
- 46.Barsun M, Jajcanin N, Vukelic B, Spoljaric J, Abramic M. Human dipeptidyl peptidase III acts as a post-proline-cleaving enzyme on endomorphins. Biological chemistry. 2007;388:343–8. doi: 10.1515/BC.2007.039. [DOI] [PubMed] [Google Scholar]
- 47.Homma S, Ishii Y, Morishima Y, Yamadori T, Matsuno Y, Haraguchi N, et al. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin Cancer Res. 2009;15:3423–32. doi: 10.1158/1078-0432.CCR-08-2822. [DOI] [PubMed] [Google Scholar]
- 48.Simaga S, Babic D, Osmak M, Ilic-Forko J, Vitale L, Milicic D, et al. Dipeptidyl peptidase III in malignant and non-malignant gynaecological tissue. Eur J Cancer. 1998;34:399–405. doi: 10.1016/s0959-8049(97)00401-2. [DOI] [PubMed] [Google Scholar]
- 49.Simaga S, Babic D, Osmak M, Sprem M, Abramic M. Tumor cytosol dipeptidyl peptidase III activity is increased with histological aggressiveness of ovarian primary carcinomas. Gynecol Oncol. 2003;91:194–200. doi: 10.1016/s0090-8258(03)00462-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.